DNA Methylation Module Network-Based Prognosis and Molecular Typing of Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection and Preprocessing
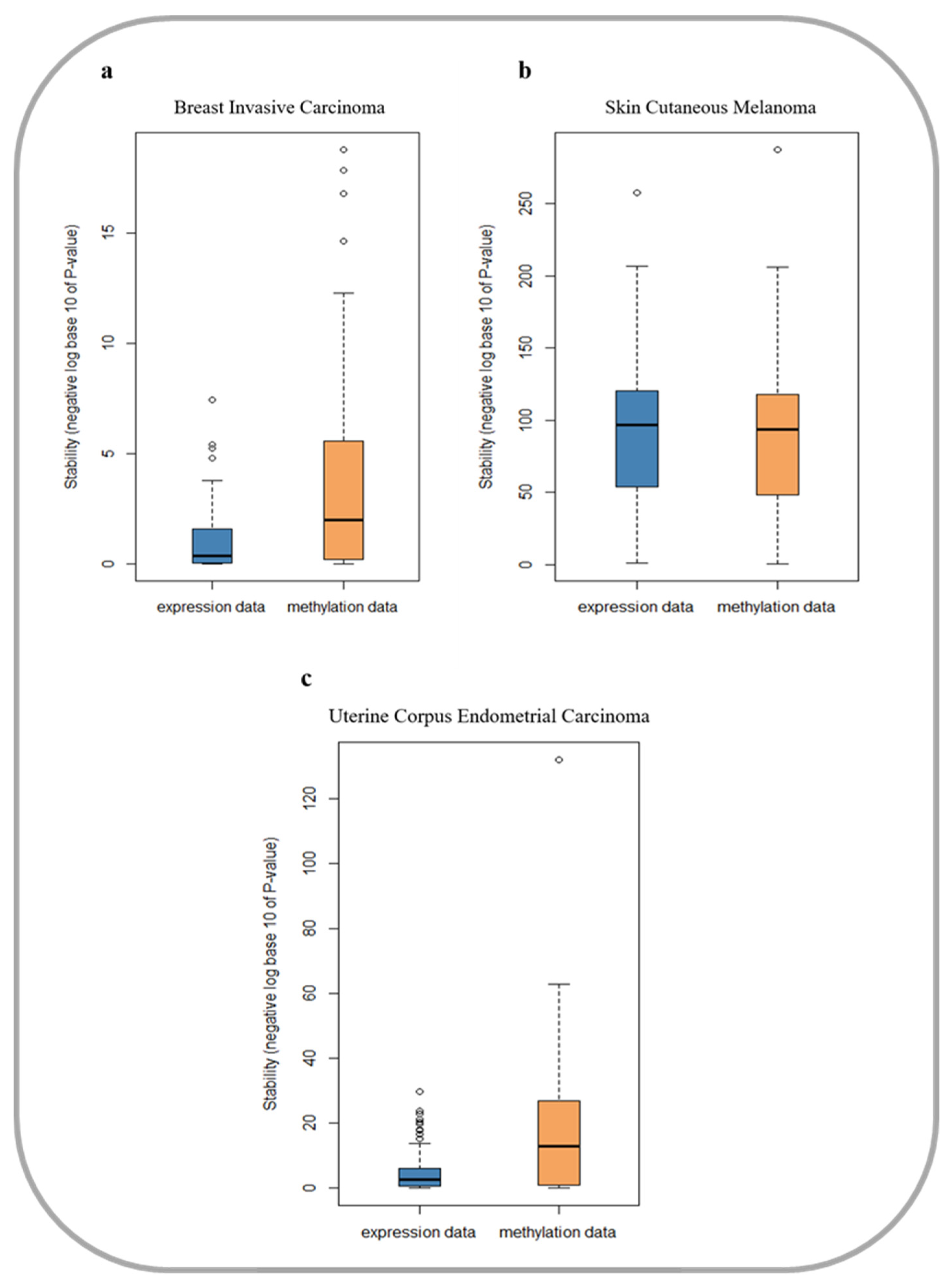
2.2. Comparison of the Stability of DNA Methylation Data and Gene Expression Data in Cancer Prognosis
2.3. Construction of the Gene Co-Methylation Networks by Rank-Based Method
2.4. Gene Module Detection in Co-Methylation Networks
2.5. Construction of the Module Networks by Permutation Method
2.6. Identifying Core Gene Modules Based on the K-Shell Method
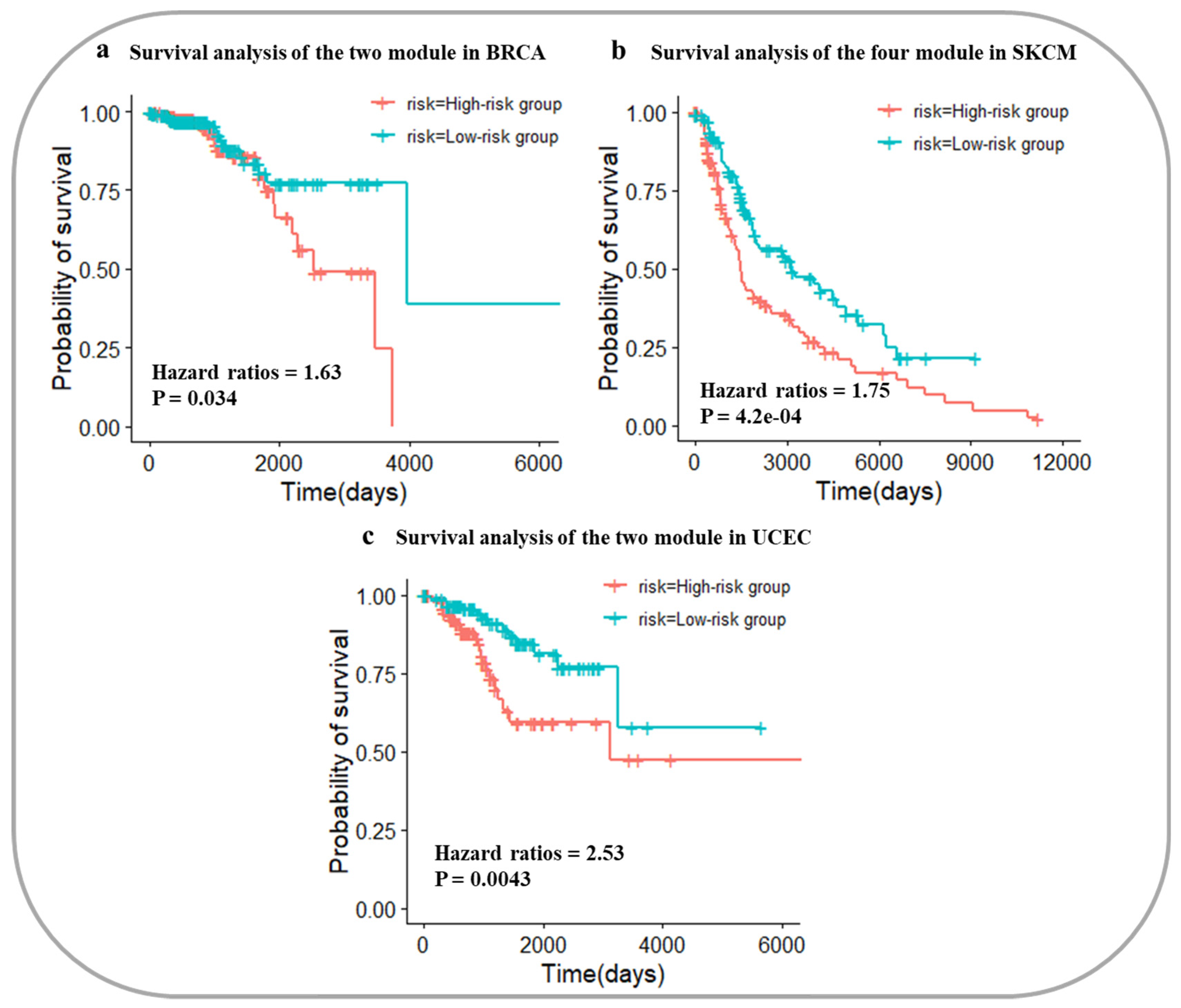
2.7. Survival Analysis Using Core Gene Modules
2.8. Cancer Molecular Typing Based on Core Gene Modules by K-Means Algorithm
3. Results and Discussion
3.1. DNA Methylation Data are More Stable in Cancer Prognosis
3.2. Module Networks of Three Cancers
3.2.1. Module Network of Breast Invasive Carcinoma
3.2.2. Module Network of Skin Cutaneous Melanoma
3.2.3. Module Network of Uterine Corpus Endometrial Carcinoma
3.3. The Core Gene Modules of Three Cancers
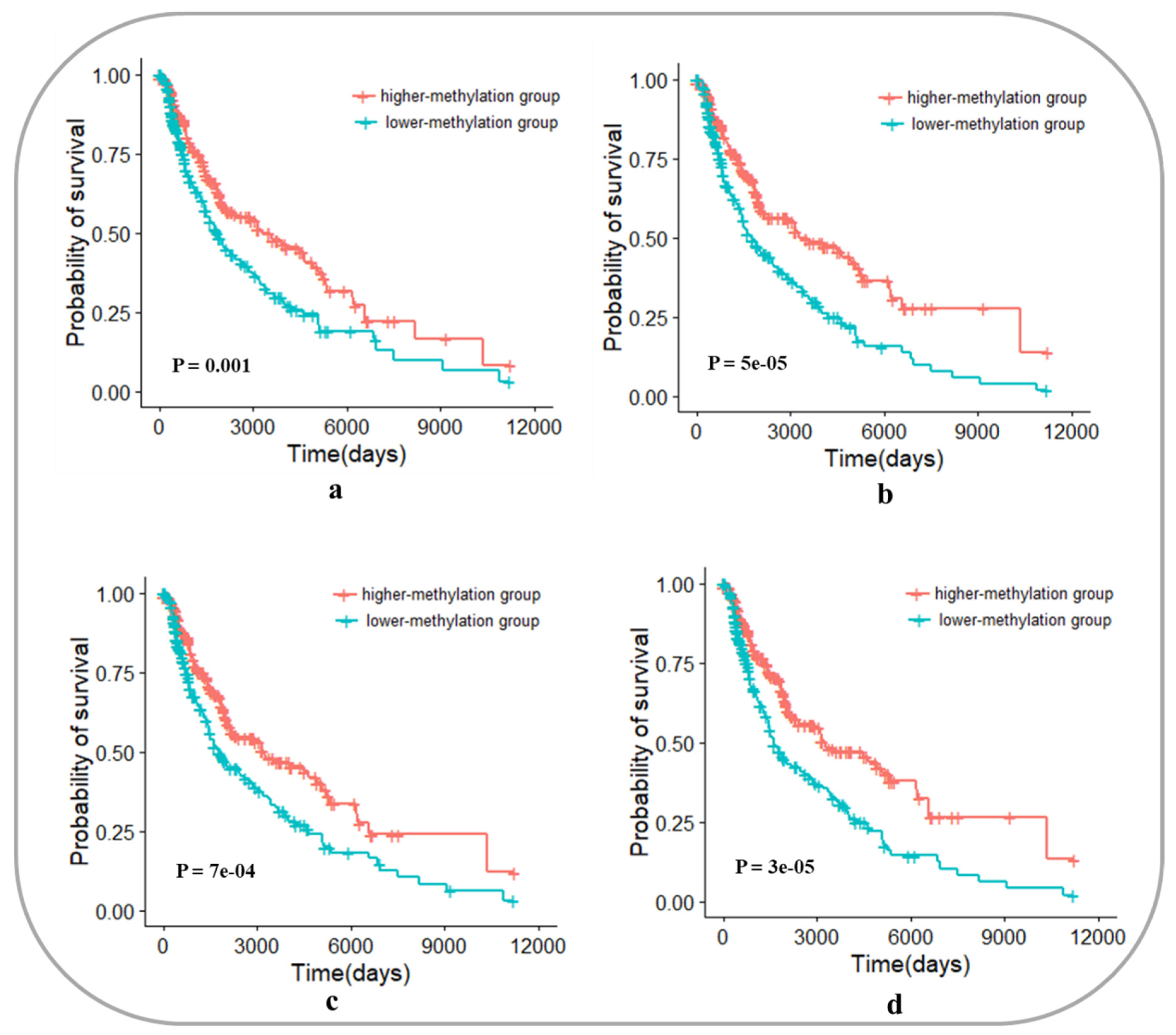
3.4. Survival Analysis of Three Cancers
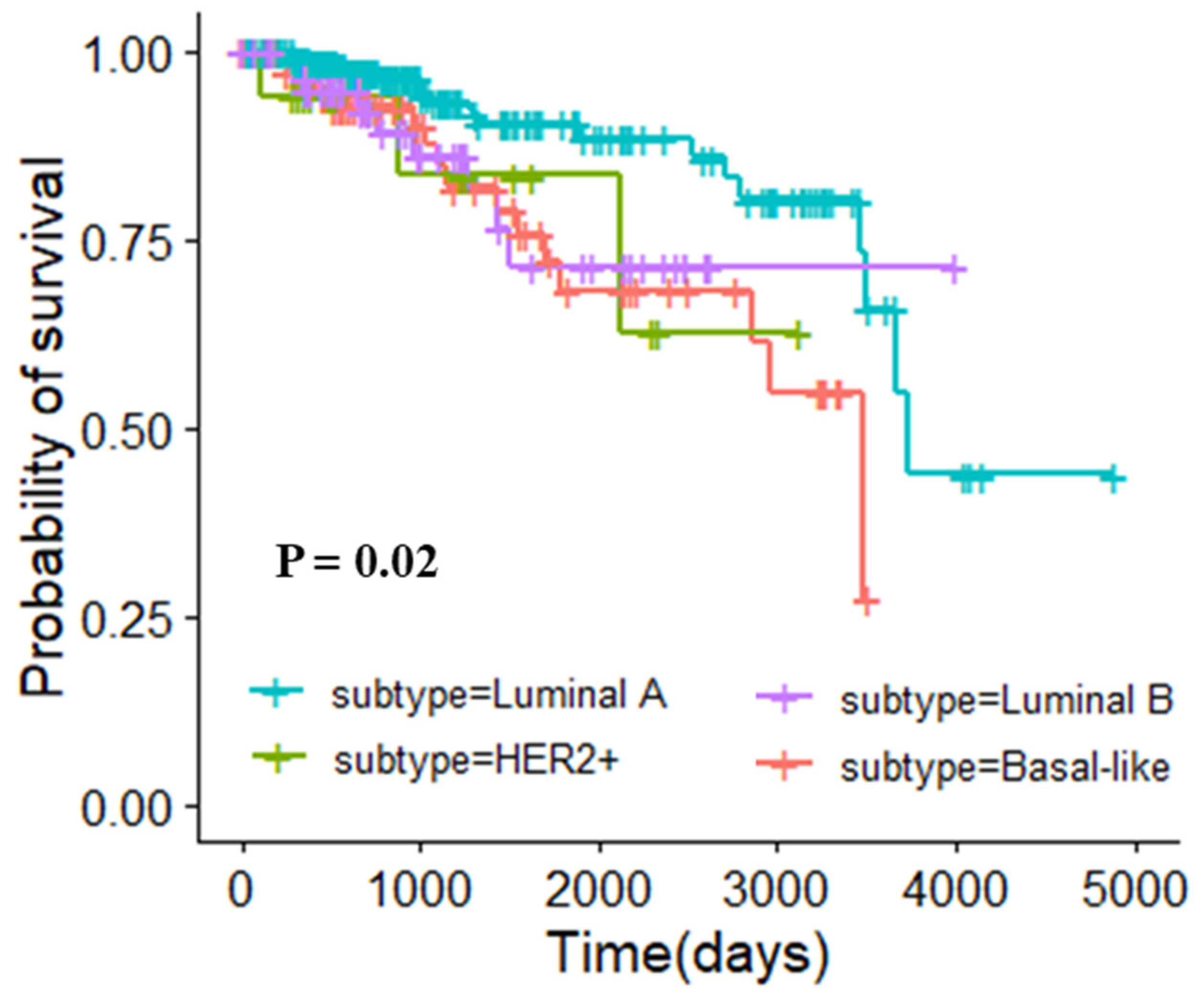
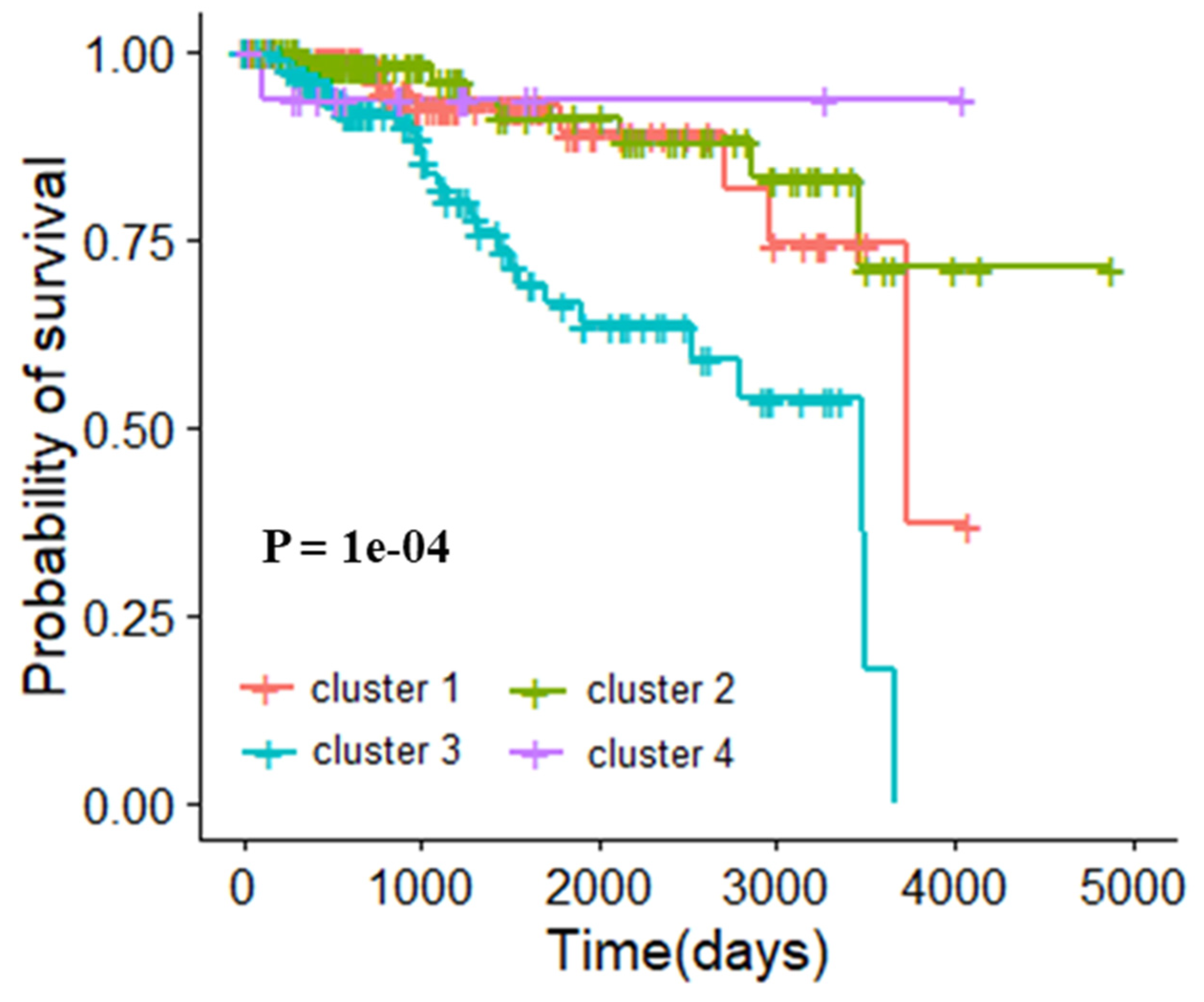
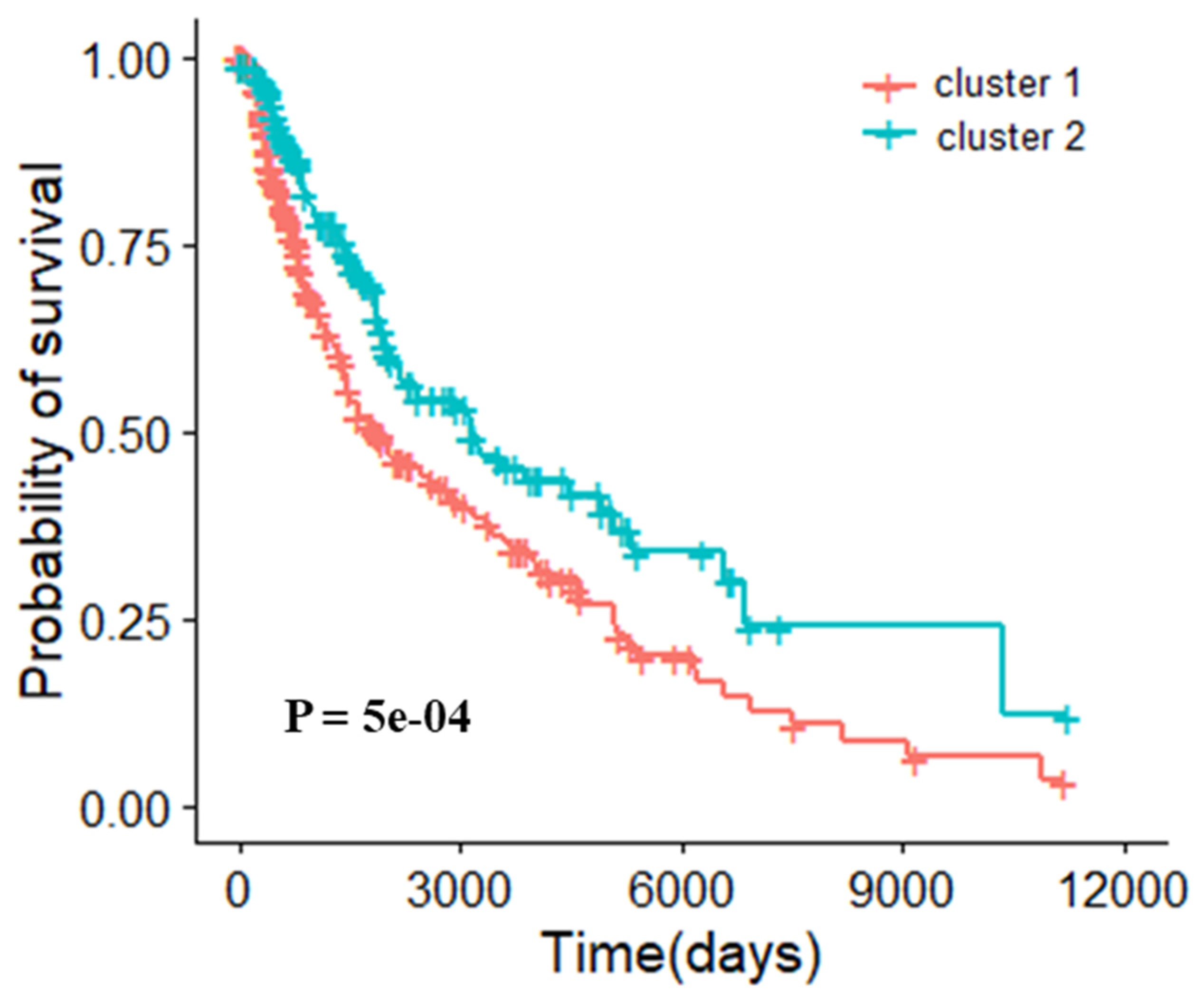
3.5. Molecular Typing Results of Three Cancers
3.5.1. Molecular Typing of Breast Invasive Carcinoma
3.5.2. Molecular Typing of Skin Cutaneous Melanoma
3.5.3. Molecular Typing of Uterine Corpus Endometrial Carcinoma
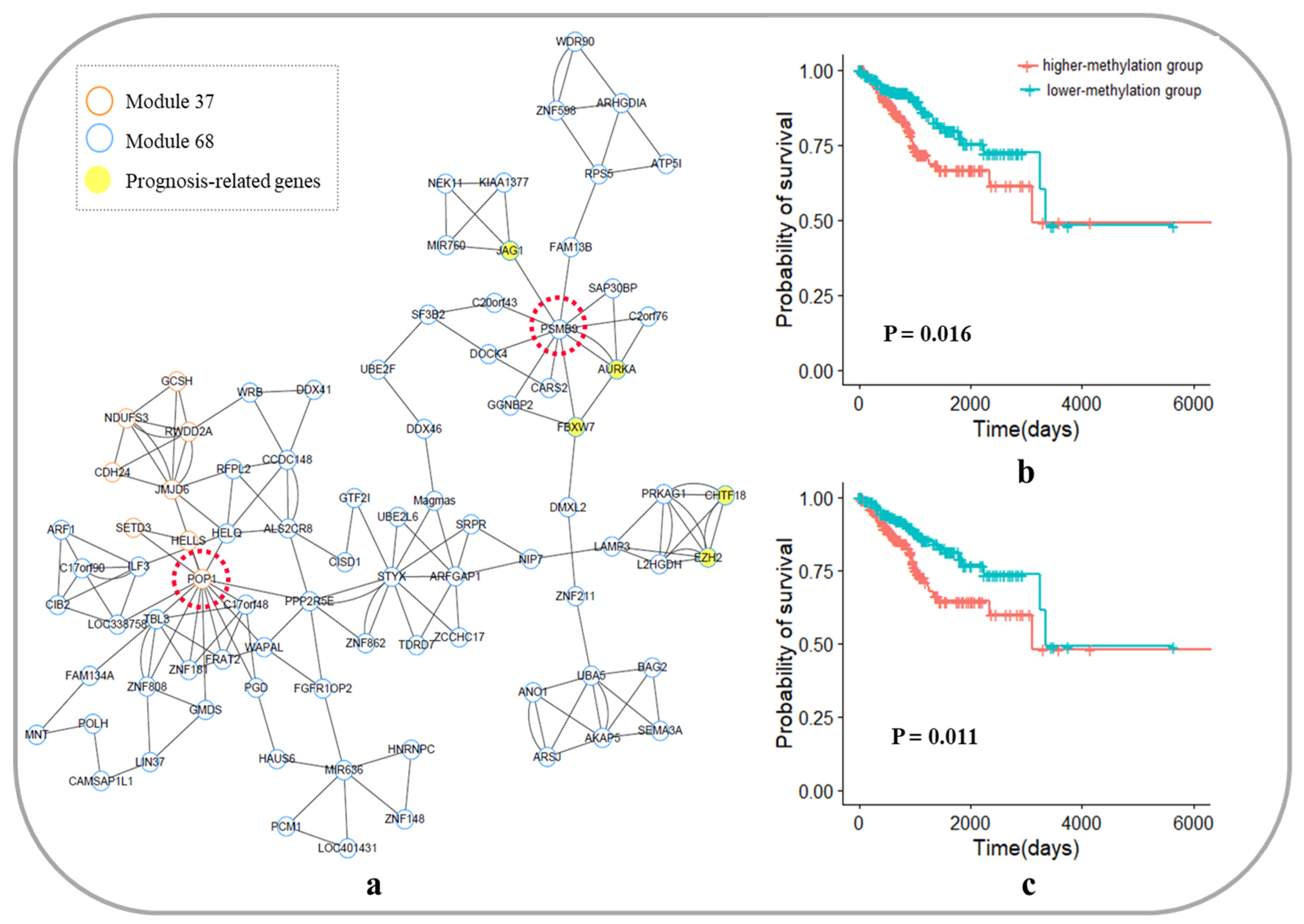
3.6. Biological Functions of the Core Module Networks
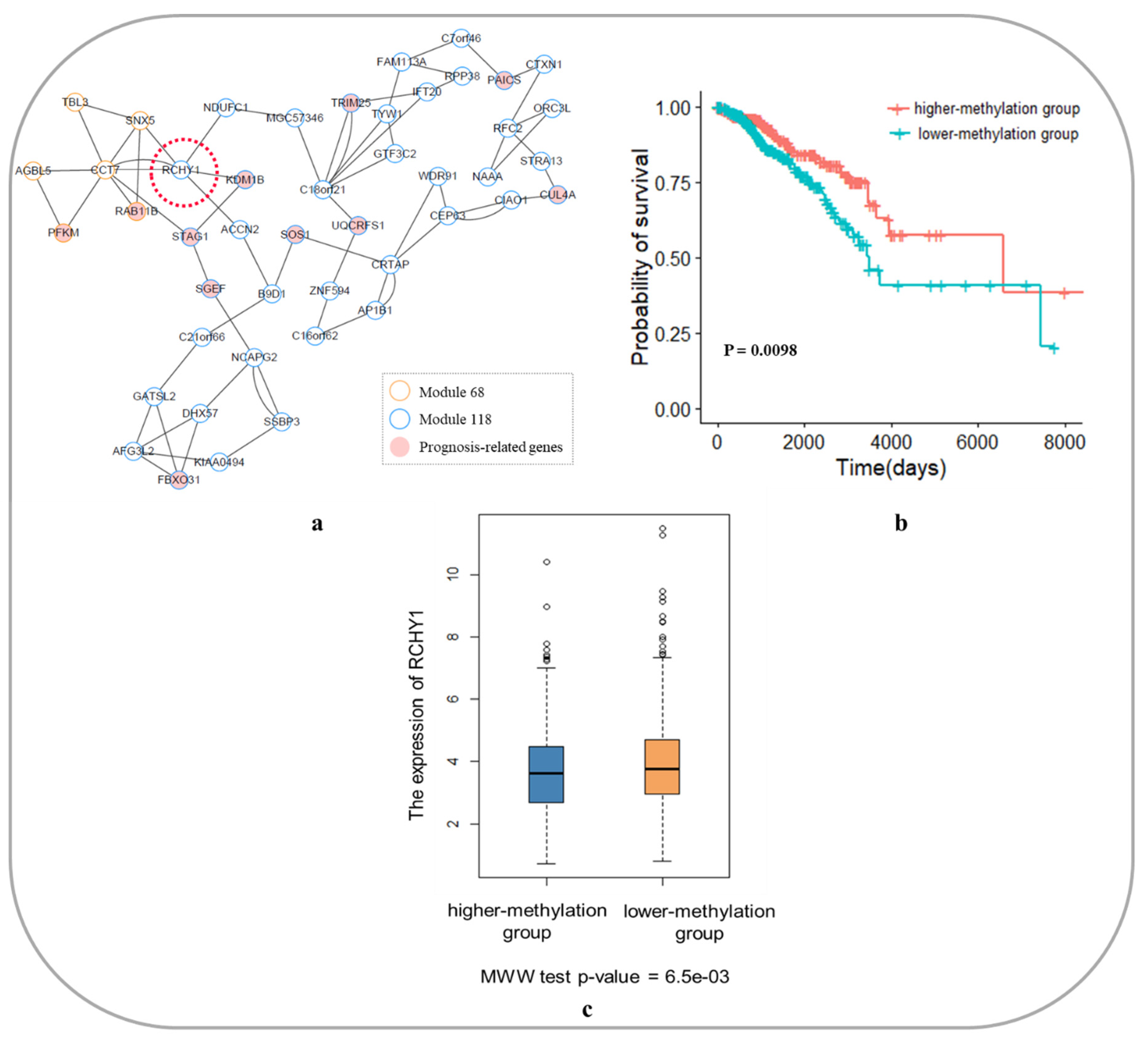
3.6.1. Core Module Network Analysis of Breast Invasive Carcinoma
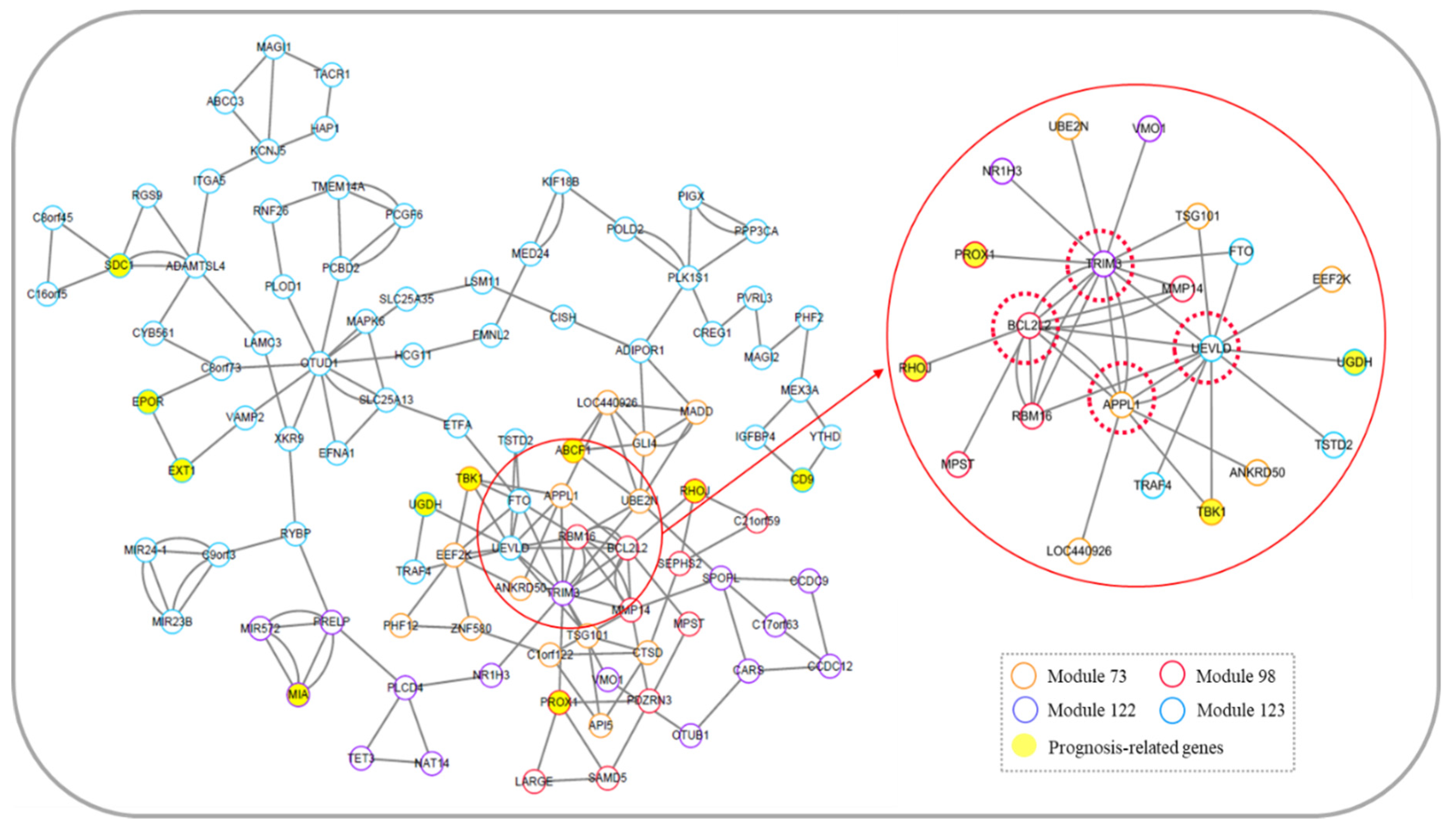
3.6.2. Core Module Network Analysis of Skin Cutaneous Melanoma
3.6.3. Core Module Network Analysis of Uterine Corpus Endometrial Carcinoma
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Montazeri, A. Quality of life data as prognostic indicators of survival in cancer patients: An overview of the literature from 1982 to 2008. Health Qual. Life Outcomes 2009, 7, 102. [Google Scholar] [CrossRef] [PubMed]
- Kuijjer, M.L.; Paulson, J.N.; Salzman, P.; Ding, W.; Quackenbush, J. Cancer subtype identification using somatic mutation data. Br. J. Cancer 2018, 118, 1492–1501. [Google Scholar] [CrossRef]
- Wang, Y.; Klijn, J.G.M.; Zhang, Y.; Sieuwerts, A.M.; Look, M.P.; Yang, F.; Talantov, D.; Timmermans, M.; Gelder, M.E.M.; Yu, J.; et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 2005, 365, 671–679. [Google Scholar] [CrossRef]
- Riester, M.; Wei, W.; Waldron, L.; Culhane, A.C.; Trippa, L.; Oliva, E.; Kim, S.; Michor, F.; Huttenhower, C.; Parmigiani, G.; et al. Risk Prediction for Late-Stage Ovarian Cancer by Meta-analysis of 1525 Patient Samples. JNCI J. Natl. Cancer Inst. 2014, 106, dju048. [Google Scholar] [CrossRef] [PubMed]
- Abraham, G.; Kowalczyk, A.; Loi, S.; Haviv, I.; Zobel, J. Prediction of breast cancer prognosis using gene set statistics provides signature stability and biological context. BMC Bioinformatics 2010, 11, 277. [Google Scholar] [CrossRef]
- Li, J.; Lenferink, A.E.G.; Deng, Y.; Collins, C.; Cui, Q.; Purisima, E.O.; O’Connor-McCourt, M.D.; Wang, E. Identification of high-quality cancer prognostic markers and metastasis network modules. Nat. Commun. 2012, 3, 655. [Google Scholar] [CrossRef]
- Seo, D.-W.; Li, H.; Guedez, L.; Wingfield, P.T.; Diaz, T.; Salloum, R.; Wei, B.; Stetler-Stevenson, W.G. TIMP-2 Mediated Inhibition of Angiogenesis: An MMP-Independent Mechanism. Cell 2003, 114, 171–180. [Google Scholar] [CrossRef]
