Transcriptome Analysis of Landrace Pig Subcutaneous Preadipocytes during Adipogenic Differentiation
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animals and Isolation of Preadipocytes
2.3. Induction of Subcutaneous Preadipocytes
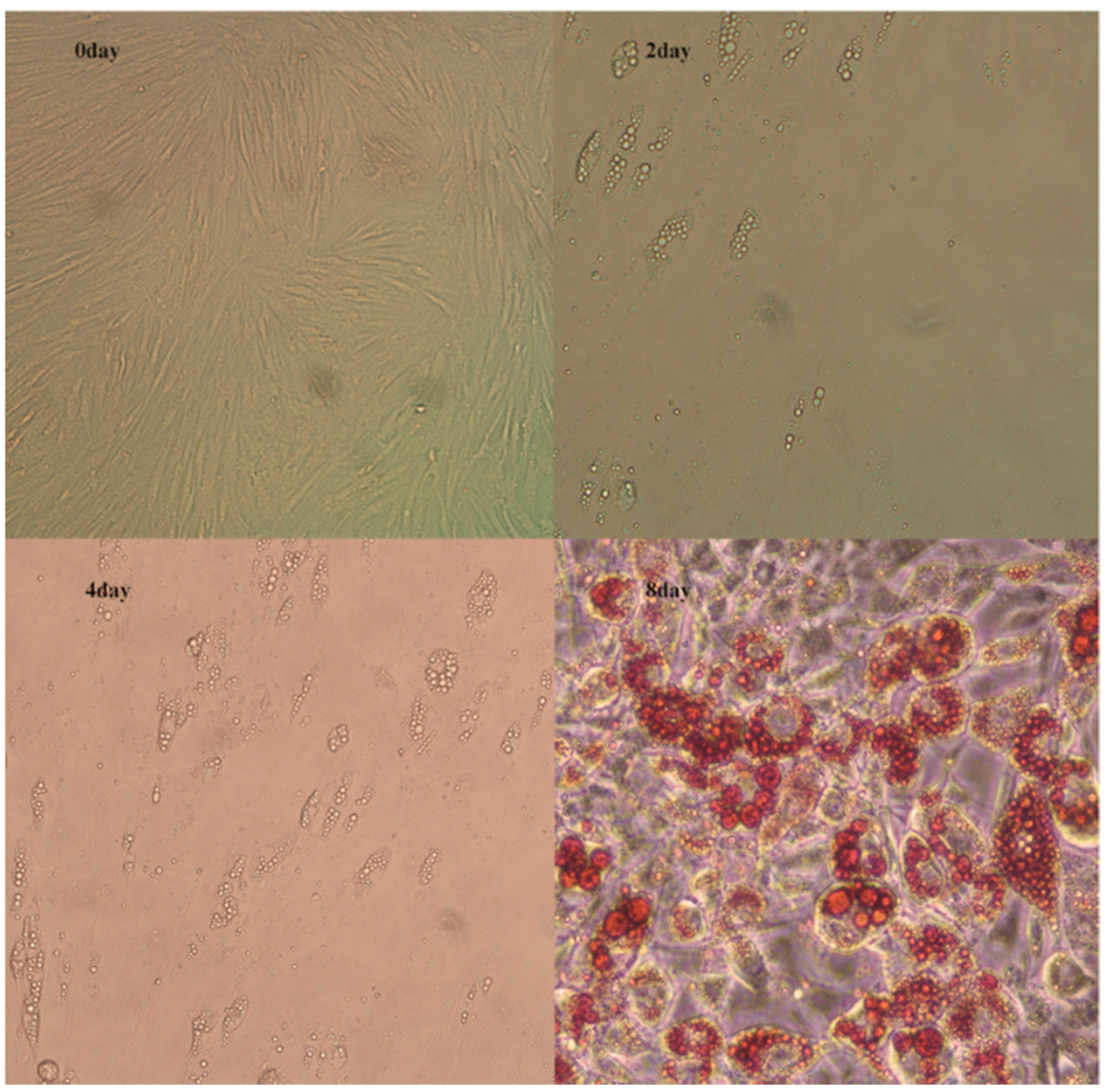
2.4. Oil Red O staining
2.5. RNA Isolation, Sequencing, and Sequence Data Processing
2.6. Short Time-Series Expression Miner Analysis
2.7. Gene Ontology and Pathway Analyses
2.8. Validation of DEGs Using Reverse Transcription Quantitative PCR
2.9. Availability of Data and Material
3. Results
3.1. Phenotypic Changes during Preadipocyte Differentiation
3.2. Sequencing Results and Quality Control
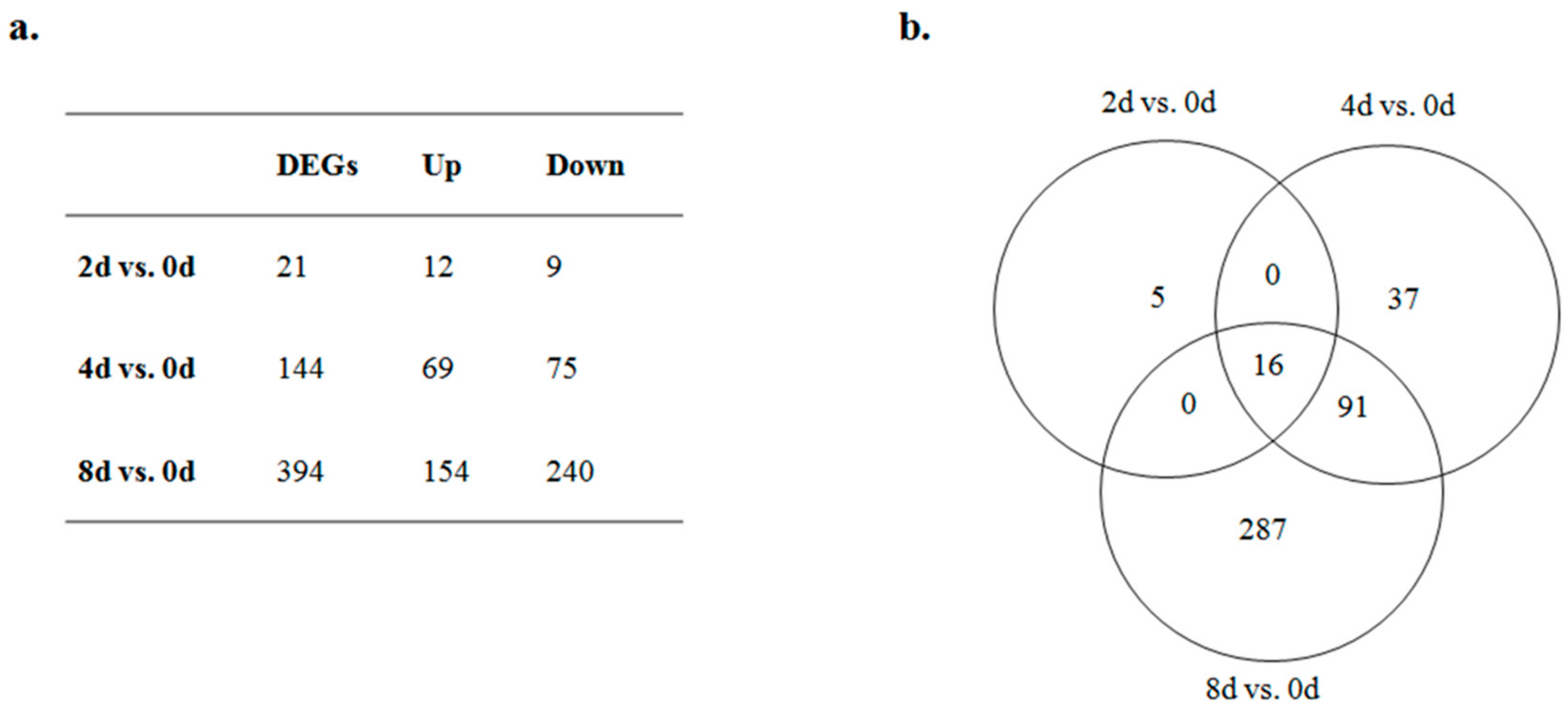
3.3. Differential Expression of mRNAs during Preadipocyte Differentiation
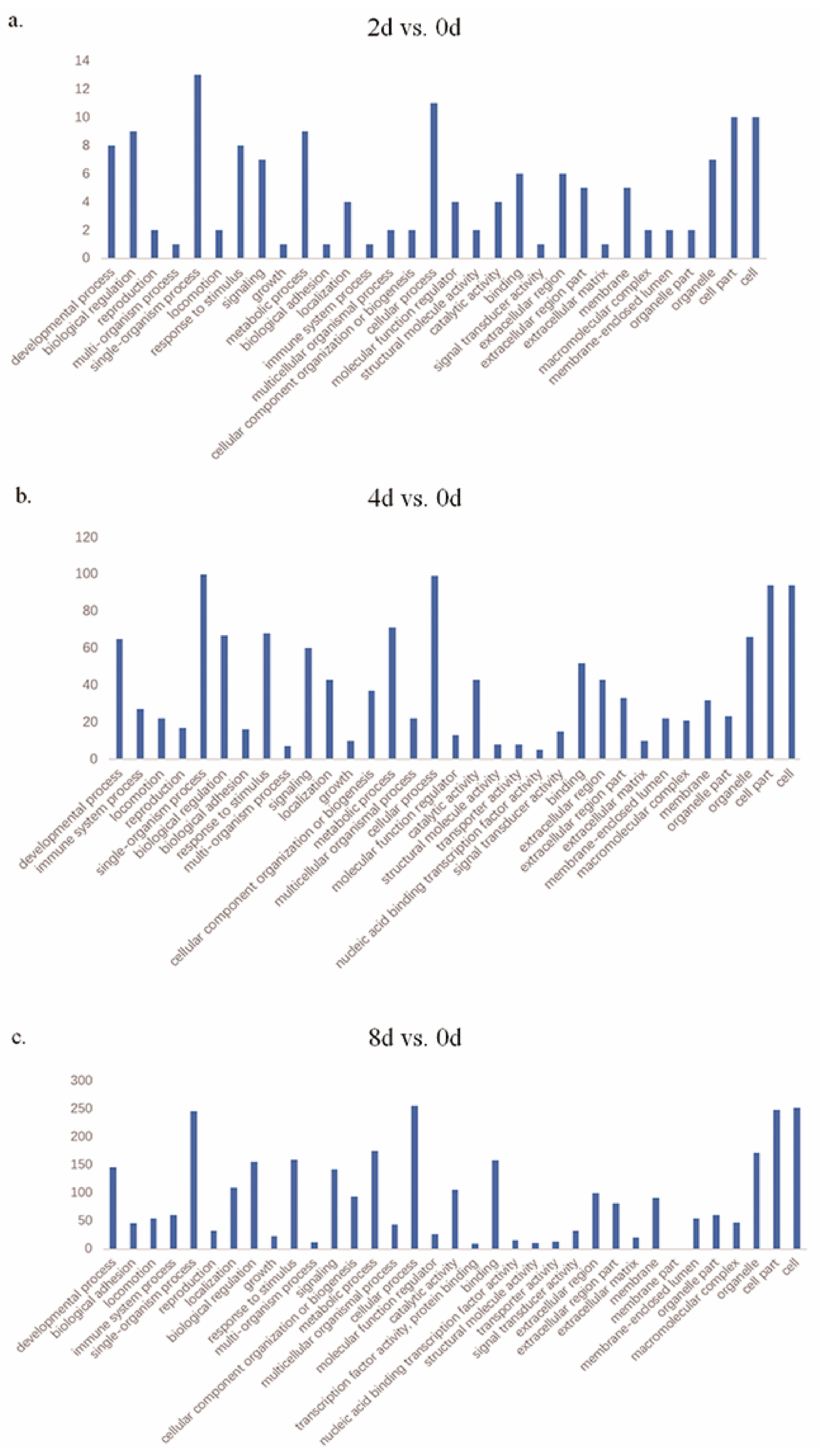
3.4. Functions of Differentially Expressed Genes
3.5. STEM Analysis
3.6. Functions of Differentially Expressed Genes Involved in Significant Gene Expression Profiles
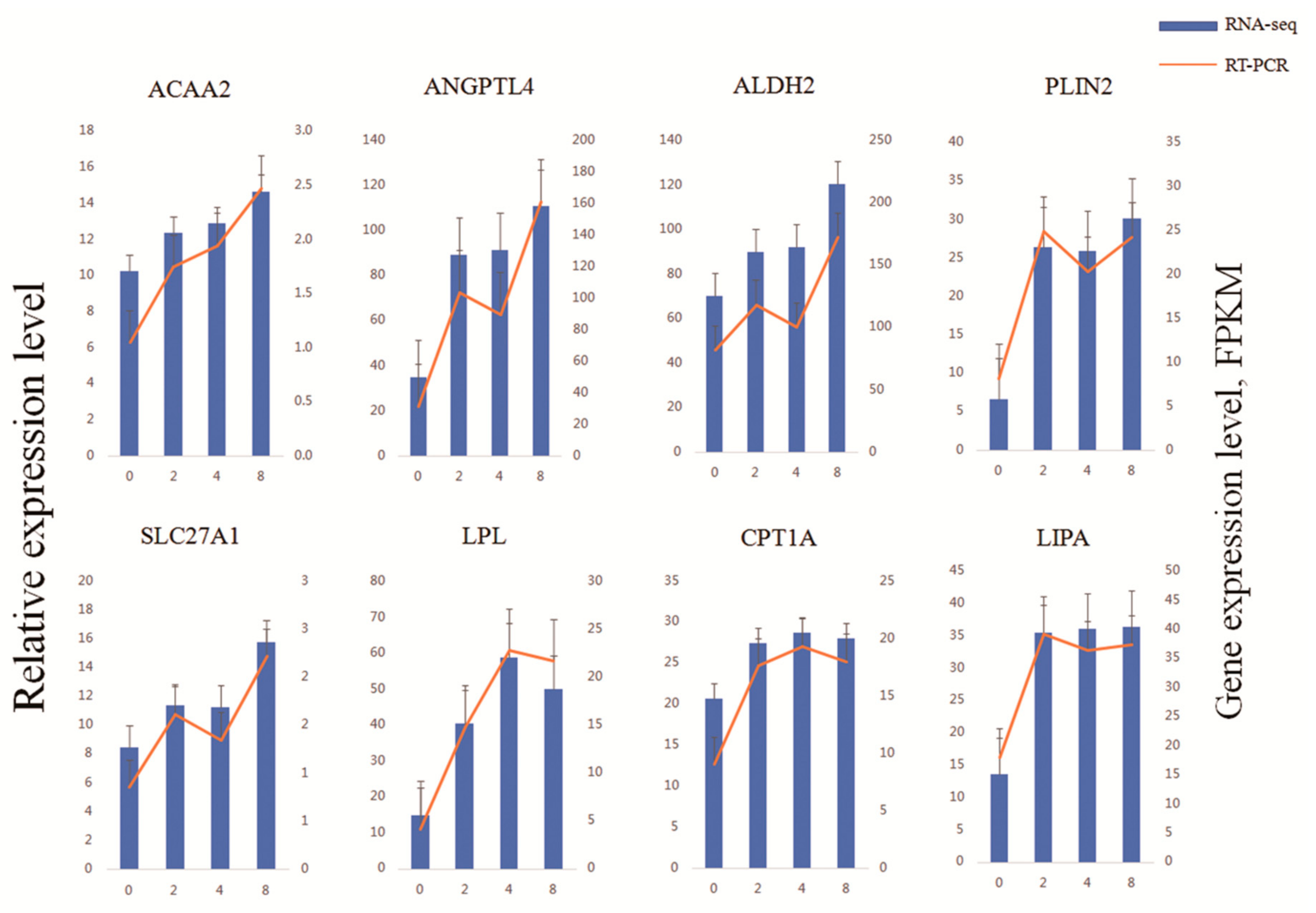
3.7. Validation of RNA-Seq-Based Gene Expression
3.8. Expression Patterns of Key Genes in Preadipocyte Differentiation
4. Discussion
4.1. Differentially Expressed Genes during Preadipocyte Differentiation
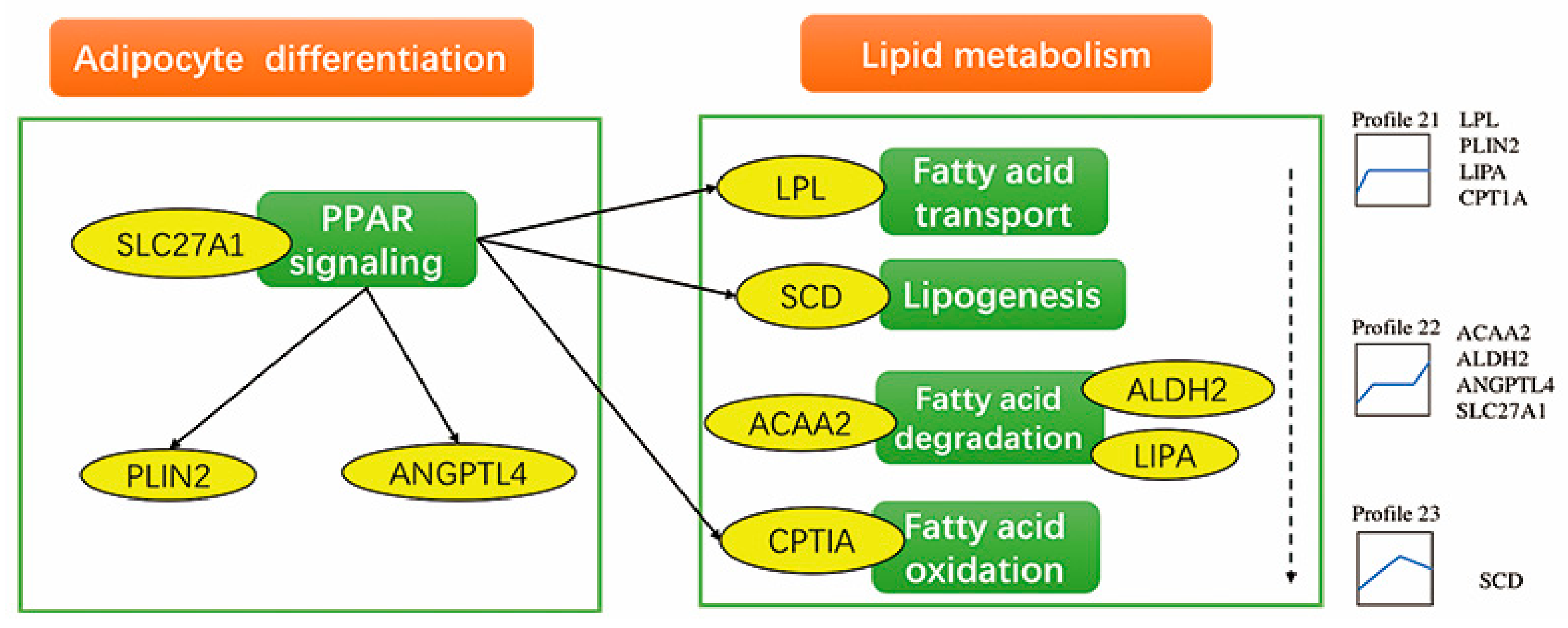
4.2. Differentially Expressed Genes and Pathways in Profile 21
4.3. Differentially Expressed Genes and Pathways in Profile 22
4.4. Other Important Differentially Expressed Genes and Pathways
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Verbeke, W.; Van Oeckel, M.J.; Warnants, N.; Viaene, J.; Boucqué, C.V. Consumer perception, facts and possibilities to improve acceptability of health and sensory characteristics of pork. Meat Sci. 1999, 53, 77–99. [Google Scholar] [CrossRef]
- Kershaw, E.E.; Flier, J.S. Adipose tissue as an endocrine organ. J. Clin. Endocrinol. Metab. 2004, 89, 2548–2556. [Google Scholar] [CrossRef] [PubMed]
- Choe, S.S.; Huh, J.Y.; Hwang, I.J.; Kim, J.I.; Kim, J.B. Adipose Tissue Remodeling: Its Role in Energy Metabolism and Metabolic Disorders. Front. Endocrinol. (Lausanne). 2016, 7, 30. [Google Scholar] [CrossRef] [PubMed]
- Gregoire, F.M.; Smas, C.M.; Sul, H.S. Understanding adipocyte differentiation. Physiol. Rev. 1998, 78, 783–809. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, Y.; Aoki, M.; Endo, T.; Sato, K. Characterization of the expression profiles of adipogenesis-related factors, ZNF423, KLFs and FGF10, during preadipocyte differentiation and abdominal adipose tissue development in chickens. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 2013, 165, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Kirkland, J.L.; Tchkonia, T.; Pirtskhalava, T.; Han, J.; Karagiannides, I. Adipogenesis and aging: does aging make fat go MAD? Exp. Gerontol. 2002, 37, 757–767. [Google Scholar] [CrossRef]
- Desvergne, B. Peroxisome Proliferator-Activated Receptors: Nuclear Control of Metabolism. Endocr. Rev. 1999, 20, 649–688. [Google Scholar]
- Ricote, M.; Li, A.C.; Willson, T.M.; Kelly, C.J.; Glass, C.K. The peroxisome proliferator-activated receptor-γ is a negative regulator of macrophage activation. Nature 1998, 391, 79–82. [Google Scholar] [CrossRef]
- Gupta, R.K.; Arany, Z.; Seale, P.; Mepani, R.J.; Ye, L.; Conroe, H.M.; Roby, Y.A.; Kulaga, H.; Reed, R.R.; Spiegelman, B.M. Transcriptional control of preadipocyte determination by Zfp423. Nature 2010, 464, 619–623. [Google Scholar] [CrossRef]
- Banerjee, S.S.; Feinberg, M.W.; Watanabe, M.; Gray, S.; Haspel, R.L.; Denkinger, D.J.; Kawahara, R.; Hauner, H.; Jain, M.K. The Krüppel-like factor KLF2 inhibits peroxisome proliferator-activated receptor-γ expression and adipogenesis. J. Biol. Chem. 2003, 278, 2581. [Google Scholar] [CrossRef]
