Seasonal Stability and Dynamics of DNA Methylation in Plants in a Natural Environment
Abstract
1. Introduction
2. Materials and Methods
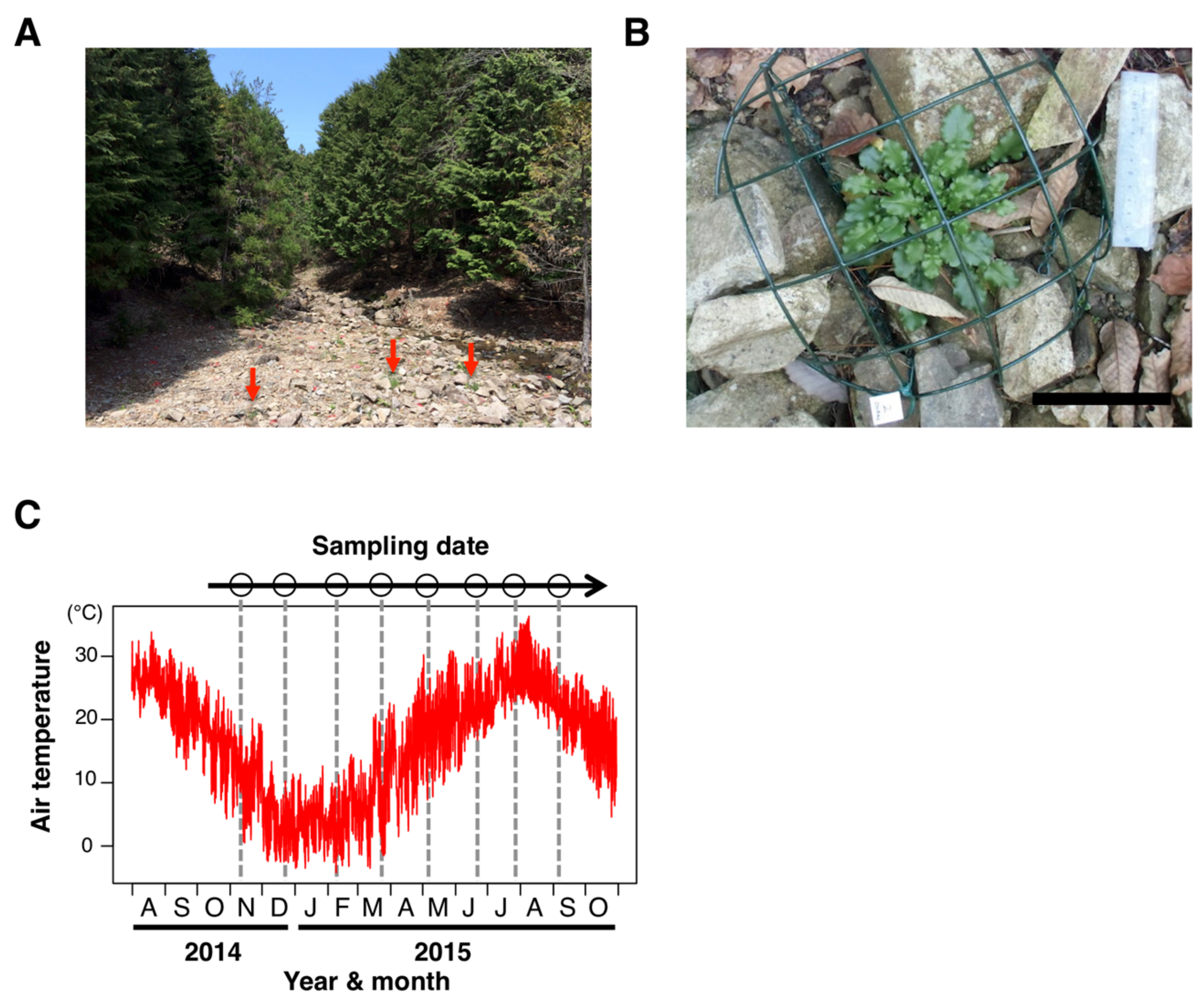
2.1. Plant Materials
2.2. DNA Extraction and WGBS Library Preparation
2.3. Processing of WGBS Data
2.4. Transcriptome Analysis
2.5. Data Availability
3. Results
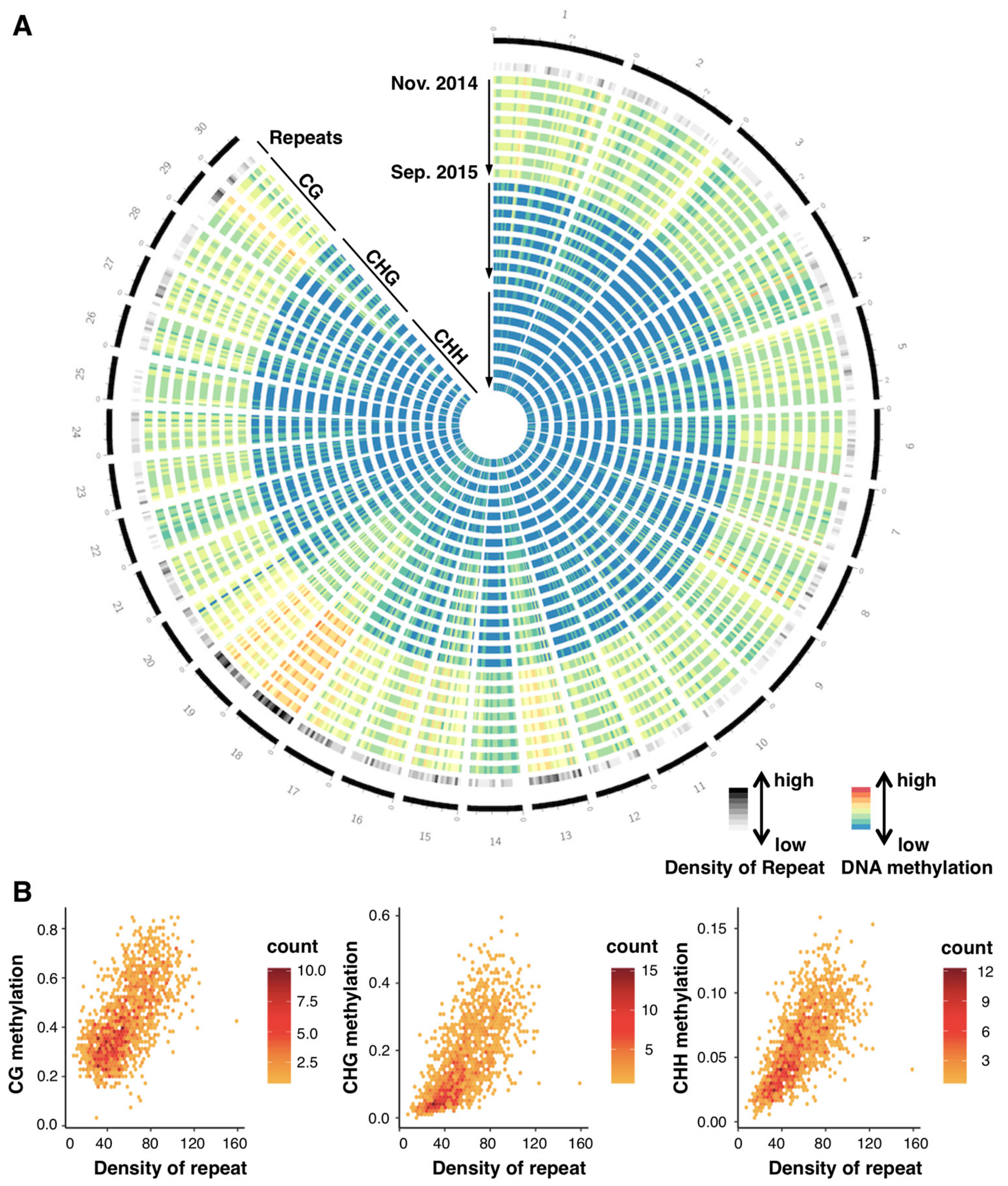
3.1. Seasonal Stability in Large-Scale Distribution of DNA Methylation
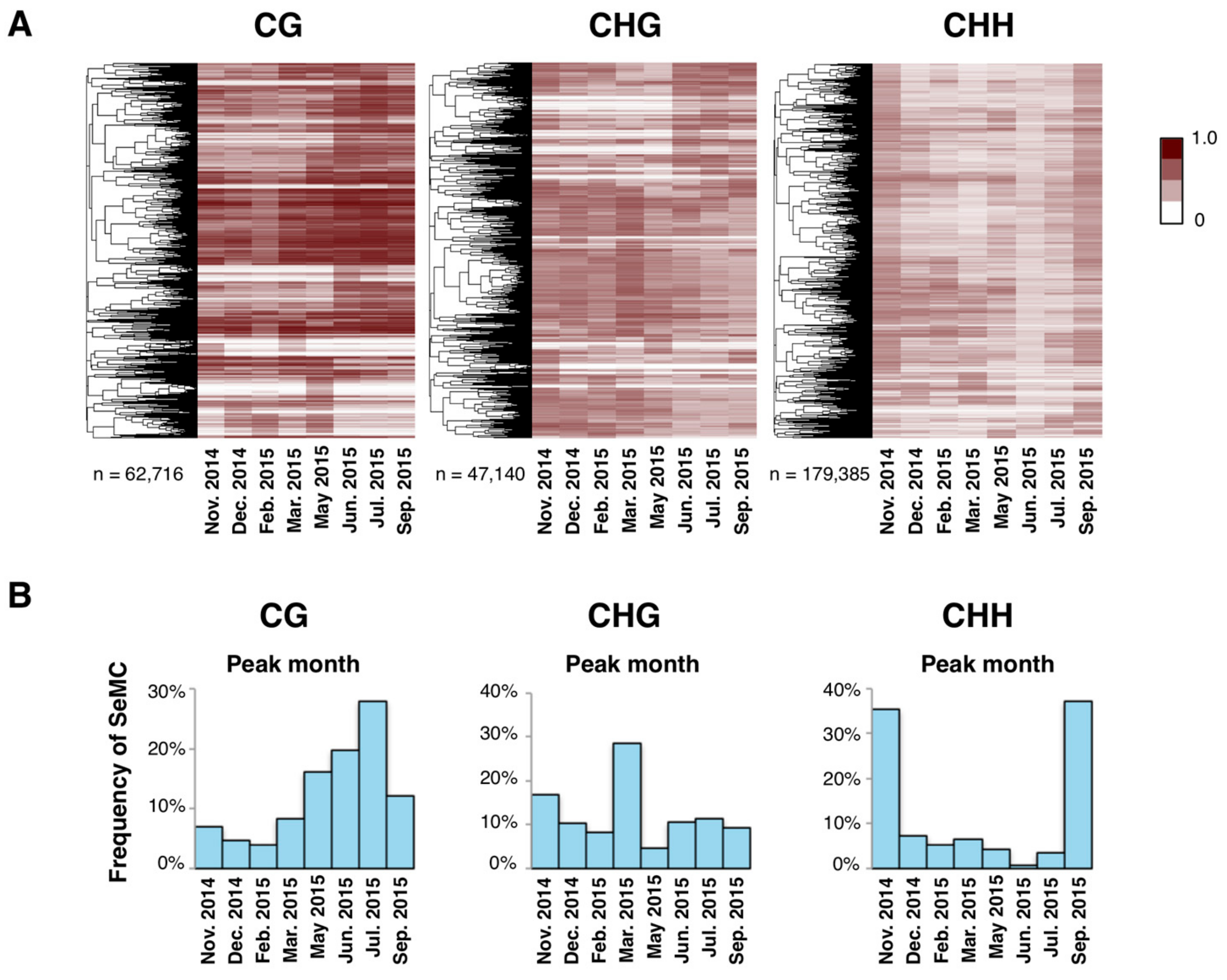
3.2. SeMCs: Seasonal Dynamics of DNA Methylation
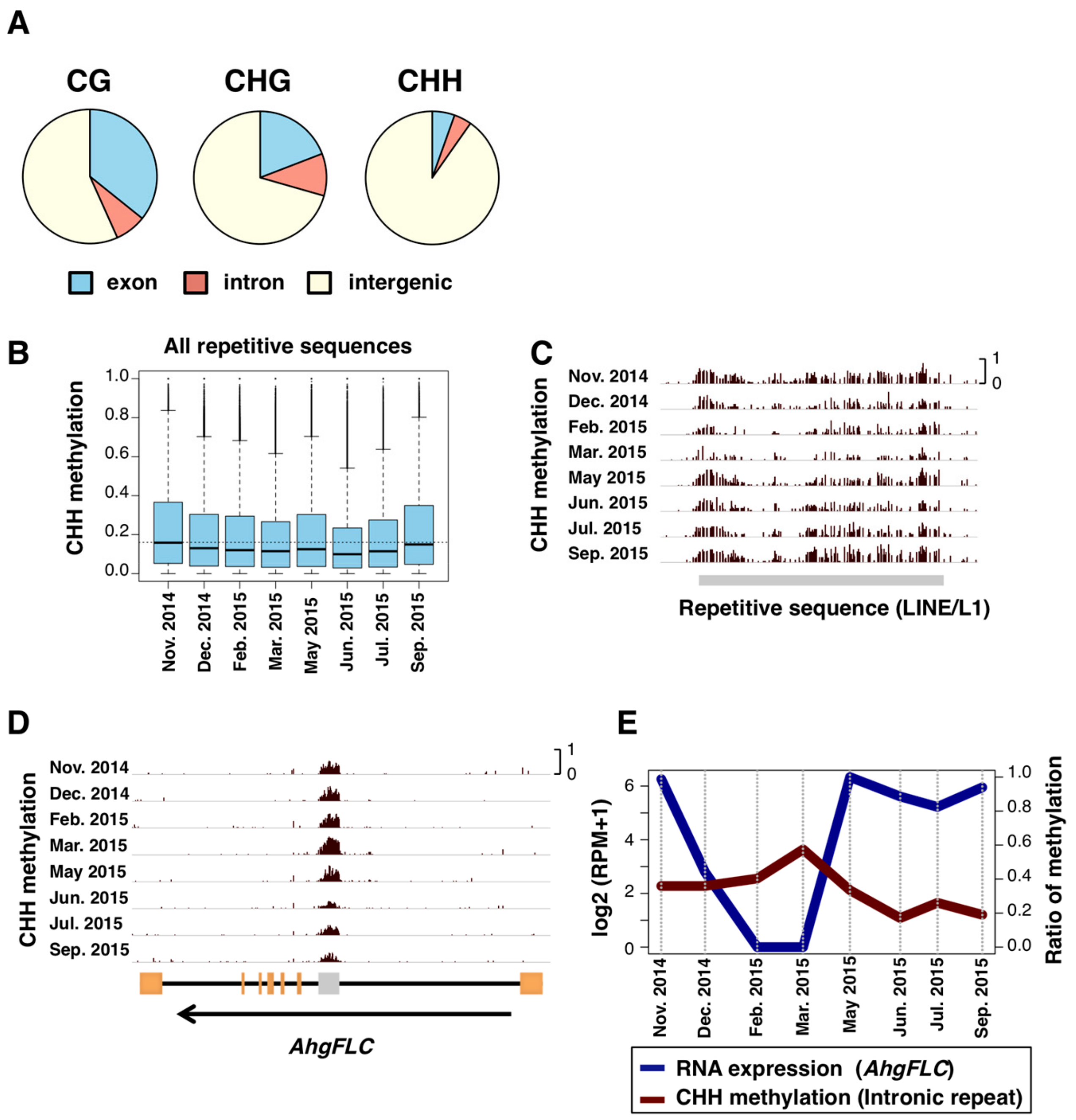
3.3. CHH SeMCs in Repetitive Sequences
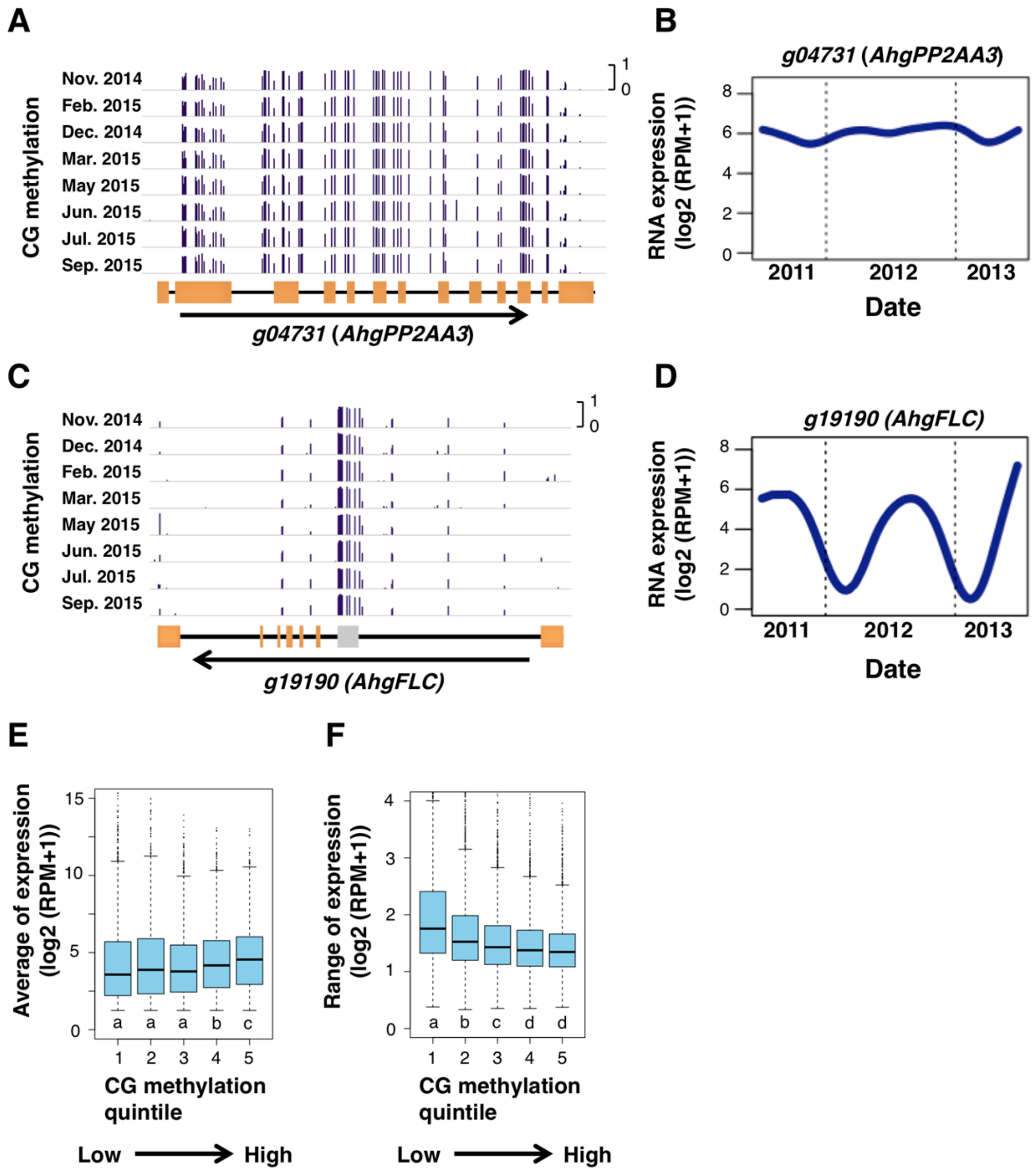
3.4. Gene-Body CG Methylation (gbM) and Constant RNA Expression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Law, J.A.; Jacobsen, S.E. Establishing, Maintaining and Modifying DNA Methylation Patterns in Plants and Animals. Nat. Rev. Genet. 2010, 11, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Cokus, S.J.; Feng, S.; Zhang, X.; Chen, Z.; Merriman, B.; Haudenschild, C.D.; Pradhan, S.; Nelson, S.F.; Pellegrini, M.; Jacobsen, S.E. Shotgun Bisulphite Sequencing of the Arabidopsis Genome Reveals DNA Methylation Patterning. Nature 2008, 452, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Lister, R.; O’Malley, R.C.; Tonti-Filippini, J.; Gregory, B.D.; Berry, C.C.; Millar, A.H.; Ecker, J.R. Highly Integrated Single-Base Resolution Maps of the Epigenome in Arabidopsis. Cell 2008, 133, 523–536. [Google Scholar] [CrossRef] [PubMed]
- Lister, R.; Pelizzola, M.; Dowen, R.H.; Hawkins, R.D.; Hon, G.; Tonti-Filippini, J.; Nery, J.R.; Lee, L.; Ye, Z.; Ngo, Q.-M.; et al. Human DNA Methylomes at Base Resolution Show Widespread Epigenomic Differences. Nature 2009, 462, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Cokus, S.J.; Zhang, X.; Chen, P.-Y.; Bostick, M.; Goll, M.G.; Hetzel, J.; Jain, J.; Strauss, S.H.; Halpern, M.E.; et al. Conservation and Divergence of Methylation Patterning in Plants and Animals. Proc. Natl. Acad. Sci. USA 2010, 107, 8689–8694. [Google Scholar] [CrossRef] [PubMed]
- Zemach, A.; McDaniel, I.E.; Silva, P.; Zilberman, D. Genome-Wide Evolutionary Analysis of Eukaryotic DNA Methylation. Science 2010, 328, 916–919. [Google Scholar] [CrossRef] [PubMed]
- Huff, J.T.; Zilberman, D. Dnmt1-Independent CG Methylation Contributes to Nucleosome Positioning in Diverse Eukaryotes. Cell 2014, 156, 1286–1297. [Google Scholar] [CrossRef]
- Niederhuth, C.E.; Bewick, A.J.; Ji, L.; Alabady, M.S.; Kim, K.D.; Li, Q.; Rohr, N.A.; Rambani, A.; Burke, J.M.; Udall, J.A.; et al. Widespread Natural Variation of DNA Methylation within Angiosperms. Genome Biol. 2016, 17. [Google Scholar] [CrossRef]