- Gu, P.; Xing, X.; Tänzer, M.; Röcken, C.; Weichert, W.; Ivanauskas, A.; Pross, M.; Peitz, U.; Malfertheiner, P.; Schmid, R.M.; et al. Frequent Loss of TIMP-3 Expression in Progression of Esophageal and Gastric Adenocarcinomas. Neoplasia 2008, 10, 563–572. [Google Scholar] [CrossRef]
- Licht, J.D. DNA Methylation Inhibitors in Cancer Therapy: The Immunity Dimension. Cell 2015, 162, 938–939. [Google Scholar] [CrossRef]
- Coppedè, F. Epigenetic biomarkers of colorectal cancer: Focus on DNA methylation. Cancer Lett. 2014, 342, 238–247. [Google Scholar] [CrossRef]
- Koch, A.; Joosten, S.C.; Feng, Z.; de Ruijter, T.C.; Draht, M.X.; Melotte, V.; Smits, K.M.; Veeck, J.; Herman, J.G.; Van Neste, L.; et al. Analysis of DNA methylation in cancer: Location revisited. Nat. Rev. Clin. Oncol. 2018, 15, 459–466. [Google Scholar] [CrossRef]
- Hartwell, L.H.; Hopfield, J.J.; Leibler, S.; Murray, A.W. From molecular to modular cell biology. Nature 1999, 402, C47–C52. [Google Scholar] [CrossRef]
- Tsunematsu, Y.; Ishikawa, N.; Wakana, D.; Goda, Y.; Noguchi, H.; Moriya, H.; Hotta, K.; Watanabe, K. Distinct mechanisms for spiro-carbon formation reveal biosynthetic pathway crosstalk. Nat. Chem. Biol. 2013, 9, 818–825. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, Y.; Gu, Y.; Zhu, J.; Ci, C.; Guo, Z.; Chen, C.; Wei, Y.; Lv, W.; Liu, H.; et al. Specific breast cancer prognosis-subtype distinctions based on DNA methylation patterns. Mol. Oncol. 2018, 12, 1047–1060. [Google Scholar] [CrossRef]
- de Almeida, B.P.; Apolónio, J.D.; Binnie, A.; Castelo-Branco, P. Roadmap of DNA methylation in breast cancer identifies novel prognostic biomarkers. BMC Cancer 2019, 19, 219. [Google Scholar] [CrossRef]
- Tomczak, K.; Czerwińska, P.; Wiznerowicz, M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Współczesna Onkol. 2015, 1A, 68–77. [Google Scholar] [CrossRef]
- Chen, Y.; Lemire, M.; Choufani, S.; Butcher, D.T.; Grafodatskaya, D.; Zanke, B.W.; Gallinger, S.; Hudson, T.J.; Weksberg, R. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 2013, 8, 203–209. [Google Scholar] [CrossRef]
- Du, P.; Zhang, X.; Huang, C.-C.; Jafari, N.; Kibbe, W.A.; Hou, L.; Lin, S.M. Comparison of β-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics 2010, 11, 587. [Google Scholar] [CrossRef]
- Hu, W.-L.; Zhou, X.-H. Identification of prognostic signature in cancer based on DNA methylation interaction network. BMC Med. Genomics 2017, 10, 63. [Google Scholar] [CrossRef]
- Azuaje, F.J.; Wang, H.; Zheng, H.; Léonard, F.; Rolland-Turner, M.; Zhang, L.; Devaux, Y.; Wagner, D.R. Predictive integration of gene functional similarity and co-expression defines treatment response of endothelial progenitor cells. BMC Syst. Biol. 2011, 5, 46. [Google Scholar] [CrossRef]
- Horan, K.; Jang, C.; Bailey-Serres, J.; Mittler, R.; Shelton, C.; Harper, J.F.; Zhu, J.-K.; Cushman, J.C.; Gollery, M.; Girke, T. Annotating Genes of Known and Unknown Function by Large-Scale Coexpression Analysis. Plant Physiol. 2008, 147, 41–57. [Google Scholar] [CrossRef]
- Akulenko, R.; Helms, V. DNA co-methylation analysis suggests novel functional associations between gene pairs in breast cancer samples. Hum. Mol. Genet. 2013, 22, 3016–3022. [Google Scholar] [CrossRef]
- Elo, L.L.; Järvenpää, H.; Orešič, M.; Lahesmaa, R.; Aittokallio, T. Systematic construction of gene coexpression networks with applications to human T helper cell differentiation process. Bioinformatics 2007, 23, 2096–2103. [Google Scholar] [CrossRef]