- Mori, T.; Sakaue, H.; Iguchi, H.; Gomi, H.; Okada, Y.; Takashima, Y.; Nakamura, K.; Nakamura, T.; Yamauchi, T.; Kubota, N.; et al. Role of Krüppel-like factor 15 (KLF15) in transcriptional regulation of adipogenesis. J. Biol. Chem. 2005, 280, 12867–12875. [Google Scholar] [CrossRef] [PubMed]
- Kaczynski, J.; Cook, T.; Urrutia, R. Sp1- and Krüppel-like transcription factors. Genome Biol. 2003, 4, 206. [Google Scholar] [CrossRef] [PubMed]
- Ohta, H.; Konishi, M.; Itoh, N. FGF10 and FGF21 as regulators in adipocyte development and metabolism. Endocr. Metab. Immune Disord. Drug Targets 2011, 11, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Al Alam, D.; El Agha, E.; Sakurai, R.; Kheirollahi, V.; Moiseenko, A.; Danopoulos, S.; Shrestha, A.; Schmoldt, C.; Quantius, J.; Herold, S.; et al. Evidence for the involvement of fibroblast growth factor 10 in lipofibroblast formation during embryonic lung development. Development 2015, 142, 4139–4150. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaka, T.; Shimano, H. Elovl6: a new player in fatty acid metabolism and insulin sensitivity. J. Mol. Med. (Berl). 2009, 87, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, T.; Nakajima, I.; Eguchi-Ogawa, T.; Nagamura, Y.; Hamasima, N.; Uenishi, H. Changes in gene expression in a porcine preadipocyte cell line during differentiation. Anim. Genet. 2012, 43, 535. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.C.; Ko, Y.H.; Mersmann, H.J.; Chen, C.L.; Ding, S.T. The expression of genes related to adipocyte differentiation in pigs. J. Anim. Sci. 2006, 84, 1059–1066. [Google Scholar] [CrossRef]
- Li, W.Z.; Zhao, S.M.; Huang, Y.; Yang, M.H.; Pan, H.B.; Zhang, X.; Ge, C.R.; Gao, S.Z. Expression of lipogenic genes during porcine intramuscular preadipocyte differentiation. Res. Vet. Sci. 2012, 93, 1190–1194. [Google Scholar] [CrossRef]
- Zhou, X.; Li, D.; Yin, J.; Ni, J.; Dong, B.; Zhang, J.; Du, M. CLA differently regulates adipogenesis in stromal vascular cells from porcine subcutaneous adipose and skeletal muscle. J. Lipid Res. 2007, 48, 1701–1709. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Glaser, C.; Heinrich, J.; Koletzko, B. Role of FADS1 and FADS2 polymorphisms in polyunsaturated fatty acid metabolism. Metabolism 2010, 59, 993–999. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Ernst, J.; Bar-Joseph, Z. STEM: A tool for the analysis of short time series gene expression data. BMC Bioinformatics 2006, 7, 191. [Google Scholar] [CrossRef]
- Ghandhi, S.A.; Sinha, A.; Markatou, M.; Amundson, S.A. Time-series clustering of gene expression in irradiated and bystander fibroblasts: An application of FBPA clustering. BMC Genom. 2011, 12, 2. [Google Scholar] [CrossRef]
- Dennis, G.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, R60. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum Information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Liu, X.; Liu, K.; Shan, B.; Wei, S.; Li, D.; Han, H.; Wei, W.; Chen, J.; Liu, H.; Zhang, L. A genome-wide landscape of mRNAs, lncRNAs, and circRNAs during subcutaneous adipogenesis in pigs. J. Anim. Sci. Biotechnol. 2018, 9, 76. [Google Scholar] [CrossRef]
- Mo, D.; Yu, K.; Chen, H.; Chen, L.; Liu, X.; He, Z.; Cong, P.; Chen, Y. Transcriptome Landscape of Porcine Intramuscular Adipocytes during Differentiation. J. Agric. Food Chem. 2017, 65, 6317–6328. [Google Scholar] [CrossRef]
- He, K.; Wang, Q.; Wang, Z.; Pan, Y. Association study between gene polymorphisms in PPAR signaling pathway and porcine meat quality traits. Mamm. Genome 2013, 24, 322–331. [Google Scholar] [CrossRef]