- Cubas, P.; Vincent, C.; Coen, E. An Epigenetic Mutation Responsible for Natural Variation in Floral Symmetry. Nature 1999, 401, 157–161. [Google Scholar] [CrossRef]
- Manning, K.; Tör, M.; Poole, M.; Hong, Y.; Thompson, A.J.; King, G.J.; Giovannoni, J.J.; Seymour, G.B. A Naturally Occurring Epigenetic Mutation in a Gene Encoding an SBP-Box Transcription Factor Inhibits Tomato Fruit Ripening. Nat. Genet. 2006, 38, 948–952. [Google Scholar] [CrossRef]
- Ong-Abdullah, M.; Ordway, J.M.; Jiang, N.; Ooi, S.-E.; Kok, S.-Y.; Sarpan, N.; Azimi, N.; Hashim, A.T.; Ishak, Z.; Rosli, S.K.; et al. Loss of Karma Transposon Methylation Underlies the Mantled Somaclonal Variant of Oil Palm. Nature 2015, 525, 533–537. [Google Scholar] [CrossRef] [PubMed]
- Dowen, R.H.; Pelizzola, M.; Schmitz, R.J.; Lister, R.; Dowen, J.M.; Nery, J.R.; Dixon, J.E.; Ecker, J.R. Widespread Dynamic DNA Methylation in Response to Biotic Stress. Proc. Natl. Acad. Sci. USA 2012, 109, E2183–E2191. [Google Scholar] [CrossRef] [PubMed]