- Presson, A.P.; Sobel, E.M.; Papp, J.C.; Suarez, C.J.; Whistler, T.; Rajeevan, M.S.; Vernon, S.D.; Horvath, S. Integrated Weighted Gene Co-expression Network Analysis with an Application to Chronic Fatigue Syndrome. BMC Syst. Biol. 2008, 2, 95. [Google Scholar] [CrossRef]
- Aggarwal, A.; Li Guo, D.; Hoshida, Y.; Tsan Yuen, S.; Chu, K.-M.; So, S.; Boussioutas, A.; Chen, X.; Bowtell, D.; Aburatani, H.; et al. Topological and Functional Discovery in a Gene Coexpression Meta-Network of Gastric Cancer. Cancer Res. 2006, 66, 232–241. [Google Scholar] [CrossRef]
- Ray, M.; Ruan, J.; Zhang, W. Variations in the transcriptome of Alzheimer’s disease reveal molecular networks involved in cardiovascular diseases. Genome Biol. 2008, 9, R148. [Google Scholar] [CrossRef]
- Ruan, J.; Dean, A.K.; Zhang, W. A general co-expression network-based approach to gene expression analysis: Comparison and applications. BMC Syst. Biol. 2010, 4, 8. [Google Scholar] [CrossRef]
- Shannon, P. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Bader, G.D.; Hogue, C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003, 27. [Google Scholar] [CrossRef]
- Kitsak, M.; Gallos, L.K.; Havlin, S.; Liljeros, F.; Muchnik, L.; Stanley, H.E.; Makse, H.A. Identification of influential spreaders in complex networks. Nat. Phys. 2010, 6, 888–893. [Google Scholar] [CrossRef]
- Ahmed, H.; Howton, T.C.; Sun, Y.; Weinberger, N.; Belkhadir, Y.; Mukhtar, M.S. Network biology discovers pathogen contact points in host protein-protein interactomes. Nat. Commun. 2018, 9, 2312. [Google Scholar] [CrossRef]
- Sotiriou, C.; Wirapati, P.; Loi, S.; Harris, A.; Fox, S.; Smeds, J.; Nordgren, H.; Farmer, P.; Praz, V.; Haibe-Kains, B.; et al. Gene expression profiling in breast cancer: Understanding the molecular basis of histologic grade to improve prognosis. JNCI J. Natl. Cancer Inst. 2006, 98, 262–272. [Google Scholar] [CrossRef]
- Zhou, X.H.; Chu, X.Y.; Xue, G.; Xiong, J.H.; Zhang, H.-Y. Identifying cancer prognostic modules by module network analysis. BMC Bioinformatics 2019, 20, 85. [Google Scholar] [CrossRef]
- Rousseeuw, P.J. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987, 20, 53–65. [Google Scholar] [CrossRef]
- Du, X.; Li, X.Q.; Li, L.; Xu, Y.Y.; Feng, Y.M. The detection of ESR1/PGR/ERBB2 mRNA levels by RT-qPCR: A better approach for subtyping breast cancer and predicting prognosis. Breast Cancer Res. Treat. 2013, 138, 59–67. [Google Scholar] [CrossRef]
- Katz, T.A.; Vasilatos, S.N.; Harrington, E.; Oesterreich, S.; Davidson, N.E.; Huang, Y. Inhibition of histone demethylase, LSD2 (KDM1B), attenuates DNA methylation and increases sensitivity to DNMT inhibitor-induced apoptosis in breast cancer cells. Breast Cancer Res. Treat. 2014, 146, 99–108. [Google Scholar] [CrossRef]
- Chen, L.; Vasilatos, S.N.; Qin, Y.; Katz, T.A.; Cao, C.; Wu, H.; Tasdemir, N.; Levine, K.M.; Oesterreich, S.; Davidson, N.E.; et al. Functional characterization of lysine-specific demethylase 2 (LSD2/KDM1B) in breast cancer progression. Oncotarget 2017, 8, 81737–81753. [Google Scholar] [CrossRef]
- Sheng, Y.; Laister, R.C.; Lemak, A.; Wu, B.; Tai, E.; Duan, S.; Lukin, J.; Sunnerhagen, M.; Srisailam, S.; Karra, M.; et al. Molecular basis of Pirh2-mediated p53 ubiquitylation. Nat. Struct. Mol. Biol. 2008, 15, 1334–1342. [Google Scholar] [CrossRef]
- Liu, L.; Lee, S.; Zhang, J.; Peters, S.B.; Hannah, J.; Zhang, Y.; Yin, Y.; Koff, A.; Ma, L.; Zhou, P. CUL4A abrogation augments DNA damage response and protection against skin carcinogenesis. Mol. Cell 2009, 34, 451–460. [Google Scholar] [CrossRef]
