Tomato is one of the most cultivated crops around the world [
24] with an annual production of around 164 million tons [
25]. Traditionally, tomato has been a research model for fruit development and since the completion of its genome sequence in 2012 [
26], it offers an excellent system to study gene regulation in relation to plant stress response. Abiotic stresses, like drought, heat, cold, salinity, and biotic stresses like bacteria or fungi infection or herbivores attack dramatically affect the yield and quality of crops. Recently, the important role of sRNAs as a versatile regulation mechanism of the plant response to stress has been evidenced. In this work high-throughput sequencing of sRNAs with SOLiD technology has been used to obtain the miRNA profile of tomato plants undergoing 5 different stress conditions (drought, heat,
P. syringae infection,
B. cinerea infection, and herbivore insect attack with CPB larvae) or chemical treatment with the plant defense inducer Hx. We have detected 104 known miRNAs belonging to 37 families, and 62 novel tomato miRNAs. For the identification of known miRNAs, strict parameters in sRNAbench tool of the sRNAtoolbox were used and no mismatches or gaps allowed. For novel miRNAs predictions, strict default parameters were also used, and only were selected as putative novel miRNAs those having reads for both the 5p-arm and 3p-arm mature miRNAs sequences able to form a duplex, and with a pre-miR sequence mapped in the tomato genome. Read counts differed notably among conserved and novel miRNAs, with the latter displaying lower expression. A few conserved miRNA families such as
miR482,
miR167, and
miR159 were the most abundantly expressed (more than 27,000 RPM), accounting for 36%, 20% and 15% of all the conserved miRNA reads, respectively. When comparing miRNAs expression patterns among stress samples and controls 40 out of the 165 miRNAs identified were found stress-responsive, which might potentially be implicated in the regulation of stress response in tomato. Supporting the reliability of the miRNA detection by deep sequencing, fold change of eleven random selected differential miRNAs was consistent with the analysis by RT-qPCR (91% miRNAs validated). In all stress conditions, most stress-responsive miRNAs were upregulated (55% in the total 41 stress-responsive miRNAs detected). None of these 41 miRNAs was differentially expressed in all stress conditions, whereas
sly-miR167c-3p expression was detected in
B. cinerea and
P. syringae infection,
sly-newmiR26-3p in drought and Hx treatment samples, and
sly-newmiR33-3p,
sly-newmiR6-3p, and
sly-newmiR8-3p both in biotic and abiotic stresses. Therefore, it seems that miRNAs expression during stress is dependent on the specific stress to which the plant is subjected.
4.4. P. syringae Stress-Responsive miRNAs
Arabidopsis plants challenged with
P. syringae showed similar expression profile of most of those highly and moderately conserved miRNAs detected in our study [
36]. Among others,
miR396,
miR390 and
miR166 families were down-regulated, and
miR167,
miR169, and
miR172 families were upregulated, while
miR168 and
miR482 were not detected in
Arabidopsis [
36].
Auxin is considered a pattern-forming phytohormone responsible of plant growth and major developmental processes, many of which are modulated by the auxin response transcription factor (ARF) family [
37] targeted by
miR167 and
miR390. While
ARF6 and
ARF8 are targets of
miR167, expression of
ARF2,
ARF3 and
ARF4 is regulated by
miRNA390 through TAS3-derived ta-siRNAs (trans-acting short-interfering RNAs, a class endogenous secondary siRNAs produced through the action of RNA-dependent-RNA-polymerase-6 upon microRNA-mediated cleavage of non-coding TAS RNAs) in
Arabidopsis [
38,
39,
40].
sly-miR167 not only was found up-regulated after bacterial challenging with
P. syringae in tomato (present work) and
Arabidopsis [
41] but also in response to infection with the fungal pathogen
Fusarium oxysporum in tomato leaves [
42]. In the case of
miR390, it does not target a protein-coding mRNA, but rather triggers the production of tasiRNAs from the TAS3 locus, which in turn causes degradation of the ARF3 and ARF4 mRNAs in a miRNA-like fashion [
43,
44].
miR167 and
miR390 have been also described to be responsive to ABA [
45,
46], a phytohormone that has been involved in the early infection stage of antibacterial defense [
47]. After
P. syringae infection we have found
miR167 and
miR390 differentially expressed, as well as
miR169, also implicated in ABA functions [
48,
49].
miRNA families that target genes involved in auxin signaling and ABA response might shape a regulatory network for the molecular adaptation of tomato plants in response to P. syringae infection.