- Kersten, S.; Desvergne, B.; Wahli, W. Roles of PPARS in health and disease. Nature 2000, 405, 421–424. [Google Scholar] [CrossRef]
- Michalik, L.; Desvergne, B.; Wahli, W. Peroxisome-proliferator-activated receptors and cancers: complex stories. Nat. Rev. Cancer 2004, 4, 61–70. [Google Scholar] [CrossRef]
- Rosen, E.D.; Sarraf, P.; Troy, A.E.; Bradwin, G.; Moore, K.; Milstone, D.S.; Spiegelman, B.M.; Mortensen, R.M. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol. Cell 1999, 4, 611–617. [Google Scholar] [CrossRef]
- Xue, G.; Hemmings, B.A. PKB/Akt-Dependent Regulation of Cell Motility. JNCI J. Natl. Cancer Inst. 2013, 105, 393–404. [Google Scholar] [CrossRef]
- Xia, X.; Serrero, G. Inhibition of adipose differentiation by phosphatidylinositol 3-kinase inhibitors. J. Cell. Physiol. 1999, 178, 9–16. [Google Scholar] [CrossRef]
- Chen, G.; Goeddel, D.V. TNF-R1 signaling: A beautiful pathway. Science 2002, 296, 1634–1635. [Google Scholar] [CrossRef]
- Duncan, R.E.; Ahmadian, M.; Jaworski, K.; Sarkadi-Nagy, E.; Sul, H.S. Regulation of Lipolysis in Adipocytes. Annu. Rev. Nutr. 2007, 27, 79–101. [Google Scholar] [CrossRef]
- Ross, S.E. Inhibition of Adipogenesis by Wnt Signaling. Science (80-. ). 2000, 289, 950–953. [Google Scholar] [CrossRef]
- Bennett, C.N.; Ross, S.E.; Longo, K.A.; Bajnok, L.; Hemati, N.; Johnson, K.W.; Harrison, S.D.; MacDougald, O.A. Regulation of Wnt Signaling during Adipogenesis. J. Biol. Chem. 2002, 277, 30998–31004. [Google Scholar] [CrossRef]
- Zhou, Y.; Song, T.; Peng, J.; Zhou, Z.; Wei, H.; Zhou, R.; Jiang, S.; Peng, J. SIRT1 suppresses adipogenesis by activating Wnt/β;-catenin signaling in vivo and in vitro. Oncotarget 2016, 7, 47. [Google Scholar] [CrossRef]
- Tian, L.; Wen, A.; Dong, S.; Yan, P. Molecular Characterization of Microtubule Affinity-Regulating Kinase4 from Sus scrofa and Promotion of Lipogenesis in Primary Porcine Placental Trophoblasts. Int. J. Mol. Sci. 2019, 20, 1206. [Google Scholar] [CrossRef]
- Fried, S.K.; Russell, C.D.; Grauso, N.L.; Brolin, R.E. Lipoprotein lipase regulation by insulin and glucocorticoid in subcutaneous and omental adipose tissues of obese women and men. J. Clin. Invest. 1993, 92, 2191–2198. [Google Scholar] [CrossRef]
- Goldberg, I.J. Lipoprotein lipase and lipolysis: central roles in lipoprotein metabolism and atherogenesis. J. Lipid Res. 1996, 37, 693–707. [Google Scholar]
- Merkel, M.; Eckel, R.H.; Goldberg, I.J. Lipoprotein lipase: genetics, lipid uptake, and regulation. J. Lipid Res. 2002, 43, 1997–2006. [Google Scholar] [CrossRef]
- Dahlhoff, M.; Camera, E.; Picardo, M.; Zouboulis, C.C.; Chan, L.; Chang, B.H.-J.; Schneider, M.R. PLIN2, the major perilipin regulated during sebocyte differentiation, controls sebaceous lipid accumulation in vitro and sebaceous gland size in vivo. Biochim. Biophys. Acta Gen. Subj. 2013, 1830, 4642–4649. [Google Scholar] [CrossRef]
- Xing, K.; Zhu, F.; Zhai, L.; Liu, H.; Wang, Z.; Hou, Z.; Wang, C. The liver transcriptome of two full-sibling Songliao black pigs with extreme differences in backfat thickness. J. Anim. Sci. Biotechnol. 2014, 5, 32. [Google Scholar] [CrossRef]
- Gandolfi, G.; Mazzoni, M.; Zambonelli, P.; Lalatta-Costerbosa, G.; Tronca, A.; Russo, V.; Davoli, R. Perilipin 1 and perilipin 2 protein localization and gene expression study in skeletal muscles of European cross-breed pigs with different intramuscular fat contents. Meat Sci. 2011, 88, 631–637. [Google Scholar] [CrossRef]
- Davoli, R.; Gandolfi, G.; Braglia, S.; Comella, M.; Zambonelli, P.; Buttazzoni, L.; Russo, V. New SNP of the porcine perilipin 2 (PLIN2) gene, association with carcass traits and expression analysis in skeletal muscle. Mol. Biol. Rep. 2011, 38, 1575–1583. [Google Scholar] [CrossRef]
