A High Protein Model Alters the Endometrial Transcriptome of Mares
Abstract
:1. Introduction
2. Material and Methods
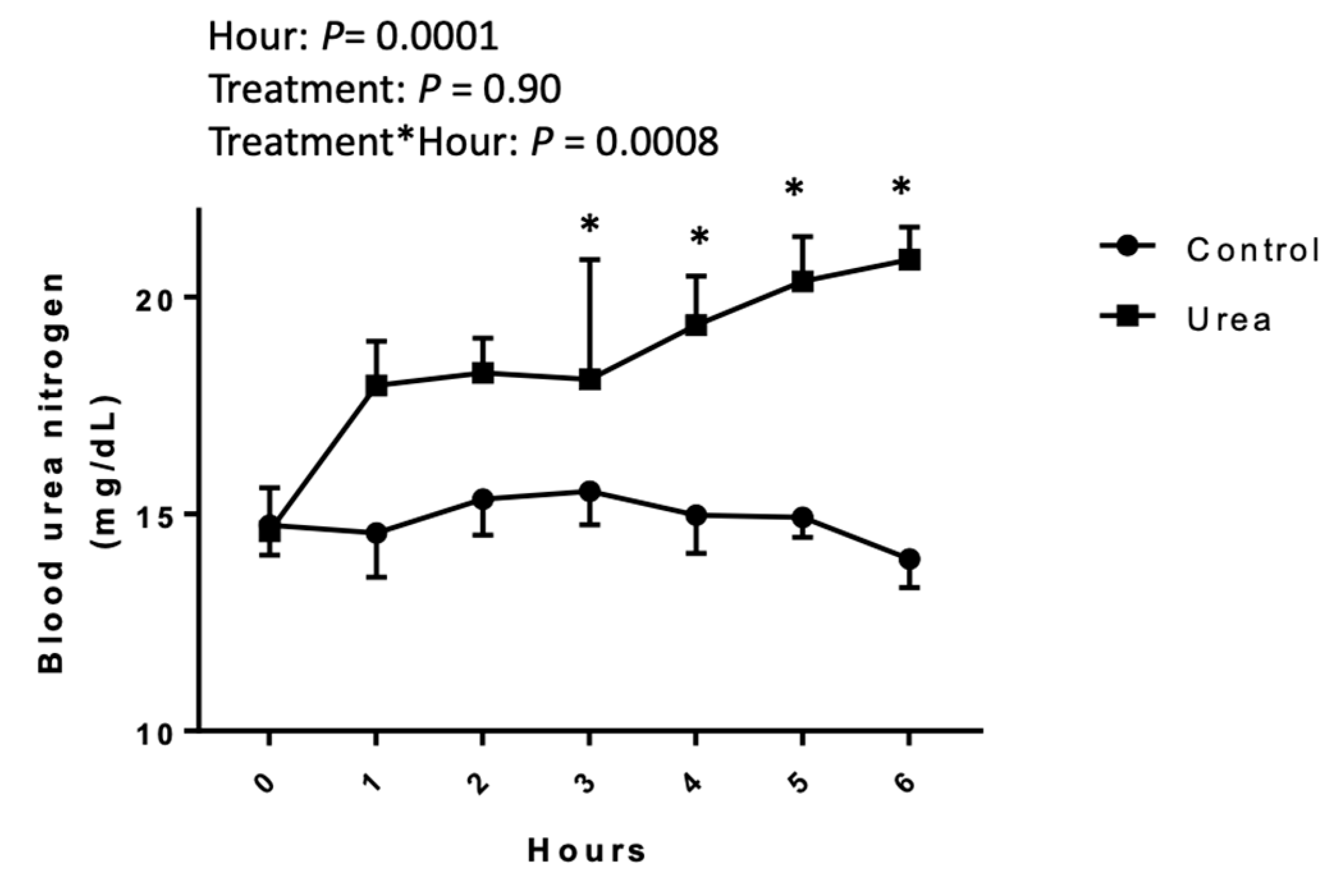
2.1. Blood Urea Nitrogen Concentration
2.2. Uterine and Vaginal pH
2.3. Endometrial Biopsies
2.4. RNA Extraction
2.5. mRNA Library Preparation and Next Generation Sequencing
2.6. Next Generation Sequencing Data Analysis
2.7. Functional Annotation and Pathway Analysis
2.8. Quantitative Real-Time PCR
2.9. Statistical Analyses
3. Results
3.1. Blood Urea Nitrogen Concentrations
3.2. Uterine pH
3.3. Vaginal pH
3.4. RNA Sequencing
3.5. Differentially Expressed Genes
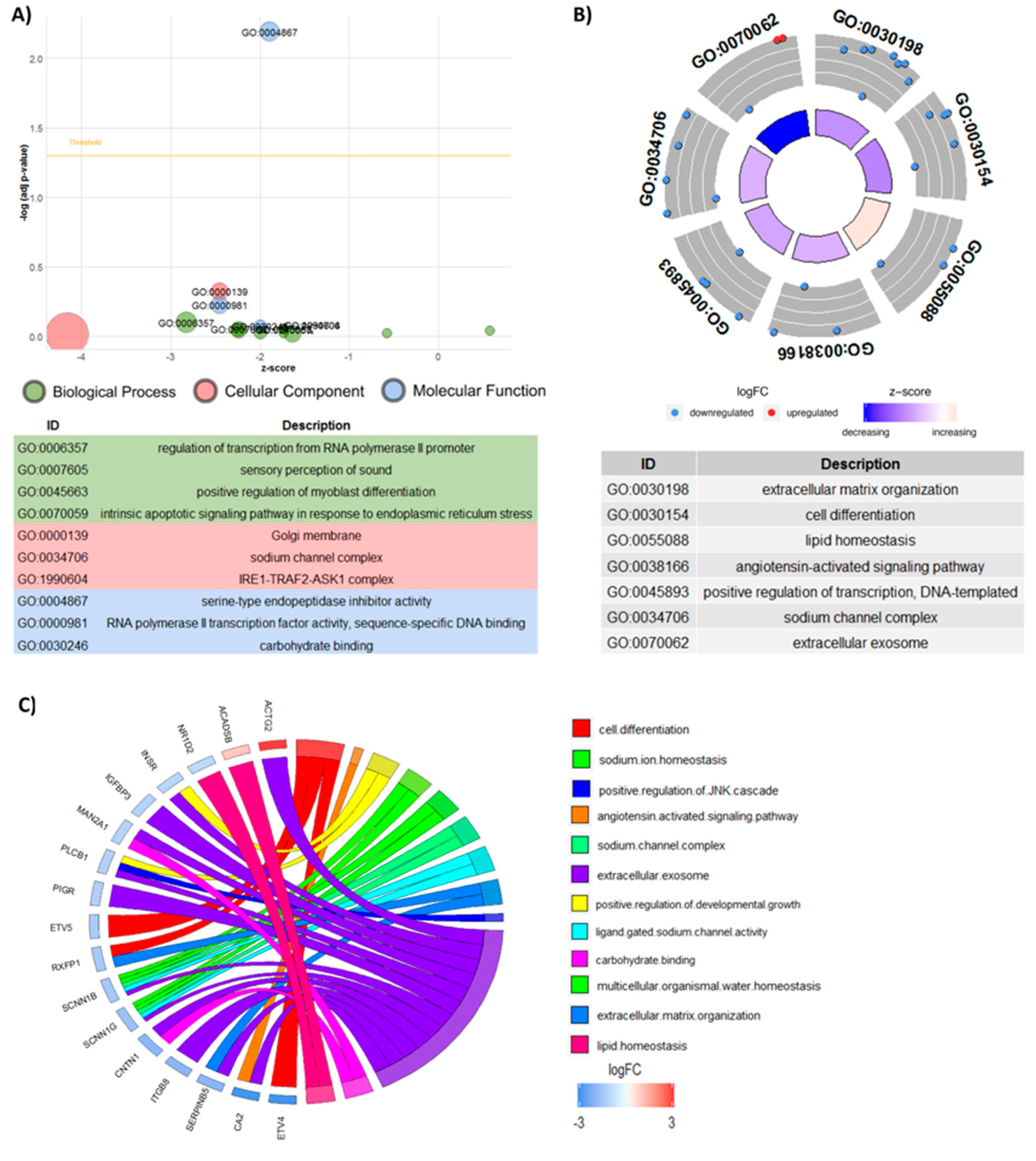
3.6. Functional Analyses
3.7. Upstream Regulators
3.8. Weighted Correlation Network Analysis (WGCNA)
3.9. Quantitative Real-Time PCR
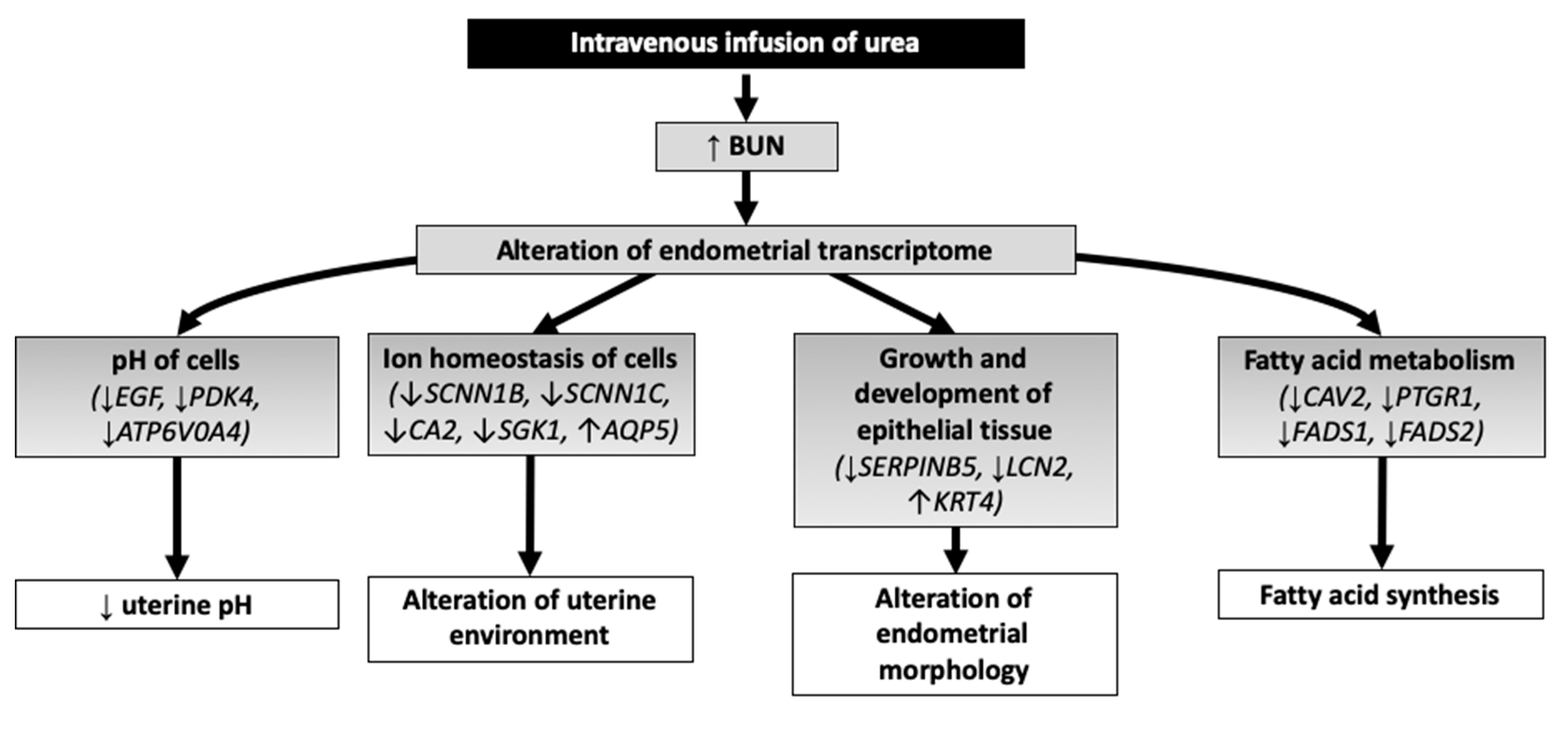
4. Discussion
4.1. pH of Cells
4.2. Solute Carriers and Ion Homeostasis of Cells
4.3. Growth and Development of Epithelial Tissue
4.4. Fatty Acid Metabolism
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ball, B.A. Embryonic loss in mares. Incidence, possible causes, and diagnostic considerations. Vet. Clin. N. Am. Equine Pract. 1988, 4, 263–290. [Google Scholar] [CrossRef]
- Elrod, C.C.; Van Amburgh, M.; Butler, W.R. Alterations of pH in response to increased dietary protein in cattle are unique to the uterus. J. Anim. Sci. 1993, 71, 702–706. [Google Scholar] [CrossRef] [PubMed]