- Chen, L.C.; Manjeshwar, S.; Lu, Y.; Moore, D.; Ljung, U.M.; Kuo, W.L.; Dairkee, S.H.; Wernick, M.; Collins, C. The human homologue for the caenorhabditis elegans cul-4 gene is amplified and overexpressed in primary breast cancers. Cancer Res. 1998, 58, 3677–3683. [Google Scholar]
- Melchor, L.; Saucedo-Cuevas, L.P.; Muñoz-Repeto, I.; Rodríguez-Pinilla, S.M.; Honrado, E.; Campoverde, A.; Palacios, J.; Nathanson, K.L.; García, M.J.; Benítez, J. Comprehensive characterization of the DNA amplification at 13q34 in human breast cancer reveals TFDP1 and CUL4A as likely candidate target genes. Breast Cancer Res. 2009, 11, R86. [Google Scholar] [CrossRef]
- Kumar, R.; Neilsen, P.M.; Crawford, J.; McKirdy, R.; Lee, J.; Powell, J.A.; Saif, Z.; Martin, J.M.; Lombaerts, M.; Cornelisse, C.J.; et al. FBXO31 Is the Chromosome 16q24.3 Senescence Gene, a Candidate Breast Tumor Suppressor, and a Component of an SCF Complex. Cancer Res. 2005, 65, 11304–11313. [Google Scholar] [CrossRef]
- Ciccone, D.N.; Su, H.; Hevi, S.; Gay, F.; Lei, H.; Bajko, J.; Xu, G.; Li, E.; Chen, T. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 2009, 461, 415–418. [Google Scholar] [CrossRef]
- Meng, M.; Chen, Y.; Jia, J.; Li, L.; Yang, S. Knockdown of PAICS inhibits malignant proliferation of human breast cancer cell lines. Biol. Res. 2018, 51, 24. [Google Scholar] [CrossRef]
- Gallenne, T.; Ross, K.N.; Visser, N.L.; Salony; Desmet, C.J.; Wittner, B.S.; Wessels, L.F.A.; Ramaswamy, S.; Peeper, D.S. Systematic functional perturbations uncover a prognostic genetic network driving human breast cancer. Oncotarget 2017, 8, 20572–20587. [Google Scholar] [CrossRef]
- Ahsan, H.; Halpern, J.; Kibriya, M.G.; Pierce, B.L.; Tong, L.; Gamazon, E.; McGuire, V.; Felberg, A.; Shi, J.; Jasmine, F.; et al. A genome-wide association study of early-onset breast cancer identifies PFKM as a novel breast cancer gene and supports a common genetic spectrum for breast cancer at any age. Cancer Epidemiol. Biomarkers Prev. 2014, 23, 658–669. [Google Scholar] [CrossRef]
- Hashimoto, A.; Oikawa, T.; Hashimoto, S.; Sugino, H.; Yoshikawa, A.; Otsuka, Y.; Handa, H.; Onodera, Y.; Nam, J.-M.; Oneyama, C.; et al. P53- and mevalonate pathway–driven malignancies require Arf6 for metastasis and drug resistance. J. Cell Biol. 2016, 213, 81–95. [Google Scholar] [CrossRef]
- Bhuin, T.; Roy, J.K. Rab11 in Disease Progression. Int. J. Mol. Cell Med. 2015, 4, 1–8. [Google Scholar]
- Goicoechea, S.M.; Zinn, A.; Awadia, S.S.; Snyder, K.; Garcia-Mata, R. A RhoG-mediated signaling pathway that modulates invadopodia dynamics in breast cancer cells. J. Cell Sci. 2017, 130, 1064–1077. [Google Scholar] [CrossRef]
- Ellerbroek, S.M.; Wennerberg, K.; Arthur, W.T.; Dunty, J.M.; Bowman, D.R.; DeMali, K.A.; Der, C.; Burridge, K. SGEF, a RhoG Guanine Nucleotide Exchange Factor that Stimulates Macropinocytosis. Mol. Biol. Cell 2004, 15, 3309–3319. [Google Scholar] [CrossRef]
- De, S.; Dermawan, J.K.T.; Stark, G.R. EGF receptor uses SOS1 to drive constitutive activation of NF B in cancer cells. Proc. Natl. Acad. Sci. USA 2014, 111, 11721–11726. [Google Scholar] [CrossRef]
- Field, L.A.; Love, B.; Deyarmin, B.; Hooke, J.A.; Shriver, C.D.; Ellsworth, R.E. Identification of differentially expressed genes in breast tumors from African American compared with Caucasian women. Cancer 2012, 118, 1334–1344. [Google Scholar] [CrossRef]