- Dubin, M.J.; Zhang, P.; Meng, D.; Remigereau, M.-S.; Osborne, E.J.; Paolo Casale, F.; Drewe, P.; Kahles, A.; Jean, G.; Vilhjálmsson, B.; et al. DNA Methylation in Arabidopsis Has a Genetic Basis and Shows Evidence of Local Adaptation. eLife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Secco, D.; Wang, C.; Shou, H.; Schultz, M.D.; Chiarenza, S.; Nussaume, L.; Ecker, J.R.; Whelan, J.; Lister, R. Stress Induced Gene Expression Drives Transient DNA Methylation Changes at Adjacent Repetitive Elements. eLife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Becker, C.; Hagmann, J.; Müller, J.; Koenig, D.; Stegle, O.; Borgwardt, K.; Weigel, D. Spontaneous Epigenetic Variation in the Arabidopsis thaliana Methylome. Nature 2011, 480, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, R.J.; Schultz, M.D.; Lewsey, M.G.; O’Malley, R.C.; Urich, M.A.; Libiger, O.; Schork, N.J.; Ecker, J.R. Transgenerational Epigenetic Instability Is a Source of Novel Methylation Variants. Science 2011, 334, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, K.K.; Kudoh, H.; Kobayashi, M.J. Plant Sexual Reproduction during Climate Change: Gene Function in Natura Studied by Ecological and Evolutionary Systems Biology. Ann. Bot. 2011, 108, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Kudoh, H. Molecular Phenology in Plants: In Natura Systems Biology for the Comprehensive Understanding of Seasonal Responses under Natural Environments. New Phytol. 2015, 210, 399–412. [Google Scholar] [CrossRef]