- Butler, W.R. Nutritional interactions with reproductive performance in dairy cattle. Anim. Reprod. Sci. 2000, 60–61, 449–457. [Google Scholar] [CrossRef]
- Butler, W.R.; Calaman, J.J.; Beam, S.W. Plasma and milk urea nitrogen in relation to pregnancy rate in lactating dairy cattle. J. Anim. Sci. 1996, 74, 858–865. [Google Scholar] [CrossRef] [PubMed]
- McEvoy, T.G.; Robinson, J.J.; Aitken, R.P.; Findlay, P.A.; Robertson, I.S. Dietary excesses of urea influence the viability and metabolism of preimplantation sheep embryos and may affect fetal growth among survivors. Anim. Reprod. Sci. 1997, 47, 71–90. [Google Scholar] [CrossRef]
- Ferguson, J.D.; Galligan, D.T.; Blanchard, T.; Reeves, M. Serum urea nitrogen and conception rate: The usefulness of test information. J. Dairy Sci. 1993, 76, 3742–3746. [Google Scholar] [CrossRef]
- Rhoads, M.L.; Rhoads, R.P.; Gilbert, R.O.; Toole, R.; Butler, W.R. Detrimental effects of high plasma urea nitrogen levels on viability of embryos from lactating dairy cows. Anim. Reprod. Sci. 2006, 91, 1–10. [Google Scholar] [CrossRef]
- Hammon, D.S.; Holyoak, G.R.; Dhiman, T.R. Association between blood plasma urea nitrogen levels and reproductive fluid urea nitrogen and ammonia concentrations in early lactation dairy cows. Anim. Reprod. Sci. 2005, 86, 195–204. [Google Scholar] [CrossRef]