- Giannini, G.; Ambrosini, M.I.; Di Marcotullio, L.; Cerignoli, F.; Zani, M.; MacKay, A.R.; Screpanti, I.; Frati, L.; Gulino, A. EGF- and cell-cycle-regulatedSTAG1/PMEPA1/ERG1.2 belongs to a conserved gene family and is overexpressed and amplified in breast and ovarian cancer. Mol. Carcinog. 2003, 38, 188–200. [Google Scholar] [CrossRef]
- Walsh, L.A.; Alvarez, M.J.; Sabio, E.Y.; Reyngold, M.; Makarov, V.; Mukherjee, S.; Lee, K.-W.; Desrichard, A.; Turcan, Ş.; Dalin, M.G.; et al. An Integrated Systems Biology Approach Identifies TRIM25 as a Key Determinant of Breast Cancer Metastasis. Cell Rep. 2017, 20, 1623–1640. [Google Scholar] [CrossRef]
- Cao, F.; Li, D.-P.; Wang, L.; Li, M.; Zhang, H.; Tao, M. TRIM25 promotes oncogenic activities through regulation of ZEB1 in breast cancer. Int. J. Clin. Exp. Pathol. 2016, 9, 9751–9760. [Google Scholar]
- Ohashi, Y.; Kaneko, S.J.; Cupples, T.E.; Young, S.R. Ubiquinol cytochrome c reductase (UQCRFS1) gene amplification in primary breast cancer core biopsy samples. Gynecol. Oncol. 2004, 93, 54–58. [Google Scholar] [CrossRef]
- Heimerl, S.; Bosserhoff, A.K.; Langmann, T.; Ecker, J.; Schmitz, G. Mapping ATP-binding cassette transporter gene expression profiles in melanocytes and melanoma cells. Melanoma Res. 2007, 17, 265–273. [Google Scholar] [CrossRef]
- Rambow, F.; Malek, O.; Geffrotin, C.; Leplat, J.-J.; Bouet, S.; Piton, G.; Hugot, K.; Bevilacqua, C.; Horak, V.; Vincent-Naulleau, S. Identification of differentially expressed genes in spontaneously regressing melanoma using the MeLiM Swine Model: Differential gene expression in swine melanoma. Pigment Cell Melanoma Res. 2008, 21, 147–161. [Google Scholar] [CrossRef]
- Miyake, M.; Inufusa, H.; Adachi, M.; Ishida, H.; Hashida, H.; Tokuhara, T.; Kakehi, Y. Suppression of pulmonary metastasis using adenovirally motility related protein-1 (MRP-1/CD9) gene delivery. Oncogene 2000, 19, 5221–5226. [Google Scholar] [CrossRef]
- Mirmohammadsadegh, A.; Marini, A.; Gustrau, A.; Delia, D.; Nambiar, S.; Hassan, M.; Hengge, U.R. Role of Erythropoietin Receptor Expression in Malignant Melanoma. J. Invest. Dermatol. 2010, 130, 201–210. [Google Scholar] [CrossRef]
- Kumar, S.M.; Zhang, G.; Bastian, B.C.; Arcasoy, M.O.; Karande, P.; Pushparajan, A.; Acs, G.; Xu, X. Erythropoietin receptor contributes to melanoma cell survival in vivo. Oncogene 2012, 31, 1649–1660. [Google Scholar] [CrossRef]
- Ropero, S.; Setien, F.; Espada, J.; Fraga, M.F.; Herranz, M.; Asp, J.; Benassi, M.S.; Franchi, A.; Patiño, A.; Ward, L.S.; et al. Epigenetic loss of the familial tumor-suppressor gene exostosin-1 (EXT1) disrupts heparan sulfate synthesis in cancer cells. Hum. Mol. Genet. 2004, 13, 2753–2765. [Google Scholar] [CrossRef]
- Stahlecker, J.; Gauger, A.; Bosserhoff, A.; Büttner, R.; Ring, J.; Hein, R. MIA as a reliable tumor marker in the serum of patients with malignant melanoma. Anticancer Res. 2000, 20, 5041–5044. [Google Scholar]
- Uslu, U.; Schliep, S.; Schliep, K.; Erdmann, M.; Koch, H.U.; Parsch, H.; Rosenheinrich, S.; Anzengruber, D.; Bosserhoff, A.K.; Schuler, G.; et al. Comparison of the Serum Tumor Markers S100 and Melanoma-inhibitory Activity (MIA) in the Monitoring of Patients with Metastatic Melanoma Receiving Vaccination Immunotherapy with Dendritic Cells. Anticancer Res. 2017, 37, 5033–5037. [Google Scholar]