- Zhang, Y.-F.; Yuan, Z.-Q.; Song, D.-G.; Zhou, X.-H.; Wang, Y.-Z. Effects of cannabinoid receptor 1 (brain) on lipid accumulation by transcriptional control of CPT1A and CPT1B. Anim. Genet. 2014, 45, 38–47. [Google Scholar] [CrossRef]
- Schoors, S.; Bruning, U.; Missiaen, R.; Queiroz, K.C.S.; Borgers, G.; Elia, I.; Zecchin, A.; Cantelmo, A.R.; Christen, S.; Goveia, J.; et al. Fatty acid carbon is essential for dNTP synthesis in endothelial cells. Nature 2015, 520, 192–197. [Google Scholar] [CrossRef]
- Kastner, R.; Dussurget, O.; Archambaud, C.; Kernbauer, E.; Soulat, D.; Cossart, P.; Decker, T. LipA, a Tyrosine and Lipid Phosphatase Involved in the Virulence of Listeria monocytogenes. Infect. Immun. 2011, 79, 2489–2498. [Google Scholar] [CrossRef]
- Reiner, Ž.; Guardamagna, O.; Nair, D.; Soran, H.; Hovingh, K.; Bertolini, S.; Jones, S.; Ćorić, M.; Calandra, S.; Hamilton, J.; et al. Lysosomal acid lipase deficiency--an under-recognized cause of dyslipidaemia and liver dysfunction. Atherosclerosis 2014, 235, 21–30. [Google Scholar] [CrossRef]
- Valayannopoulos, V.; Mengel, E.; Brassier, A.; Grabowski, G. Lysosomal acid lipase deficiency: Expanding differential diagnosis. Mol. Genet. Metab. 2017, 120, 62–66. [Google Scholar] [CrossRef]
- Wang, Q.Q.; Yang, B.; Xia, L.L.; Sun, X.X.; Geng, T.Y.; Gong, D.Q. Cloning of Goose Acetyl-Coenzyme A Acyltransferase 2 and Its Expression Pattern in the Development of Fatty Liver. Chinese J. Anim. Vet. Sci. 2016. [Google Scholar]
- Sodhi, S.S.; Ghosh, M.; Song, K.D.; Sharma, N.; Kim, J.H.; Kim, N.E.; Lee, S.J.; Kang, C.W.; Oh, S.J.; Jeong, D.K. An approach to identify SNPs in the gene encoding acetyl-CoA acetyltransferase-2 (ACAT-2) and their proposed role in metabolic processes in pig. PLoS ONE 2014, 9, e102432. [Google Scholar] [CrossRef]
- Miltiadou, D.; Hager-Theodorides, A.C.; Symeou, S.; Constantinou, C.; Psifidi, A.; Banos, G.; Tzamaloukas, O. Variants in the 3’ untranslated region of the ovine acetyl -coenzyme A acyltransferase 2 gene are associated with dairy traits and exhibit differential allelic expression. J. Dairy Sci. 2017, 100, 6285–6297. [Google Scholar] [CrossRef]
- Wang, C.; Niu, Y.; Chi, D.; Zeng, Y.; Liu, H.; Dai, Y.; Li, J. Influence of Delipation on the Energy Metabolism in Pig Parthenogenetically Activated Embryos. Reprod. Domest. Anim. 2015, 50, 826–833. [Google Scholar] [CrossRef]
- Lichtenstein, L.; Berbeée, J.F.P.; van Dijk, S.J.; van Dijk, K.W.; Bensadoun, A.; Kema, I.P.; Voshol, P.J.; Muüller, M.; Rensen, P.C.N.; Kersten, S. Angptl4 Upregulates Cholesterol Synthesis in Liver via Inhibition of LPL- and HL-Dependent Hepatic Cholesterol Uptake. Arterioscler. Thromb. Vasc. Biol. 2007, 27, 2420–2427. [Google Scholar] [CrossRef]
- Köster, A.; Chao, Y.B.; Mosior, M.; Ford, A.; Gonzalez-DeWhitt, P.A.; Hale, J.E.; Li, D.; Qiu, Y.; Fraser, C.C.; Yang, D.D.; et al. Transgenic Angiopoietin-Like (Angptl)4 Overexpression and Targeted Disruption of Angptl4 and Angptl3: Regulation of Triglyceride Metabolism. Endocrinology 2005, 146, 4943–4950. [Google Scholar] [CrossRef]
- Koliwad, S.K.; Kuo, T.; Shipp, L.E.; Gray, N.E.; Backhed, F.; So, A.Y.-L.; Farese, R.V.; Wang, J.-C. Angiopoietin-like 4 (ANGPTL4, Fasting-induced Adipose Factor) Is a Direct Glucocorticoid Receptor Target and Participates in Glucocorticoid-regulated Triglyceride Metabolism. J. Biol. Chem. 2009, 284, 25593–25601. [Google Scholar] [CrossRef]
- Mikhak, B.; Weinsheimer, S.; Pawlikowska, L.; Poon, A.; Kwok, P.-Y.; Lawton, M.T.; Chen, Y.; Zaroff, J.G.; Sidney, S.; McCulloch, C.E.; et al. Angiopoietin-Like 4 (ANGPTL4) Gene Polymorphisms and Risk of Brain Arteriovenous Malformations. Cerebrovasc. Dis. 2011, 31, 338–345. [Google Scholar] [CrossRef]