- Klein, C.; Scoggin, K.E.; Ealy, A.D.; Troedsson, M.H. Transcriptional profiling of equine endometrium during the time of maternal recognition of pregnancy. Biol. Reprod. 2010, 83, 102–113. [Google Scholar] [CrossRef]
- Rhoads, M.L.; Gilbert, R.O.; Lucy, M.C.; Butler, W.R. Effects of urea infusion on the uterine luminal environment of dairy cows. J. Dairy Sci. 2004, 87, 2896–2901. [Google Scholar] [CrossRef]
- Smith, J.; Hagemann, L.; Peterson, A.; Beaumont, S.; Berg, M.; Donnison, M.; Ledgard, A.; McDonald, R.; Verkerk, G. Effect of infusion of urea or ammonia to dairy heifers on the composition of follicular fluid and uterine secretions. In Proceedings of the 14th International Congress Animal Reproduction, Stockholm, Sweden, 2–6 July 2000; p. 37. [Google Scholar]
- Jordan, E.R.; Chapman, T.E.; Holtan, D.W.; Swanson, L.V. Relationship of dietary crude protein to composition of uterine secretions and blood in high-producing postpartum dairy cows. J. Dairy Sci. 1983, 66, 1854–1862. [Google Scholar] [CrossRef]
- Elrod, C.; Butler, W. Reduction of fertility and alteration of uterine pH in heifers fed excess ruminally degradable protein. J. Anim. Sci. 1993, 71, 694–701. [Google Scholar] [CrossRef] [PubMed]
- Gunaretnam, I.; Pretheeban, T.; Rajamahendran, R. Effects of ammonia and urea in vitro on mRNA of candidate bovine endometrial genes. Anim. Reprod. Sci. 2013, 141, 42–51. [Google Scholar] [CrossRef] [PubMed]