- Kudoh, H.; Honjo, M.N.; Nishio, H.; Sugisaka, J. The Long-Term “In Natura” Study Sites of Arabidopsis halleri for Plant Transcription and Epigenetic Modification Analyses in Natural Environments. In Methods in Molecular Biology; Springer: New York, NY, USA, 2018; pp. 41–57. [Google Scholar]
- Aikawa, S.; Kobayashi, M.J.; Satake, A.; Shimizu, K.K.; Kudoh, H. Robust Control of the Seasonal Expression of the Arabidopsis FLC Gene in a Fluctuating Environment. Proc. Natl. Acad. Sci. USA 2010, 107, 11632–11637. [Google Scholar] [CrossRef]
- Nagano, A.J.; Kawagoe, T.; Sugisaka, J.; Honjo, M.N.; Iwayama, K.; Kudoh, H. Annual Transcriptome Dynamics in Natural Environments Reveals Plant Seasonal Adaptation. Nat. Plants 2019, 5, 74–83. [Google Scholar] [CrossRef]
- Stroud, H.; Do, T.; Du, J.; Zhong, X.; Feng, S.; Johnson, L.; Patel, D.J.; Jacobsen, S.E. Non-CG Methylation Patterns Shape the Epigenetic Landscape in Arabidopsis. Nat. Struct. Mol. Biol. 2013, 21, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Kawashima, T.; Berger, F. Epigenetic Reprogramming in Plant Sexual Reproduction. Nat. Rev. Genet. 2014, 15, 613–624. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Ecker, J.R. Non-CG Methylation in the Human Genome. Annu. Rev. Genom. Hum. Genet. 2015, 16, 55–77. [Google Scholar] [CrossRef] [PubMed]