sly-miR168 expression was up-regulated in tomato plants undergoing
P. syringae infection. The
miR482/2118 superfamily is unusually diverse (at least 31 isoforms), and variable both in sequence (22 nucleotides rather than 21 nucleotides long) and in expression level. The abundance of
miR482 members varies greatly among species and families of plants, having the Solanum genera remarkably high levels of
miR482. All variants of this superfamily target the mRNA sequences of genes coding for disease resistance proteins (Resistance-like genes) with nucleotide binding site (NBS) and leucine-rich repeat (LRR) motifs [
7] for direct degradation as well as by generating secondary small interfering RNAs (siRNAs) in
P. syringae infected
Nicotiana benthamiana plants [
50], and also in the
Arabidopsis 22-nt
miR472 related to
miR482 [
51]. In contrast,
sly-miR482d-5p was described to target a PPO encoding gene in potato plants [
52] and
sly-miR482d-5p targets predicted in the present study were not related to defense functions.
In tomato plants challenged with
P. syringae,
sly-miR168 expression was found reduced. It has been described that
miR168, by regulating ARGONAUTE 1 (AGO1) homeostasis, exerts a regulatory feedback control over other miRNAs biogenesis [
53]. Thus,
miR168 functioning as an initial regulator modulates the levels of miRNAs coordinating the cross talk of stress response pathways and development programs.
miRNA regulated transcription factors are major nodes coordinating plant growth and differentiation related processes, stress responses, and signaling pathways crosstalk. It is well known that
miR396 targets plant GROWTH-REGULATING FACTORS (GRFs) implicated in the regulation of leaf growth,
miR172 binds to the 3′-end of APETHALA 2 (AP2) domain transcription factors, which are involved in flowering time control, and
miR166 target HOMEODOMAIN LEUCINE ZIPPER (
HD-ZIP) transcription factors family genes that regulate plant shoot apical meristems development [
54]. Moreover, members of the NF-Y family of transcription factors that play crucial roles in development and in response to adverse environmental conditions are targeted by
miR169 [
48]. Tomato plants challenged with
P. syringae showed altered profiles of all those miRNAs indicating that physiological adaptation to pathogen attack requires an integrated expression of genes responsible of immune defense and growth.
4.5. CPB Stress-Responsive miRNAs
Very few studies addressed miRNA identification by high-throughput sequencing in plants attacked by insects. We have detected two conserved miRNAs (
sly-miR171a and
sly-miR9471a-3p) and four novel miRNAs differentially expressed in CPB infested tomato plants.
miR171 family was significantly increased in wounded
Nicotiana attenuata leaves treated with
Manduca sexta oral secretions whereas
miR171 potential targets GRAS proteins were down-regulated [
55]. Among other miRNAs,
miR171 was considered as a JA-independent miRNA [
55]. In contrast, Gao et al. [
56] profiling miRNAs under wound treatment in
Aquilaria sinensis found consistently lower expression levels of
miR171 family members in wounded stems compared to healthy stems. Similarly, in our study,
sly-miR171a was repressed in tomato plants damaged by CPB. Regarding
sly-miR9471a-3p, this miRNA was among the most abundant miRNAs families detected in
Alternaria alternata infected tomato plants, although no differential expression was observed comparing to control plants [
57].
4.6. miRNAs Expressed in Response to the Priming Agent Hx
As a result of exposure to stress, plants often become more resistant to future exposure through a memory acquisition process named priming, which can also be mediated by natural compounds like Hx [
58]. Plant defense priming allows plants to respond to biotic and abiotic stress better than unprimed plants avoiding the fitness costs associated with permanent full defense activation [
59]. No differentially expressed conserved miRNAs following Hx treatment were identified and twelve novel Hx responsive miRNAs were found.
Little is known about the molecular mechanisms underlying priming process, but sustained impaired levels of signaling molecules after the initial stress that boost extensive transcriptional reprogramming of defense genes upon further challenge, as well as histone acetylation and DNA methylation epigenetic modifications have been proposed as critical regulators of defense priming [
60,
61,
62]. A role for miRNAs in stress memory has been recently described based on their specific functions as translational inhibitors. Induction of isoforms of the microRNA
miR156 has been reported following heat stress, and repression of their target genes was needed for the maintained enhanced expression of memory genes and for physiological heat stress memory [
63]. In addition, Soto-Suárez et al. [
64] provided evidence that miRNAs might be sustaining defense priming by demonstrating that reduced
miR396 levels and up-regulation of its target genes sensitized plants to mount more robust defense responses during pathogen infection even when in the absence of pathogen challenge, the transcriptome and development of modified plants with diminished
miR396 activity could not be distinguished from those in wild-type plants.