- De Wit, A.A.; Cesar, M.L.; Kruip, T.A. Effect of urea during in vitro maturation on nuclear maturation and embryo development of bovine cumulus-oocyte-complexes. J. Dairy Sci. 2001, 84, 1800–1804. [Google Scholar] [CrossRef]
- Ocon, O.M.; Hansen, P.J. Disruption of bovine oocytes and preimplantation embryos by urea and acidic pH. J. Dairy Sci. 2003, 86, 1194–1200. [Google Scholar] [CrossRef]
- Harper, M.; Swinker, A.; Staniar, W.; Welker, A. Ration evaluation of Chesapeake Bay watershed horse farms from a nutrient management perspective. J. Equine Vet. Sci. 2009, 5, 401–402. [Google Scholar] [CrossRef]
- Honoré, E.K.; Uhlinger, C.A. Equine feeding practices in central North Carolina: A preliminary survey. J. Equine Vet. Sci. 1994, 14, 424–429. [Google Scholar] [CrossRef]
- Mok, C.H.; Levesque, C.L.; Urschel, K.L. Using the indicator amino acid oxidation technique to study threonine requirements in horses receiving a predominantly forage diet. J. Anim. Physiol. Anim. Nutr. 2018, 102, 1366–1381. [Google Scholar] [CrossRef]
- Tabacco, A.; Meiattini, F. Urea Determination Method. U.S. Patent 4,507,388, 26 March 1985. [Google Scholar]
- Herrera, M.; Herrera, J.M.; Cantatore, S.; Aguilar, J.; Felipe, A.; Fumuso, E. Comparative histomorphological study of endometrium in mares. Anat. Histol. Embryol. 2018, 47, 153–158. [Google Scholar] [CrossRef]
- Andrews, S. FASTQC. A Quality Control Tool for High throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 10 February 2019).