- McQueen, A.E.; Kanamaluru, D.; Yan, K.; Gray, N.E.; Wu, L.; Li, M.-L.; Chang, A.; Hasan, A.; Stifler, D.; Koliwad, S.K.; et al. The C-terminal fibrinogen-like domain of angiopoietin-like 4 stimulates adipose tissue lipolysis and promotes energy expenditure. J. Biol. Chem. 2017, 292, 16122–16134. [Google Scholar] [CrossRef]
- Yang, L.Y.; Yu, C.G.; Wang, X.H.; Yuan, S.S.; Zhang, L.J.; Lang, J.N.; Zhao, D.; Feng, Y.M. Angiopoietin-Like Protein 4 Is a High-Density Lipoprotein (HDL) Component for HDL Metabolism and Function in Nondiabetic Participants and Type-2 Diabetic Patients. J. Am. Heart Assoc. 2017. [Google Scholar] [CrossRef]
- Ren, Z.Q.; Wu, W.J.; Liu, W.H.; Zheng, R.; Li, J.L.; Zuo, B.; Xu, D.Q.; Li, F.E.; Lei, M.G.; Ni, D.B.; et al. Differential expression and effect of the porcine ANGPTL4 gene on intramuscular fat. Genet. Mol. Res. 2014, 13, 2949–2958. [Google Scholar] [CrossRef]
- Deng, C.; Tang, S.; Huang, X.; Gao, J.; Tian, J.; Zhou, X.; Xie, Y.; Liao, M.; Mo, Z.; Wang, Q. Identification of three novel loci of ALDH2 Gene for Serum Folate levels in a Male Chinese Population by Genome-Wide Association Study. Gene 2018, 674, 121–126. [Google Scholar] [CrossRef]
- Xu, L.; Zhao, G.; Wang, J.; Shen, C.; Li, X.; Lu, F.; Jiang, H.; Liu, G.; Hu, K.; Tang, Y. Impact of Genetic Variation in Aldehyde Dehydrogenase 2 and Alcohol Consumption on Coronary Artery Lesions in Chinese Patients with Stable Coronary Artery Disease. Int. Heart J. 2018, 59, 4. [Google Scholar] [CrossRef]
- Yu, Y.-H.; Liao, P.-R.; Guo, C.-J.; Chen, C.-H.; Mochly-Rosen, D.; Chuang, L.-M. PKC-ALDH2 Pathway Plays a Novel Role in Adipocyte Differentiation. PLoS ONE 2016, 11, e0161993. [Google Scholar] [CrossRef]
- Shu, G.; Zhu, X.T.; Wang, X.Q.; Song, Y.Z.; Bin, Y.F.; Zhang, Y.L.; Gao, P.; Jiang, Q.Y. Identification and gene expression of porcine fatty acid transport protein 1 isoforms. J. Anim. Physiol. Anim. Nutr. 2010, 93, 439–446. [Google Scholar] [CrossRef]
- Martin, G.; Nemoto, M.; Gelman, L.; Geffroy, S.; Najib, J.; Fruchart, J.C.; Roevens, P.; de Martinville, B.; Deeb, S.; Auwerx, J. The human fatty acid transport protein-1 (SLC27A1; FATP-1) cDNA and gene: organization, chromosomal localization, and expression. Genomics 2000, 66, 296–304. [Google Scholar] [CrossRef]
- Auinger, A.; Helwig, U.; Pfeuffer, M.; Rubin, D.; Luedde, M.; Rausche, T.; Eddine El Mokhtari, N.; Fölsch, U.R.; Schreiber, S.; Frey, N.; et al. A variant in the heart-specific fatty acid transport protein 6 is associated with lower fasting and postprandial TAG, blood pressure and left ventricular hypertrophy. Br. J. Nutr. 2012, 107, 1422–1428. [Google Scholar] [CrossRef]
- Gimeno, R.E.; Ortegon, A.M.; Patel, S.; Punreddy, S.; Ge, P.; Sun, Y.; Lodish, H.F.; Stahl, A. Characterization of a heart-specific fatty acid transport protein. J. Biol. Chem. 2003, 278, 16039–16044. [Google Scholar]
- Watkins, P.A.; Pevsner, J.; Steinberg, S.J. Human very long-chain acyl-CoA synthetase and two human homologs: Initial characterization and relationship to fatty acid transport protein. Prostaglandins Leukot. Essent. Fat. Acids 1999, 60, 323–328. [Google Scholar] [CrossRef]
- Nafikov, R.A.; Schoonmaker, J.P.; Korn, K.T.; Noack, K.; Garrick, D.J.; Koehler, K.J.; Minickbormann, J.; Reecy, J.M.; Spurlock, D.E.; Beitz, D.C. Association of polymorphisms in solute carrier family 27, isoform A6 ( SLC27A6 ) and fatty acid-binding protein-3 and fatty acid-binding protein-4 ( FABP3 and FABP4 ) with fatty acid composition of bovine milk. J. Dairy Sci. 2013, 96, 6007–6021. [Google Scholar] [CrossRef]