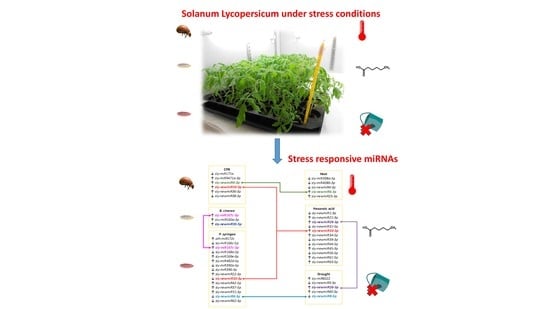
4.7. miRNAs Diferentially Expressed across Stresses
To the best of our knowledge, there is no research work on global expression profiling of tomato miRNAs in response to
P. syringae infection. We have detected eight conserved differentially expressed miRNAs one of them also responsive to
B. cinerea infection (
sly-miR167c-3p), and seven novel miRNAs two of which were responsive to other stresses as well (
sly-newmiR8-3p also down-regulated in response to drought, and
sly-newmiR33-3p responsive to CPB infestation or Hx treatment) (
Figure 5). Among the eight known
P. syringae-responsive miRNAs identified, five miRNAs (
ath-miR172c,
sly-miR166c-5p,
sly-miR168a-5p,
sly-miR396-3p and
sly-miR390a-5p) belong to families highly conserved in plants, two are moderately conserved (
sly-miR167c-3p and
sly-miR169e-3p), and
sly-miR482d-5p is a non-conserved plant miRNA. Highly and moderately conserved miRNA families have high expression levels and play relevant functions in plant development regulating gene expression of multiple targets in numerous plant species [
13]. Our analysis revealed several miRNA families that target genes involved in a number of essential pathways mainly regulating auxin signaling (
miR167 and
miR390), development and reprograming processes (
miR396,
miR172,
miR169 and
miR166), and stress defense (
miR167,
miR169 and
miR390), especially ABA response.
In our work, we analyzed the miRNA profile of tomato plants treated with the natural defense priming inducer Hx that protects tomato against
B. cinerea and
P. syringae with high efficiency [
65]. Interestingly, although no differentially expressed conserved miRNAs following Hx treatment were detected, two of the twelve novel Hx responsive miRNAs found (
sly-newmiR33-3p and
sly-newmiR26-3p) showed varied expression in tomato plants subjected to other stresses (CPB infestation and
P. syringae infection, or drought, respectively,
Figure 5). These results suggest that Hx treatment might exert its priming effect by triggering persistent expression of a complex network of miRNAs (not all of them necessarily involved in known stress signaling cascades) that upon exposure to a subsequent stress regulate an enhanced, more efficient or more rapid response. Further research will be needed to uncover the role of each Hx responsive novel miRNAs and their target genes, and decipher how they contribute to Hx defense priming.
4.8. Validation of Stress-Responsive miRNAs and Their Targets
A total of 181 targets genes were predicted (87 for conserved miRNAs and 94 for novel miRNAs) using the mature sequences of the 41 differentially expressed miRNAs. Most of the targets (82%) were predicted to be negatively regulated in a miRNA cleavage manner, therefore an opposite expression profile is expected for those miRNA-target pairs. We have selected 5 of these targets for validation with RT-qPCR focusing on P. syringae infection and CPB larvae infestation, two stress conditions for which there is a lack of information about their miRNA expression profile. We have selected targets for miRNAs that were validated with deep sequencing and RT-qPCR and that were either stress specific conserved miRNAs, sly-miR171a for CPB (GRAS and ABC transporter) and sly-miR172c for (PPR protein), stress specific novel miRNA as sly-newmiR22-3p for P. syringae infection (GDPDL3 and RLP1) or multi-stress miRNA, as sly-miR167c-3p for P. syringae and B. cinerea infection (Kinesin).