- Krueger, F. Trim Galore: A Wrapper Tool Around Cutadapt and FastQC to Consistently Apply Quality and Adapter Trimming to FastQ files, with Some Extra Functionality for MspI-Digested RRBS-Type (Reduced Representation Bisufite-Seq) Libraries. Available online: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 23 February 2019).
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Thomas, P. Protein networks and pathway analysis. Methods Mol. Biol. 2009, 563, 123–140. [Google Scholar] [PubMed]
- Kramer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.; Karlen, Y.; Bakker, O.; Van den Hoff, M.J.; Moorman, A.F. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- De Spiegelaere, W.; Dern-Wieloch, J.; Weigel, R.; Schumacher, V.; Schorle, H.; Nettersheim, D.; Bergmann, M.; Brehm, R.; Kliesch, S.; Vandekerckhove, L.; et al. Reference gene validation for RT-qPCR, a note on different available software packages. PLoS ONE 2015, 10, e0122515. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Moolenaar, W.H.; Tsien, R.Y.; Van der Saag, P.T.; de Laat, S.W. Na+/H+ exchange and cytoplasmic pH in the action of growth factors in human fibroblasts. Nature 1983, 304, 645–648. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Cohen, D.M. Urea stress is more akin to EGF exposure than to hypertonic stress in renal medullary cells. Am. J. Physiol. Ren. Physiol. 2002, 283, F388–F398. [Google Scholar] [CrossRef] [Green Version]
- Squirrell, J.M.; Lane, M.; Bavister, B.D. Altering intracellular pH disrupts development and cellular organization in preimplantation hamster embryos. Biol. Reprod. 2001, 64, 1845–1854. [Google Scholar] [CrossRef] [PubMed]
- Edwards, L.J.; Williams, D.A.; Gardner, D.K. Intracellular pH of the mouse preimplantation embryo: Amino acids act as buffers of intracellular pH. Hum. Reprod. 1998, 13, 3441–3448. [Google Scholar] [CrossRef]
- Ferreira, F.A.; Gomez, R.G.; Joaquim, D.C.; Watanabe, Y.F.; de Castro e Paula, L.A.; Binelli, M.; Rodrigues, P.H. Short-term urea feeding decreases in vitro hatching of bovine blastocysts. Theriogenology 2011, 76, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Fahey, J.; Boland, M.; O’Callaghan, D. The effects of dietary urea on embryo development in superovulated donor ewes and on early embryo survival and development in recipient ewes. Anim. Sci. 2001, 72, 395–400. [Google Scholar] [CrossRef]
- Groebner, A.E.; Rubio-Aliaga, I.; Schulke, K.; Reichenbach, H.D.; Daniel, H.; Wolf, E.; Meyer, H.H.; Ulbrich, S.E. Increase of essential amino acids in the bovine uterine lumen during preimplantation development. Reproduction 2011, 141, 685–695. [Google Scholar] [CrossRef] [Green Version]
- Forde, N.; Spencer, T.E.; Bazer, F.W.; Song, G.; Roche, J.F.; Lonergan, P. Effect of pregnancy and progesterone concentration on expression of genes encoding for transporters or secreted proteins in the bovine endometrium. Physiol. Genom. 2010, 41, 53–62. [Google Scholar] [CrossRef] [Green Version]
- Samborski, A.; Graf, A.; Krebs, S.; Kessler, B.; Bauersachs, S. Deep sequencing of the porcine endometrial transcriptome on day 14 of pregnancy. Biol. Reprod. 2013, 88, 84. [Google Scholar] [CrossRef]