- Saghai-Maroof, M.A.; Soliman, K.M.; Jorgensen, R.A.; Allard, R.W. Ribosomal DNA Spacer-Length Polymorphisms in Barley: Mendelian Inheritance, Chromosomal Location, and Population Dynamics. Proc. Natl. Acad. Sci. USA 1984, 81, 8014–8018. [Google Scholar] [CrossRef]
- Fu, Y.; Kawabe, A.; Etcheverry, M.; Ito, T.; Toyoda, A.; Fujiyama, A.; Colot, V.; Tarutani, Y.; Kakutani, T. Mobilization of a Plant Transposon by Expression of the Transposon-Encoded Anti-Silencing Factor. Embo J. 2013, 32, 2407–2417. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Briskine, R.V.; Paape, T.; Shimizu-Inatsugi, R.; Nishiyama, T.; Akama, S.; Sese, J.; Shimizu, K.K. Genome Assembly and Annotation of Arabidopsis halleri, a Model for Heavy Metal Hyperaccumulation and Evolutionary Ecology. Mol. Ecol. Resour. 2016, 17, 1025–1036. [Google Scholar] [CrossRef]
- Krueger, F.; Andrews, S.R. Bismark: A Flexible Aligner and Methylation Caller for Bisulfite-Seq Applications. Bioinformatics 2011, 27, 1571–1572. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Smit, A.F.A.; Hubley, R. RepeatModeler Open-1.0. 2008–2015. Available online: http://www.repeatmasker.org (accessed on 12 September 2016).