- Vitoux, D.; Mourah, S.; Kerob, D.; Verola, O.; Basset-Seguin, N.; Baccard, M.; Schartz, N.; Ollivaud, L.; Archimbaud, A.; Servant, J.-M.; et al. Highly Sensitive Multivariable Assay Detection of Melanocytic Differentiation Antigens and Angiogenesis Biomarkers in Sentinel Lymph Nodes With Melanoma Micrometastases. Arch. Dermatol. 2009, 145, 1105–1113. [Google Scholar] [CrossRef][Green Version]
- Monzani, E.; Facchetti, F.; Galmozzi, E.; Corsini, E.; Benetti, A.; Cavazzin, C.; Gritti, A.; Piccinini, A.; Porro, D.; Santinami, M.; et al. Melanoma contains CD133 and ABCG2 positive cells with enhanced tumourigenic potential. Eur. J. Cancer 2007, 43, 935–946. [Google Scholar] [CrossRef]
- Ho, H.; Soto Hopkin, A.; Kapadia, R.; Vasudeva, P.; Schilling, J.; Ganesan, A.K. RhoJ modulates melanoma invasion by altering actin cytoskeletal dynamics. Pigment Cell Melanoma Res. 2013, 26, 218–225. [Google Scholar] [CrossRef]
- Ruiz, R.; Jahid, S.; Harris, M.; Marzese, D.M.; Espitia, F.; Vasudeva, P.; Chen, C.-F.; de Feraudy, S.; Wu, J.; Gillen, D.L.; et al. The RhoJ-BAD signaling network: An Achilles’ heel for BRAF mutant melanomas. PLOS Genet. 2017, 13, e1006913. [Google Scholar] [CrossRef]
- Wang, C.; Tseng, T.; Jhang, Y.; Tseng, J.; Hsieh, C.; Wu, W.; Lee, S. Loss of cell invasiveness through PKC-mediated syndecan-1 downregulation in melanoma cells under anchorage independency. Exp. Dermatol. 2014, 23, 843–849. [Google Scholar] [CrossRef]
- Tseng, T.; Uen, W.; Tseng, J.; Lee, S. Enhanced chemosensitization of anoikis-resistant melanoma cells through syndecan-2 upregulation upon anchorage independency. Oncotarget 2017, 8, 61528–61537. [Google Scholar] [CrossRef][Green Version]
- Vu, H.L.; Aplin, A.E. Targeting TBK1 Inhibits Migration and Resistance to MEK Inhibitors in Mutant NRAS Melanoma. Mol. Cancer Res. 2014, 12, 1509–1519. [Google Scholar] [CrossRef]
- Deen, A.J.; Arasu, U.T.; Pasonen-Seppänen, S.; Hassinen, A.; Takabe, P.; Wojciechowski, S.; Kärnä, R.; Rilla, K.; Kellokumpu, S.; Tammi, R.; et al. UDP-sugar substrates of HAS3 regulate its O-GlcNAcylation, intracellular traffic, extracellular shedding and correlate with melanoma progression. Cell. Mol. Life Sci. 2016, 73, 3183–3204. [Google Scholar] [CrossRef]
- Umene, K.; Yanokura, M.; Banno, K.; Irie, H.; Adachi, M.; Iida, M.; Nakamura, K.; Nogami, Y.; Masuda, K.; Kobayashi, Y.; et al. Aurora kinase A has a significant role as a therapeutic target and clinical biomarker in endometrial cancer. Int. J. Oncol. 2015, 46, 1498–1506. [Google Scholar] [CrossRef]
- Townsend, M.H.; Ence, Z.E.; Felsted, A.M.; Parker, A.C.; Piccolo, S.R.; Robison, R.A.; O’Neill, K.L. Potential new biomarkers for endometrial cancer. Cancer Cell Int. 2019, 19, 19. [Google Scholar] [CrossRef]
- Price, J.C.; Pollock, L.M.; Rudd, M.L.; Fogoros, S.K.; Mohamed, H.; Hanigan, C.L.; Le Gallo, M.; NIH Intramural Sequencing Center (NISC) Comparative Sequencing Program; Zhang, S.; Cruz, P.; et al. Sequencing of Candidate Chromosome Instability Genes in Endometrial Cancers Reveals Somatic Mutations in ESCO1, CHTF18, and MRE11A. PLoS ONE 2013, 8, e63313. [Google Scholar] [CrossRef]
- Oki, S.; Sone, K.; Oda, K.; Hamamoto, R.; Ikemura, M.; Maeda, D.; Takeuchi, M.; Tanikawa, M.; Mori-Uchino, M.; Nagasaka, K.; et al. Oncogenic histone methyltransferase EZH2: A novel prognostic marker with therapeutic potential in endometrial cancer. Oncotarget 2017, 8, 40402–40411. [Google Scholar] [CrossRef]
- Alldredge, J.K.; Eskander, R.N. EZH2 inhibition in ARID1A mutated clear cell and endometrioid ovarian and endometrioid endometrial cancers. Gynecol. Oncol. Res. Pract. 2017, 4, 17. [Google Scholar] [CrossRef]