- Zhu, C.; Jiang, Z.; Bazer, F.W.; Johnson, G.A.; Burghardt, R.C.; Wu, G. Aquaporins in the female reproductive system of mammals. Front. Biosci. 2015, 20, 838–871. [Google Scholar] [Green Version]
- Ruan, Y.C.; Chen, H.; Chan, H.C. Ion channels in the endometrium: Regulation of endometrial receptivity and embryo implantation. Hum. Reprod. Update 2014, 20, 517–529. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.M.; Zhang, D.; Wang, T.T.; Sheng, J.Z.; Huang, H.F. Ion/water channels for embryo implantation barrier. Physiology 2014, 29, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Canessa, C.M.; Schild, L.; Buell, G.; Thorens, B.; Gautschi, I.; Horisberger, J.D.; Rossier, B.C. Amiloride-sensitive epithelial Na+ channel is made of three homologous subunits. Nature 1994, 367, 463–467. [Google Scholar] [CrossRef] [PubMed]
- Boggula, V.R.; Hanukoglu, I.; Sagiv, R.; Enuka, Y.; Hanukoglu, A. Expression of the epithelial sodium channel in the endometrium—Implications for fertility in a patient with pseudohypoaldosteronism. J. Steroid Biochem. Mol. Biol. 2018, 183, 137–141. [Google Scholar] [CrossRef]
- Lang, F.; Artunc, F.; Vallon, V. The physiological impact of the serum and glucocorticoid-inducible kinase SGK1. Curr. Opin. Nephrol. Hypertens. 2009, 18, 439–448. [Google Scholar] [CrossRef]
- Salker, M.S.; Christian, M.; Steel, J.H.; Nautiyal, J.; Lavery, S.; Trew, G.; Webster, Z.; Al-Sabbagh, M.; Puchchakayala, G.; Foller, M.; et al. Deregulation of the serum- and glucocorticoid-inducible kinase SGK1 in the endometrium causes reproductive failure. Nat. Med. 2011, 17, 1509–1513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindsay, L.A.; Murphy, C.R. Redistribution of aquaporins 1 and 5 in the rat uterus is dependent on progesterone: A study with light and electron microscopy. Reproduction 2006, 131, 369–378. [Google Scholar] [CrossRef]
- Klein, C.; Troedsson, M.H.; Rutllant, J. Expression of aquaporin water channels in equine endometrium is differentially regulated during the oestrous cycle and early pregnancy. Reprod. Domest. Anim. 2013, 48, 529–537. [Google Scholar] [CrossRef]
- Lindsay, L.A.; Murphy, C.R. Aquaporins are upregulated in glandular epithelium at the time of implantation in the rat. J. Mol. Histol. 2007, 38, 87–95. [Google Scholar] [CrossRef]
- Supuran, C.T.; Scozzafava, A.; Casini, A. Carbonic anhydrase inhibitors. Med. Res. Rev. 2003, 23, 146–189. [Google Scholar] [CrossRef]
- Hu, J.; Spencer, T.E. Carbonic anhydrase regulate endometrial gland development in the neonatal uterus. Biol. Reprod. 2005, 73, 131–138. [Google Scholar] [CrossRef]
- Brown, K.A.; Boerboom, D.; Bouchard, N.; Dore, M.; Lussier, J.G.; Sirois, J. Human chorionic gonadotropin-dependent induction of an equine aldo-keto reductase (AKR1C23) with 20alpha-hydroxysteroid dehydrogenase activity during follicular luteinization in vivo. J. Mol. Endocrinol. 2006, 36, 449–461. [Google Scholar] [CrossRef]
- Ogle, T.F.; Beyer, B.K. Steroid-binding specificity of the progesterone receptor from rat placenta. J. Steroid Biochem. 1982, 16, 147–150. [Google Scholar] [CrossRef]