- Storey, J.D. A Direct Approach to False Discovery Rates. J. R. Stat. Soc. Ser. B Stat. Methodol. 2002, 64, 479–498. [Google Scholar] [CrossRef]
- De Hoon, M.J.L.; Imoto, S.; Nolan, J.; Miyano, S. Open Source Clustering Software. Bioinformatics 2004, 20, 1453–1454. [Google Scholar] [CrossRef] [PubMed]
- Saldanha, A.J. Java Treeview—Extensible Visualization of Microarray Data. Bioinformatics 2004, 20, 3246–3248. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An Information Aesthetic for Comparative Genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative Genomics Viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [PubMed]
- Barturen, G.; Rueda, A.; Oliver, J.L.; Hackenberg, M. Methylextract. Zenodo 2013. [Google Scholar] [CrossRef]
- Huson, D.H.; Scornavacca, C. Dendroscope 3: An Interactive Tool for Rooted Phylogenetic Trees and Networks. Syst. Biol. 2012, 61, 1061–1067. [Google Scholar] [CrossRef] [PubMed]
- Townsley, B.T.; Covington, M.F.; Ichihashi, Y.; Zumstein, K.; Sinha, N.R. BrAD-Seq: Breath Adapter Directional Sequencing: A Streamlined, Ultra-Simple and Fast Library Preparation Protocol for Strand Specific mRNA Library Construction. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-Optimal Probabilistic RNA-Seq Quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Seymour, D.K.; Koenig, D.; Hagmann, J.; Becker, C.; Weigel, D. Evolution of DNA Methylation Patterns in the Brassicaceae Is Driven by Differences in Genome Organization. PLoS Genet. 2014, 10, e1004785. [Google Scholar] [CrossRef] [PubMed]
- Fedoroff, N.V. Transposable Elements, Epigenetics, and Genome Evolution. Science 2012, 338, 758–767. [Google Scholar] [CrossRef] [PubMed]
- Tenaillon, M.I.; Hollister, J.D.; Gaut, B.S. A Triptych of the Evolution of Plant Transposable Elements. Trends Plant. Sci. 2010, 15, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Czechowski, T. Genome-Wide Identification and Testing of Superior Reference Genes for Transcript Normalization in Arabidopsis. Plant. Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; De Jonge, J.; Forsberg, S.K.G.; Pettersson, M.E.; Sheng, Z.; Hennig, L.; Carlborg, Ö. Natural CMT2 Variation Is Associated With Genome-Wide Methylation Changes and Temperature Seasonality. PLoS Genet. 2014, 10, e1004842. [Google Scholar] [CrossRef] [PubMed]
- Zemach, A.; Kim, M.Y.; Hsieh, P.-H.; Coleman-Derr, D.; Eshed-Williams, L.; Thao, K.; Harmer, S.L.; Zilberman, D. The Arabidopsis Nucleosome Remodeler DDM1 Allows DNA Methyltransferases to Access H1-Containing Heterochromatin. Cell 2013, 153, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Popova, O.V.; Dinh, H.Q.; Aufsatz, W.; Jonak, C. The RdDM Pathway Is Required for Basal Heat Tolerance in Arabidopsis. Mol. Plant. 2013, 6, 396–410. [Google Scholar] [CrossRef] [PubMed]
- Romon, M.; Soustre-Gacougnolle, I.; Schmitt, C.; Perrin, M.; Burdloff, Y.; Chevalier, E.; Mutterer, J.; Himber, C.; Zervudacki, J.; Montavon, T.; et al. RNA Silencing Is Resistant to Low-Temperature in Grapevine. PLoS ONE 2013, 8, e82652. [Google Scholar] [CrossRef] [PubMed]
- Sung, S.; He, Y.; Eshoo, T.W.; Tamada, Y.; Johnson, L.; Nakahigashi, K.; Goto, K.; Jacobsen, S.E.; Amasino, R.M. Epigenetic Maintenance of the Vernalized State in Arabidopsis thaliana Requires LIKE HETEROCHROMATIN PROTEIN 1. Nat. Genet. 2006, 38, 706–710. [Google Scholar] [CrossRef]
- Castaings, L.; Bergonzi, S.; Albani, M.C.; Kemi, U.; Savolainen, O.; Coupland, G. Evolutionary Conservation of Cold-Induced Antisense RNAs of FLOWERING LOCUS C in Arabidopsis thaliana Perennial Relatives. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Gazzani, S.; Gendall, A.R.; Lister, C.; Dean, C. Analysis of the Molecular Basis of Flowering Time Variation in Arabidopsis Accessions. Plant. Physiol. 2003, 132, 1107–1114. [Google Scholar] [CrossRef]