- Ramsay, T.G.; White, M.E.; Wolverton, C.K. Insulin-like growth factor 1 induction of differentiation of porcine preadipocytes. J. Anim. Sci. 1989, 67, 2452–2459. [Google Scholar] [CrossRef]
- Harasim, E.; Kalinowska, A.; Chabowski, A.; Stepek, T. The role of fatty-acid transport proteins (FAT/CD36, FABPpm, FATP) in lipid metabolism in skeletal muscles. Postep. Hig. Med. Dosw. 2008, 62, 433–441. [Google Scholar]
- Unlu, A.H.; Tajeri, S.; Bilgic, H.B.; Eren, H.; Karagenc, T.; Langsley, G. The secreted Theileria annulata Ta9 protein contributes to activation of the AP-1 transcription factor. PLoS ONE 2018, 13, e0196875. [Google Scholar] [CrossRef]
- Zhou, C.; Martinez, E.; Di Marcantonio, D.; Solanki-Patel, N.; Aghayev, T.; Peri, S.; Ferraro, F.; Skorski, T.; Scholl, C.; Fröhling, S.; et al. JUN is a key transcriptional regulator of the unfolded protein response in acute myeloid leukemia. Leukemia 2017, 31, 1196–1205. [Google Scholar]
- Pisanu, M.E.; Noto, A.; De Vitis, C.; Morrone, S.; Scognamiglio, G.; Botti, G.; Venuta, F.; Diso, D.; Jakopin, Z.; Padula, F.; et al. Blockade of Stearoyl-CoA-desaturase 1 activity reverts resistance to cisplatin in lung cancer stem cells. Cancer Lett. 2017, 406, 93–104. [Google Scholar] [CrossRef]
- Noto, A.; De Vitis, C.; Pisanu, M.E.; Roscilli, G.; Ricci, G.; Catizone, A.; Sorrentino, G.; Chianese, G.; Taglialatela-Scafati, O.; Trisciuoglio, D.; et al. Stearoyl-CoA-desaturase 1 regulates lung cancer stemness via stabilization and nuclear localization of YAP/TAZ. Oncogene 2017, 36, 4573–4584. [Google Scholar] [CrossRef]
- Loebhennard, C.; Mcintyre, J.O. (R)-3-hydroxybutyrate dehydrogenase: selective phosphatidylcholine binding by the C-terminal domain. Biochemistry 2000, 39, 11928–11938. [Google Scholar] [CrossRef]
- Chelius, D.; Loeb-Hennard, C.; Fleischer, S.; Mcintyre, J.O.; Marks, A.R.; De, S.; Hahn, S.; Jehl, M.M.; Moeller, J.; Philipp, R. Phosphatidylcholine activation of human heart (R)-3-Hydroxybutyrate dehydrogenase mutants lacking active center sulfhydryls: Site-directed mutagenesis of a new recombinant fusion protein. Biochemistry 2000, 39, 9687–9697. [Google Scholar] [CrossRef]
- Kubo, F. Wnt2b inhibits differentiation of retinal progenitor cells in the absence of Notch activity by downregulating the expression of proneural genes. Development 2005, 132, 2759–2770. [Google Scholar] [CrossRef]
- MacDonald, B.T.; Tamai, K.; He, X. Wnt/β-Catenin Signaling: Components, Mechanisms, and Diseases. Dev. Cell 2009, 17, 9–26. [Google Scholar] [CrossRef]


| Sample | Before Filtering | After Filtering | Unique Mapped Reads | Multiple Mapped Reads | Mapping Ratio | ||
|---|---|---|---|---|---|---|---|
| Clean Reads Number | Clean Q30 Bases (%) | HQ Clean Reads Number | Clean Q30 Bases (%) | ||||
| A0d | 36,419,702 | 33,808,409 (92.83%) | 35,253,720 | 33,364,121 (94.64%) | 32,064,753 (90.95%) | 274,708 (0.78%) | 91.73% |
| B0d | 31,200,494 | 29,172,462 (93.50%) | 30,099,248 | 28,606,325 (95.04%) | 27,329,518 (90.80%) | 174,318 (0.58%) | 91.38% |
| C0d | 26,486,876 | 24,717,553 (93.32%) | 25,638,168 | 24,333,185 (94.91%) | 23,365,096 (91.13%) | 190,142 (0.74%) | 91.88% |
| A2d | 28,087,146 | 26,264,290 (93.51%) | 27,127,016 | 25,784,229 (95.05%) | 24,631,732 (90.80%) | 214,138 (0.79%) | 91.59% |
| B2d | 27,412,180 | 25,644,094 (93.55%) | 26,554,122 | 25,234,382 (95.03%) | 24,196,924 (91.12%) | 151,568 (0.57%) | 91.69% |
| C2d | 26,603,022 | 24,828,600 (93.33%) | 25,547,810 | 24,262,755 (94.97%) | 23,048,014 (90.22%) | 146,522 (0.57%) | 90.79% |
| A4d | 30,269,944 | 28,196,453 (93.15%) | 29,243,938 | 27,734,951 (94.84%) | 26,631,402 (91.07%) | 215,232 (0.74%) | 91.80% |