- Nakayama, K.; Rahman, M.T.; Rahman, M.; Nakamura, K.; Ishikawa, M.; Katagiri, H.; Sato, E.; Ishibashi, T.; Iida, K.; Ishikawa, N.; et al. CCNE1 amplification is associated with aggressive potential in endometrioid endometrial carcinomas. Int. J. Oncol. 2016, 48, 506–516. [Google Scholar] [CrossRef]
- Kuhn, E.; Wu, R.-C.; Guan, B.; Wu, G.; Zhang, J.; Wang, Y.; Song, L.; Yuan, X.; Wei, L.; Roden, R.B.S.; et al. Identification of Molecular Pathway Aberrations in Uterine Serous Carcinoma by Genome-wide Analyses. JNCI J. Natl. Cancer Inst. 2012, 104, 1503–1513. [Google Scholar] [CrossRef]
- Mitsuhashi, Y.; Horiuchi, A.; Miyamoto, T.; Kashima, H.; Suzuki, A.; Shiozawa, T. Prognostic significance of Notch signalling molecules and their involvement in the invasiveness of endometrial carcinoma cells: Notch signalling in endometrial carcinoma. Histopathology 2012, 60, 826–837. [Google Scholar] [CrossRef]

| Gene Symbol | Full Name | Gene Function | References |
|---|---|---|---|
| CUL4A | cullin 4A | CUL4A is the ubiquitin ligase component of a multimeric complex involved in the degradation of DNA damage-response proteins. Overexpression of CUL4A is associated with poor prognosis in BRCA. | [39,40,41] |
| FBXO31 | F-box protein 31 | This gene is a member of the F-box family. The FBXO31 dysregulation is associated with the development of BRCA and is a candidate tumor suppressor gene. | [42] |
| KDM1B | lysine demethylase 1B | This gene is a flavin-dependent histone demethylase that regulates histone lysine methylation. Abnormal DNA methylation of KDM1B is associated with poor prognosis in BRCA. | [37,43] |
| PAICS | phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazolesuccinocarboxamide synthase | PAICS is an enzyme required for biosynthesis of purine. It is associated with poor prognosis of BRCA. | [44,45] |
| PFKM | phosphofructokinase, muscle | The gene is associated with increased risk of BRCA. | [46] |
| RAB11B | RAB11B, member RAS oncogene family | RAB11B is a member of the Ras superfamily of small GTP-binding proteins. It is associated with metastasis of breast cancer. | [47,48] |
| SGEF | Rho guanine nucleotide exchange factor 26 | The gene encodes a member of the Rho-guanine nucleotide exchange factor family and is associated with cancer invasion. | [49,50] |
| SOS1 | SOS Ras/Rac guanine nucleotide exchange factor 1 | The protein encoded by gene SOS1 is a guanine nucleotide exchange factor of RAS protein, a membrane protein that binds to guanine nucleotides and participates in signal transduction pathways. It is associated with invasion and metastasis of breast cancer. | [51,52] |
| STAG1 | stromal antigen 1 | This gene is a member of the SCC3 family and is expressed in the nucleus. Its overexpression might be regarded as a tumor marker in BRCA. | [53] |
| TRIM25 | tripartite motif containing 25 | Expression of the TRIM25 is upregulated in response to estrogen, and the TRIM25 is as a diver of poor outcome in BRCA. | [54,55] |
| UQCRFS1 | ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 | The gene is a key subunit of the cytochrome bc1 complex and is associated with the development of breast cancer. | [56] |
| Gene Symbol | Full Name | Gene Function | References |
|---|---|---|---|
| ABCF1 | ATP binding cassette subfamily F member 1 | The protein encoded by this gene is a member of the ATP-binding cassette (ABC) transporters superfamily, and it is involved in the development of SKCM. | [57] |
| CD9 | CD9 molecule | This gene encodes a transmembrane 4 superfamily member whose expression plays a key role in inhibiting the metastasis of SKCM. | [58,59] |