- Michaels, S.D.; He, Y.; Scortecci, K.C.; Amasino, R.M. Attenuation of FLOWERING LOCUS C Activity as a Mechanism for the Evolution of Summer-Annual Flowering Behavior in Arabidopsis. Proc. Natl. Acad. Sci. USA 2003, 100, 10102–10107. [Google Scholar] [CrossRef]
- Quadrana, L.; Bortolini Silveira, A.; Mayhew, G.F.; LeBlanc, C.; Martienssen, R.A.; Jeddeloh, J.A.; Colot, V. The Arabidopsis thaliana Mobilome and Its Impact at the Species Level. eLife 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Irwin, J.; Dean, C. Remembering the Prolonged Cold of Winter. Curr. Biol. 2013, 23, R807–R811. [Google Scholar] [CrossRef] [PubMed]
- Buzas, D.M.; Robertson, M.; Finnegan, E.J.; Helliwell, C.A. Transcription-Dependence of Histone H3 Lysine 27 Trimethylation at the Arabidopsis Polycomb Target Gene FLC. Plant J. 2011, 65, 872–881. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, S.; Miura-Kamio, A.; Nakamura, Y.; Lu, F.; Cui, X.; Cao, X.; Kimura, H.; Saze, H.; Kakutani, T. Autocatalytic Differentiation of Epigenetic Modifications within the Arabidopsis Genome. Embo J. 2010, 29, 3496–3506. [Google Scholar] [CrossRef] [PubMed]
- Zilberman, D. An Evolutionary Case for Functional Gene Body Methylation in Plants and Animals. Genome Biol. 2017, 18. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yazaki, J.; Sundaresan, A.; Cokus, S.; Chan, S.W.-L.; Chen, H.; Henderson, I.R.; Shinn, P.; Pellegrini, M.; Jacobsen, S.E.; et al. Genome-Wide High-Resolution Mapping and Functional Analysis of DNA Methylation in Arabidopsis. Cell 2006, 126, 1189–1201. [Google Scholar] [CrossRef] [PubMed]
- Aceituno, F.F.; Moseyko, N.; Rhee, S.Y.; Gutiérrez, R.A. The Rules of Gene Expression in Plants: Organ Identity and Gene Body Methylation Are Key Factors for Regulation of Gene Expression in Arabidopsis thaliana. BMC Genom. 2008, 9, 438. [Google Scholar] [CrossRef]
- Coleman-Derr, D.; Zilberman, D. Deposition of Histone Variant H2A.Z within Gene Bodies Regulates Responsive Genes. PLoS Genet. 2012, 8, e1002988. [Google Scholar] [CrossRef]
- Richards, E.J. Inherited Epigenetic Variation—Revisiting Soft Inheritance. Nat. Rev. Genet. 2006, 7, 395–401. [Google Scholar] [CrossRef]
- Weigel, D.; Colot, V. Epialleles in Plant Evolution. Genome Biol. 2012, 13, 249. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ito, T.; Nishio, H.; Tarutani, Y.; Emura, N.; Honjo, M.N.; Toyoda, A.; Fujiyama, A.; Kakutani, T.; Kudoh, H. Seasonal Stability and Dynamics of DNA Methylation in Plants in a Natural Environment. Genes 2019, 10, 544. https://doi.org/10.3390/genes10070544
Ito T, Nishio H, Tarutani Y, Emura N, Honjo MN, Toyoda A, Fujiyama A, Kakutani T, Kudoh H. Seasonal Stability and Dynamics of DNA Methylation in Plants in a Natural Environment. Genes. 2019; 10(7):544. https://doi.org/10.3390/genes10070544
Chicago/Turabian StyleIto, Tasuku, Haruki Nishio, Yoshiaki Tarutani, Naoko Emura, Mie N. Honjo, Atsushi Toyoda, Asao Fujiyama, Tetsuji Kakutani, and Hiroshi Kudoh. 2019. "Seasonal Stability and Dynamics of DNA Methylation in Plants in a Natural Environment" Genes 10, no. 7: 544. https://doi.org/10.3390/genes10070544
APA StyleIto, T., Nishio, H., Tarutani, Y., Emura, N., Honjo, M. N., Toyoda, A., Fujiyama, A., Kakutani, T., & Kudoh, H. (2019). Seasonal Stability and Dynamics of DNA Methylation in Plants in a Natural Environment. Genes, 10(7), 544. https://doi.org/10.3390/genes10070544