- Kozai, K.; Hojo, T.; Tokuyama, S.; Szostek, A.Z.; Takahashi, M.; Sakatani, M.; Nambo, Y.; Skarzynski, D.J.; Okuda, K. Expression of aldo-keto reductase 1C23 in the equine corpus luteum in different luteal phases. J Reprod. Dev. 2014, 60, 150–154. [Google Scholar] [CrossRef]
- El-Sheikh Ali, H.; Legacki, E.; Loux, S.; Esteller-Vico, A.; Dini, P.; Scoggin, K.; Conley, A.; Stanley, S.; Ball, B. Equine placentitis is associated with a downregulation in myometrial progestin signaling. Biol. Reprod. 2019, 101, 162–176. [Google Scholar]
- Bauersachs, S.; Mitko, K.; Ulbrich, S.E.; Blum, H.; Wolf, E. Transcriptome studies of bovine endometrium reveal molecular profiles characteristic for specific stages of estrous cycle and early pregnancy. Exp. Clin. Endocrinol. Diabetes 2008, 116, 371–384. [Google Scholar] [CrossRef]
- Haining, R.E.; Cameron, I.T.; Van Papendorp, C.; Davenport, A.P.; Prentice, A.; Thomas, E.J.; Smith, S.K. Epidermal growth factor in human endometrium: Proliferative effects in culture and immunocytochemical localization in normal and endometriotic tissues. Hum. Reprod. 1991, 6, 1200–1205. [Google Scholar] [CrossRef]
- Macdonald, L.J.; Sales, K.J.; Grant, V.; Brown, P.; Jabbour, H.N.; Catalano, R.D. Prokineticin 1 induces Dickkopf 1 expression and regulates cell proliferation and decidualization in the human endometrium. Mol. Hum. Reprod. 2011, 17, 626–636. [Google Scholar] [CrossRef]
- Huang, Y.; Cai, L.W.; Yang, R. Expression of maspin in the early pregnant mouse endometrium and its role during embryonic implantation. Comp. Med. 2012, 62, 179–184. [Google Scholar]
- Le Jan, S.; Amy, C.; Cazes, A.; Monnot, C.; Lamande, N.; Favier, J.; Philippe, J.; Sibony, M.; Gasc, J.M.; Corvol, P.; et al. Angiopoietin-like 4 is a proangiogenic factor produced during ischemia and in conventional renal cell carcinoma. Am. J. Pathol. 2003, 162, 1521–1528. [Google Scholar] [CrossRef]
- Lord, E.; Murphy, B.D.; Desmarais, J.A.; Ledoux, S.; Beaudry, D.; Palin, M.F. Modulation of peroxisome proliferator-activated receptor δ and γ transcripts in swine endometrial tissue during early gestation. Reproduction 2006, 131, 929–942. [Google Scholar] [CrossRef]
- Merkl, M.; Ulbrich, S.E.; Otzdorff, C.; Herbach, N.; Wanke, R.; Wolf, E.; Handler, J.; Bauersachs, S. Microarray analysis of equine endometrium at days 8 and 12 of pregnancy. Biol. Reprod. 2010, 83, 874–886. [Google Scholar] [CrossRef]
- Chapman, D.; Quinn, P.J. A method for the modulation of membrane fluidity: Homogeneous catalytic hydrogenation of phospholipids and phospholipids and phospholipid-water model biomembranes. Proc. Natl. Acad. Sci. USA 1976, 73, 3971–3975. [Google Scholar] [CrossRef]
- Mattos, R.; Staples, C.R.; Thatcher, W.W. Effects of dietary fatty acids on reproduction in ruminants. Rev. Reprod. 2000, 5, 38–45. [Google Scholar] [CrossRef]
- Erkenbrack, E.M.; Maziarz, J.D.; Griffith, O.W.; Liang, C.; Chavan, A.R.; Nnamani, M.C.; Wagner, G.P. The mammalian decidual cell evolved from a cellular stress response. PLoS Biol. 2018, 16, e2005594. [Google Scholar] [CrossRef]
- Shin, D.; Howng, S.Y.; Ptacek, L.J.; Fu, Y.H. miR-32 and its target SLC45A3 regulate the lipid metabolism of oligodendrocytes and myelin. Neuroscience 2012, 213, 29–37. [Google Scholar] [CrossRef] [Green Version]