| B4d | 28,011,286 | 26,212,961 (93.58%) | 27,141,124 | 25,808,495 (95.09%) | 24,797,742 (91.37%) | 167,664 (0.62%) | 91.98% |
| C4d | 33,020,078 | 30,497,344 (92.36%) | 30,913,550 | 29,185,483 (94.41%) | 46,107,395 (90.56%) | 416,494 (0.82%) | 91.38% |
| A8d | 29,540,224 | 27,590,569 (93.40%) | 28,581,192 | 27,146,416 (94.98%) | 25,946,229 (90.78%) | 196,022 (0.69%) | 91.47% |
| B8d | 38,772,832 | 36,198,316 (93.36%) | 37,484,166 | 35,598,712 (94.97%) | 34,076,825 (90.91%) | 274,396 (0.73%) | 91.64% |
| C8d | 26,907,478 | 25,118,131 (93.35%) | 26,004,832 | 24,696,789 (94.97%) | 23,582,654 (90.69%) | 206,442 (0.79%) | 91.48% |
| Gene ID | Gene Symbol | 2d vs. 0d | 4d vs. 0d | 8d vs. 0d | |||
|---|---|---|---|---|---|---|---|
| Log2(FC) | FDR | Log2(FC) | FDR | Log2(FC) | FDR | ||
| ENSSSCG00000002777 | HSD11B2 | 3.036220 | 2.88 × 10−3 | 3.036220 | 2.88 × 10−3 | 3.292283 | 1.19 × 10−7 |
| ENSSSCG00000003894 | CYP4B1 | 2.178599 | 5.92 × 10−6 | 2.178599 | 5.92 × 10−6 | 2.290617 | 4.15 × 10−15 |
| ENSSSCG00000006474 | NES | −2.321415 | 9.39 × 10−9 | −2.321415 | 9.39 × 10−9 | −1.585476 | 4.76 × 10−4 |
| ENSSSCG00000010450 | LIPA | 1.014733 | 3.90 × 10−3 | 1.014733 | 3.90 × 10−3 | 1.052272 | 1.48 × 10−5 |
| ENSSSCG00000013599 | ANGPTL4 | 1.517481 | 2.04 × 10−3 | 1.517481 | 2.04 × 10−3 | 2.361101 | 3.22 × 10−12 |
| ENSSSCG00000015396 | SEMA3D | −1.735399 | 8.38 × 10−4 | −1.735399 | 8.38 × 10−4 | −1.620849 | 4.72 × 10−3 |
| ENSSSCG00000016634 | CAV1 | −1.864162 | 5.15 × 10−8 | −1.864162 | 5.15 × 10−8 | −1.566771 | 9.61 × 10−5 |
| ENSSSCG00000021646 | KLF9 | 1.326810 | 2.95 × 10−5 | 1.326810 | 2.95 × 10−5 | 1.523982 | 1.62 × 10−8 |
| ENSSSCG00000023760 | CLEC14A | −1.274697 | 9.98 × 10−3 | −1.274697 | 9.98 × 10−3 | −1.801044 | 7.92 × 10−8 |
| ENSSSCG00000027550 | PLCD1 | 1.203787 | 6.90 × 10−4 | 1.203787 | 6.90 × 10−4 | 1.260086 | 1.73 × 10−6 |
| ENSSSCG00000033922 | ALKAL1 | −3.057428 | 8.46 × 10−4 | −3.057428 | 8.46 × 10−4 | −3.117238 | 2.35 × 10−4 |
| ENSSSCG00000034943 | GDF6 | −2.768335 | 3.54 × 10−3 | −2.768335 | 3.54 × 10−3 | −2.175725 | 1.52 × 10−2 |
| ENSSSCG00000035863 | PLIN2 | 1.320747 | 3.95 × 10−4 | 1.320747 | 3.95 × 10−4 | 1.577466 | 4.80 × 10−5 |
| ENSSSCG00000038783 | IGFBP3 | −2.894488 | 3.73 ×10−11 | −2.894488 | 3.73 × 10−11 | −2.872955 | 1.14 × 10−21 |
| ENSSSCG00000039244 | PHYH | 1.429808 | 3.98 × 10−3 | 1.429808 | 3.98 × 10−3 | 1.278978 | 1.16 × 10−3 |
| ENSSSCG00000040631 | LPL | 2.462269 | 3.44 × 10−6 | 2.462269 | 3.44 × 10−6 | 2.391050 | 2.09 × 10−7 |
| GO ID | Description | p-Value | Genes | |
|---|---|---|---|---|
| 2d vs. 0d | GO:0030154 | Cell differentiation | 0.046435 | ANGPTL4, CAV1, ALKAL1, GDF6, IGFBP3, LPL |
| GO:0008283 | Cell proliferation | 0.001630 | KLF9, LIPA, NES, CAV1, PLCD1, IGFBP3 | |
| GO:0006629 | Lipid metabolic process | 0.000184 | LIPA, CAV1, PLCD1, PHYH, LPL | |
| 4d vs. 0d | GO:0030154 | Cell differentiation | 0.000001 | ADGRF1, MYOM1, CDH2, CDC20, RORA, IVL, S100A10, COL11A1, LFNG, SFRP2, CPT1A, ANGPTL4, ICAM1, KIF20A, AGR2, RELN, COL3A1, CAV1, ANLN, FST, BATF2, CTSB, SLC2A4, KRT4 CCNB1, MMP8, ZBTB16 SNAI1, ALKAL1, CDKN2B, GDF6, INHBA, COL1A1, PTHLH, RGS2, TRIB1, CHAC1, RND1, IGFBP3, NPNT, SOD2, IL34, LPL |
| GO:0008283 | Cell proliferation | 0.000195 | CDH2, RORA, NES, SFRP2, SPRY1, LIPA, LAMC2, CAV1, KLF9, F3, KRT4, OVOL1, PLCD1, CCNB1, ZBTB16, GPNMB, CDKN2B, INHBA, TRIB1, IGFBP3, HMOX1, SOD2, IL34 | |
| GO:0006629 | Lipid metabolic process | 0.008311 | RORA, ABCA1, GPCPD1, AGT, LIPA, SCD, CPT1A, CAV1, PLCD1, SNAI1, PHYH, LPL | |
| 8d vs. 0d | GO:0030154 | Cell differentiation | 0.000001 | ADGRF1, MYOM1, CDH2, CDC20, RORA, IVL, S100A10, COL11A1LFNG, SFRP2, CPT1A, ANGPTL4, ICAM1, KIF20A, AGR2, RELN, COL3A1, CAV1, ANLN, FST, BATF2, CTSB, SLC2A4, KRT4, CCNB1, MMP8, ZBTB16, SNAI1, ALKAL1, CDKN2B, GDF6, INHBA, COL1A1, PTHLH, RGS2, TRIB1, CHAC1, RND1, IGFBP3, NPNT, SOD2, IL34, LPL |