| EPOR | erythropoietin receptor | The gene encodes an erythropoietin receptor and is a member of the cytokine receptor family. Its dysregulation is associated with metastasis and prognosis of cutaneous melanoma. | [60,61] |
| EXT1 | exostosin glycosyltransferase 1 | This gene encodes an endoplasmic reticulum-resident type II transmembrane glycosyltransferase. It is associated with the development of SKCM. | [62] |
| MIA | MIA SH3 domain containing | It is a cartilage-derived retinoic acid-sensitive protein. This gene is associated with metastasis of cutaneous melanoma and is highly sensitive tumor markers for monitoring of patients with SKCM. | [63,64] |
| PROX1 | prospero homeobox 1 | The protein encoded by this gene is a member of the homeobox transcription factor family. It is related to the prognosis of SKCM. | [65,66] |
| RHOJ | ras homolog family member J | This gene encodes a Rho family GTP-binding protein that is involved in the invasion and metastasis of cutaneous melanoma. | [67,68] |
| SDC1 | syndecan 1 | The protein encoded by this gene is a transmembrane (type I) heparan sulfate proteoglycan, and it is associated with invasion of SKCM. | [69,70] |
| TBK1 | TANK binding kinase 1 | The gene is a member of the atypical IκB kinase family and is associated with invasion and migration of SKCM. | [71] |
| UGDH | UDP-glucose 6-dehydrogenase | The protein encoded by this gene converts UDP-glucose into UDP-glucuronic acid, thereby participating in the biosynthesis of glycosaminoglycans. It is related to the development of SKCM. | [72] |
| Gene Symbol | Full Name | Gene Function | References |
|---|---|---|---|
| AURKA | aurora kinase A | The protein encoded by this gene is a cell cycle-regulated kinase. It has a significant relationship with the prognosis of UCEC and can be used as a clinical biomarker for UCEC. | [73,74] |
| CHTF18 | chromosome transmission fidelity factor 18 | The protein encoded by the gene is a component of a replication factor C (RFC) complex. The mutation of this gene is associated with the pathogenesis of UCEC. | [75] |
| EZH2 | enhancer of zeste 2 polycomb repressive complex 2 subunit | This gene encodes a member of the Polycomb-group (PcG) family and is related to the prognosis of UCEC. It can be used as a prognostic marker for UCEC. | [76,77] |
| FBXW7 | F-box and WD repeat domain containing 7 | This gene encodes a member of the F-box protein family. It is associated with the development and prognosis of UCEC. | [78,79] |
| JAG1 | jagged canonical Notch ligand 1 | This gene encodes a jagged 1 protein. It is closely related to the invasion and prognosis of UCEC. | [80] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, Z.-J.; Zhou, X.-H.; Zhang, H.-Y. DNA Methylation Module Network-Based Prognosis and Molecular Typing of Cancer. Genes 2019, 10, 571. https://doi.org/10.3390/genes10080571
Cui Z-J, Zhou X-H, Zhang H-Y. DNA Methylation Module Network-Based Prognosis and Molecular Typing of Cancer. Genes. 2019; 10(8):571. https://doi.org/10.3390/genes10080571
Chicago/Turabian StyleCui, Ze-Jia, Xiong-Hui Zhou, and Hong-Yu Zhang. 2019. "DNA Methylation Module Network-Based Prognosis and Molecular Typing of Cancer" Genes 10, no. 8: 571. https://doi.org/10.3390/genes10080571
APA StyleCui, Z.-J., Zhou, X.-H., & Zhang, H.-Y. (2019). DNA Methylation Module Network-Based Prognosis and Molecular Typing of Cancer. Genes, 10(8), 571. https://doi.org/10.3390/genes10080571