- Marshall, M.K.; Doerrler, W.; Feingold, K.R.; Grunfeld, C. Leukemia inhibitory factor induces changes in lipid metabolism in cultured adipocytes. Endocrinology 1994, 135, 141–147. [Google Scholar] [CrossRef]
- Stewart, C.L.; Kaspar, P.; Brunet, L.J.; Bhatt, H.; Gadi, I.; Kontgen, F.; Abbondanzo, S.J. Blastocyst implantation depends on maternal expression of leukaemia inhibitory factor. Nature 1992, 359, 76–79. [Google Scholar] [CrossRef]
- Parton, R.G. Ultrastructural localization of gangliosides; GM1 is concentrated in caveolae. J. Histochem. Cytochem. 1994, 42, 155–166. [Google Scholar] [CrossRef]
- Rothberg, K.G.; Heuser, J.E.; Donzell, W.C.; Ying, Y.S.; Glenney, J.R.; Anderson, R.G. Caveolin, a protein component of caveolae membrane coats. Cell 1992, 68, 673–682. [Google Scholar] [CrossRef]



| Upstream Regulator | Expr Log Ratio | Molecule Type | p-Value of Overlap | Target Molecules in Dataset |
|---|---|---|---|---|
| ETV4 | −5.332 | transcription regulator | 0.024 | ETV4, MET |
| EGF | −3.834 | growth factor | 0.000 | ACSL4, IDS, IGFBP3, LCN2, MAP3K5, PDK4, SLC37A1, SP4, SPHK1, SPRY2 |
| EHF | −1.688 | transcription regulator | 0.022 | ANGPTL4, EHF, MET |
| IRS2 | −1.253 | enzyme | 0.028 | PPARGC1A, THRSP |
| SGK1 | −0.939 | kinase | 0.000 | KCNA3, LIF, SCNN1B, SCNN1G |
| Gene | Correlation (R) | P-Value | Corrected P-Value |
|---|---|---|---|
| ANGPTL4 | 0.538 | 0.047 | 0.846 |
| AQP5 | 0.840 | 0.000 | 0.000 |
| CA2 | 0.538 | 0.046 | 0.828 |
| EGF | 0.611 | 0.020 | 0.360 |
| ENPP1 | 0.746 | 0.002 | 0.036 |
| ERRFI1 | 0.836 | 0.000 | 0.000 |
| ETV1 | 0.382 | 0.177 | 1.000 |
| FADS1 | 0.710 | 0.004 | 0.072 |
| IGFBP3 | 0.723 | 0.003 | 0.054 |
| INSR | 0.419 | 0.136 | 1.000 |
| ITGB8 | 0.677 | 0.008 | 0.144 |
| KCNA3 | 0.393 | 0.165 | 1.000 |
| LAMC2 | 0.402 | 0.154 | 1.000 |
| PIGR | 0.601 | 0.023 | 0.414 |
| PRLR | 0.864 | 0.000 | 0.000 |
| SERPINA14 | 0.688 | 0.007 | 0.126 |
| SGK1 | 0.174 | 0.551 | 1.000 |
| SPINK7 | 0.858 | 0.000 | 0.000 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boakari, Y.L.; El-Sheikh Ali, H.; Dini, P.; Loux, S.; Fernandes, C.B.; Scoggin, K.; Esteller-Vico, A.; Lawrence, L.; Ball, B. A High Protein Model Alters the Endometrial Transcriptome of Mares. Genes 2019, 10, 576. https://doi.org/10.3390/genes10080576
Boakari YL, El-Sheikh Ali H, Dini P, Loux S, Fernandes CB, Scoggin K, Esteller-Vico A, Lawrence L, Ball B. A High Protein Model Alters the Endometrial Transcriptome of Mares. Genes. 2019; 10(8):576. https://doi.org/10.3390/genes10080576
Chicago/Turabian StyleBoakari, Yatta L., Hossam El-Sheikh Ali, Pouya Dini, Shavahn Loux, Claudia B. Fernandes, Kirsten Scoggin, Alejandro Esteller-Vico, Laurie Lawrence, and Barry Ball. 2019. "A High Protein Model Alters the Endometrial Transcriptome of Mares" Genes 10, no. 8: 576. https://doi.org/10.3390/genes10080576
APA StyleBoakari, Y. L., El-Sheikh Ali, H., Dini, P., Loux, S., Fernandes, C. B., Scoggin, K., Esteller-Vico, A., Lawrence, L., & Ball, B. (2019). A High Protein Model Alters the Endometrial Transcriptome of Mares. Genes, 10(8), 576. https://doi.org/10.3390/genes10080576