| GO:0008283 | Cell proliferation | 0.000195 | CDH2, RORA, NES, SFRP2, SPRY1, LIPA, LAMC2, CAV1, KLF9, F3, KRT4, OVOL1, PLCD1, CCNB1, ZBTB16, GPNMB, CDKN2B, INHBA, TRIB1, IGFBP3, HMOX1, SOD2, IL34 | |
| GO:0006629 | Lipid metabolic process | 0.008311 | RORA, ABCA1, GPCPD1, AGT, LIPA, SCD, CPT1A, CAV1, PLCD1, SNAI1, PHYH, LPL |
| Pathway | p-Value | Genes | |
|---|---|---|---|
| 2d vs. 0d | Steroid hormone biosynthesis | 0.00259 | HSD11B2, AKR1C1 |
| 4d vs. 0d | PPAR signaling pathway | 0.00017 | SCD, ANGPTL4, GK |
| Steroid hormone biosynthesis | 0.01279 | HSD11B2, AKR1C1 | |
| Cell cycle | 0.01253 | CDC20, CCNB1, WEE1 | |
| 8d vs. 0d | PPAR signaling pathway | 0.00077 | CD36, ANGPTL4, GK |
| Steroid hormone biosynthesis | 0.02929 | HSD11B2, AKR1C1 | |
| p53 signaling pathway | 0.04143 | CDK1, CCNB1 | |
| PI3K/Akt signaling pathway | 0.03306 | ITGA3, COL6A3, AREG, THBS3, TLR2, HGF, FGF1, ANGPT2, SPP1, MYC | |
| Wnt signaling pathway | 0.03833 | SFRP5, SFRP2, PORCN, MYC, WNT2B | |
| Cellular senescence | 0.02123 | CDK1, CCNB1, CXCL8, CCNB3, MYC | |
| TNF signaling pathway | 0.00215 | CCL2, MAP3K8, CX3CL1, CSF2, FOS |
| GO ID | Description | p-Value | Genes | |
|---|---|---|---|---|
| Profile 9 | GO:0008283 | Cell proliferation | 0.01528 | DPT, ESM1, SERPINF1, AREG, NOV, F2, HGF |
| GO:0030154 | Cell differentiation | 0.03364 | SERPINF1, L1CAM, SEMA3C, NEUROG2, NOV, ANKRD1 ITGA3, FLRT2, SLIT2, F2 | |
| Profile 21 | GO:0006629 | Lipid metabolic process | 0.00096 | PLCD1, PHYH, CPT1A, PLBD2, LPL, EEF1A2, LIPA |
| Profile 22 | GO:0006629 | Lipid metabolic process | 0.00520 | SLC27A1, AGT, ACAA2, CYP1B1, EGR1, PECR |
| GO:0008283 | Cell proliferation | 0.04071 | TRIB1, JUNB, JUN, CYP1B1, TXNRD1, KLF4, ATOH8 | |
| GO:0030154 | Cell differentiation | 0.00321 | ANGPTL4, MYBPH, TRIB1, JUNB, TXNIP, JUN, KLF4, IL18R1, ZNF365, FOS, ATOH8, EGR1, NCKIPSD, ZFP36 | |
| Profile 24 | GO:0030154 | Cell differentiation | 0.00321 | CCL2, RND1, FOXA2, RGS2, PGLYRP1, SOD2 IL34, SLC2A4 |
| Pathway | p-Value | Genes | |
|---|---|---|---|
| Profile 21 | PPAR signaling pathway | 0.00003 | PLIN2, LPL, CPT1A, LIPA |
| Fatty acid degradation | 0.01736 | ADH1C, CPT1A, LIPA | |
| Profile 22 | TNF signaling pathway | 0.00009 | JUN, IL18R1, CX3CL1 JUNB, FOS |
| PPAR signaling pathway | 0.00413 | SLC27A1, ANGPTL4 | |
| Fatty acid degradation | 0.01447 | ALDH2, ACAA2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, X.; Chen, S.; Tan, Z.; Wang, Y.; Zhang, F.; Yang, T.; Liu, Y.; Ao, H.; Xing, K.; Wang, C. Transcriptome Analysis of Landrace Pig Subcutaneous Preadipocytes during Adipogenic Differentiation. Genes 2019, 10, 552. https://doi.org/10.3390/genes10070552
Zhao X, Chen S, Tan Z, Wang Y, Zhang F, Yang T, Liu Y, Ao H, Xing K, Wang C. Transcriptome Analysis of Landrace Pig Subcutaneous Preadipocytes during Adipogenic Differentiation. Genes. 2019; 10(7):552. https://doi.org/10.3390/genes10070552
Chicago/Turabian StyleZhao, Xitong, Shaokang Chen, Zhen Tan, Yuan Wang, Fengxia Zhang, Ting Yang, Yibing Liu, Hong Ao, Kai Xing, and Chuduan Wang. 2019. "Transcriptome Analysis of Landrace Pig Subcutaneous Preadipocytes during Adipogenic Differentiation" Genes 10, no. 7: 552. https://doi.org/10.3390/genes10070552
APA StyleZhao, X., Chen, S., Tan, Z., Wang, Y., Zhang, F., Yang, T., Liu, Y., Ao, H., Xing, K., & Wang, C. (2019). Transcriptome Analysis of Landrace Pig Subcutaneous Preadipocytes during Adipogenic Differentiation. Genes, 10(7), 552. https://doi.org/10.3390/genes10070552