sly-miR172c expression was upregulated in tomato plants after infection with
P. syringae (3.89-fold). A pentatricopeptide repeat-containing protein (PPR) that belongs to the Tandem Repeats protein group is one of targets predicted for this miRNA. In
Arabidopsis, PPR proteins is one of the largest protein families with at least 466 genes in its genome [
66,
67]. Many repeat protein in plants, as PPR, LRR or WD40 have a role in primary metabolism but also act as regulators in plant secondary metabolism including abiotic and biotic stress response [
68,
69]. In
Solanum tuberosum,
PPR expression was downregulated upon
Ralstonia solanacearum infection [
70] and in
Arabidopsis, the absence of this protein led to an increase in the sensibility to a necrotrophic fungal pathogen and hypersensitivity to abiotic stresses such as salinity, glucose, and ABA [
71]. In this work,
PPR expression was downregulated in tomato leaves after
P. syringae infection (3.42-fold).
sly-miR67c-3p expression was upregulated upon
P. syringae and
B. cinerea infection (8.91-fold, 8.57-fold, respectively) and there is only one target predicted for this miRNA, a Kinesin light chain-like protein. Kinesin is a motor protein that moves along the microtubules of the cytoskeleton. The actin cytoskeleton in plants has been proposed to be associated to plant cell shape, plant development, and stress response [
72]. Both biotic and abiotic stresses might be affected by the continuous activity and reorganization of the host actin cytoskeleton [
72,
73]. In
Arabidopsis, Shimono et al. [
74] showed that kinesin mutant plants had less symptoms after
P. syringae infection and they speculate with the possibility of and integrative function for kinesin, supporting cellular traffic during pathogen invasion, and immune signaling. In this work, we have found a reduction of the kinesin gene expression after
P. syringae infection (−7.96-fold) and
B. cinerea infection (−1.74-fold).
The expression of
sly-miR171a miRNA was reduced in tomato plants (1.71-fold) after CPB larvae attack. Among the five predicted targets for this miRNA, three of them are members of the GRAS family of transcription factors (subfamily HAM): GRAS8, GRAS24 and GRAS40. In tomato, Huang et al. [
75] by 5’-RACE analysis demonstrated the regulation of GRAS24 and GRAS40 by
sly-miR171 and in
Arabidopsis, it had been previously reported that their closest homologous (AtSCL6, 22, 27) were post-transcriptionally regulated by ath-
miR171 [
76,
77]. Huang et al., [
78] established the implication of GRAS24 in the regulation of gibberellin and auxin homeostasis. Maryose et al. [
79] showed that transcript levels of eight tomato GRAS genes increased in response to mechanical stress. Also, a GRAS homologue in
Nicotiana attenuate and
Solanum nigra has been found to be induced upon
M. sexta attack [
80]. In this work we have analyzed by RT-qPCR the expression of the three GRAS genes predicted as targets of
sly-miR171a.
GRAS8 and
GRAS24 gene expression was not affected after CPB larvae attack (data not shown) but
GRAS40 expression was increased by a 2-fold, which could account for its participation in tomato response to herbivore attack.
The other putative target of
sly-miR171a is a multidrug resistance protein of the ABC transporter family, which use the energy released by ATP hydrolysis to drive the exchange of compounds across biological membranes even against electrochemical gradients [
81]. Oral secretions of herbivore chewing insects contains compounds that can be recognized by plants triggering a defense response [
82,
83] in which plants accumulate secondary metabolites and inhibitory proteins to stop pathogen invasion and insect attack. There are evidences indicating that ABC transporters mediate the secretion of plant defense compounds into both the rhizosphere and the apoplast, or onto the plant surface [
84]. In
Nicotiana tabacum, Bienert et al. [
85] showed that the ABCG5/PDR5 transporter expression in leaves was very low but it was induced after wounding by the herbivore
M. sexta. In this work, the tomato ABC transporter augmented its expression (1.98-fold) in leaves following CPB larvae attack, which would be in agreement with an implication of the pair
sly-miR171a–ABC transporter in tomato response to herbivore attack.
sly-newmiR22-3p expression was found increased after
P. syringae infection (2.46-fold). Two of its predicted targets were a Receptor-like protein kinase (RLP1), which has a LRR motif, and a Glycerophosphoryl diester phosphodiesterase family protein (GDPDL3). The plant receptor-like protein kinases with leucine-rich repeat motif (LRR-RLK) is one of largest plant protein families and they are implicated in hormone and stress response pathways and in plant developmental processes [
69]. Some members of LRR-RLK family and GDPL proteins participate in the formation of plant cell wall that acts as a barrier for both biotic and abiotic stresses [
86,
87]. In this work, we have detected about 2-fold reduction of
RLP1 and
GDPDL3 expression (−2.11-fold, −2.52-fold, respectively), which would account for the implication of this new tomato miRNA and its targets in the response to
P. syringae infection.