Abstract
One of the most commonly encountered species in the small basidiomycetous sub-phylum Wallemiomycotina is Wallemia mellicola, a xerotolerant fungus with a widespread distribution. To investigate the population characteristics of the species, whole genomes of twenty-five strains were sequenced. Apart from identification of four strains of clonal origin, the distances between the genomes failed to reflect either the isolation habitat of the strains or their geographical origin. Strains from different parts of the world appeared to represent a relatively homogenous and widespread population. The lack of concordance between individual gene phylogenies and the decay of linkage disequilibrium indicated that W. mellicola is at least occasionally recombining. Two versions of a putative mating-type locus have been found in all sequenced genomes, each present in approximately half of the strains. W. mellicola thus appears to be capable of (sexual) recombination and shows no signs of allopatric speciation or specialization to specific habitats.
1. Introduction
Towards the end of the 19th century, fish inspector Wallem was trying to tackle the problem of salted drying fish being spoiled by microbial growth [1]. From his samples in 1887, mycologist Johan Olav Olsen isolated and described the fungus Wallemia ichthyophaga [2]. More than a century later, and after several nomenclature changes, the only recognized species of Wallemia was Wallemia sebi. In 2005, the name W. ichthyophaga was resurrected for a group of Wallemia spp. strains able to grow only in media with substantially lowered water activity and an additional species—W. muriae—was described [3]. In 2015, a multi-locus phylogenetic analysis led to the description of additional species, W. mellicola, W. canadensis, W. tropicalis [4], followed by a description of W. hederae the following year [5] and finally W. peruviensis a year later [6]. In the resulting taxonomy W. sebi s. str. and W. mellicola were the most commonly isolated and most ubiquitous species of the genus. In addition to differences in molecular taxonomic markers, W. mellicola can be recognized by the larger size of conidia compared to W. sebi, while it is also less salt-tolerant and chaotolerant [4].
Due to their unusual morphology, Wallemia spp. long evaded reliable positioning into the fungal tree of life. The use of molecular phylogenetics showed that the genus is distant from all other known fungi, but its exact phylogenetic position remained uncertain. The first comprehensive molecular study by Zalar et al. [3] placed the Wallemia spp. into a new order (Wallemiales) and class (Wallemiomycetes) at the base of the Basidiomycota phylogenetic tree. Additional molecular analyses based on six genes confirmed a basal position of Wallemiomycetes to all of Pucciniomycotina, Ustilaginomycotina and Agaricomycotina [7]. Following the genome sequencing of W. mellicola and W. ichthyophaga, the analyses based on larger datasets positioned Wallemiomycetes as a sister group of Agaricomycotina [8,9]. Finally, the class Wallemiomycetes was accommodated in a new sub-phylum Wallemiomycotina, which was estimated to have emerged almost half a billion years ago, while its position in this study (as a sister group of just Agaricomycotina or basal to all three major subphyla of Basidiomycota) was again unclear and depended on the dataset used for inferring the phylogenetic relationships [10].
Wallemia spp. used to be known mainly as contaminants of food preserved with low-water-activity [3,11,12]. Later it became clear that they are frequent in both indoor and outdoor environments. They have been found in indoor air and house dust [13,14] and were reported to represent a large share of the microbiome of some species of house dust mites [15]. In natural environments Wallemia spp. are isolated particularly often from habitats characterized by low water activity [5]. While only a few isolates are known for some of the species of the genus, W. mellicola is encountered much more frequently. It can be found in different habitats around the world, among them air and house dust, hypersaline water of solar salterns, soil, salted, food preserved with low water activity, plant surface and pollen, straw and seeds [1]. These habitats reflect the extremotolerant character of Wallemia spp. Although tolerance of low water activity, especially if induced by high concentrations of salt, is rare among basidiomycetes, Wallemia spp. are among the most xerotolerant fungal taxa described to date, and some of them are even xerophilic—requiring low water activity to grow—an exceedingly rare trait in the fungal kingdom [3,5,16]. While W. mellicola is not the most extreme of Wallemia spp. in terms of halotolerance, the upper salinity levels supporting its growth are still high: 4.1 M NaCl and 1.4 M MgCl2 [1]. However, even though its growth optimum is at water activity of 0.97 to 0.92, W. mellicola also grows well in regular mycological media without additional osmolytes and is therefore considered to be xerotolerant/halotolerant rather than xerophilic/halophilic [4].
Strains of W. mellicola are known to produce secondary metabolites, namely tricyclic dihydroxysesquiterpenes wallimidione, walleminone, walleminol, and two azasteroids with antitumor activity, UCA 1064-A and UCA 1064-B [17]. Unusually, the production of wallimidione increases with increasing concentration of salt up to 2.6 M NaCl. This trait raises questions about the safety of salt-preserved food contaminated with mycotoxigenic Wallemia mellicola and other Wallemia spp. [17]. Walleminol (known also as walleminol A) was detected in food [18]. There are also sporadic reports of human infections by Wallemia spp. [19], although these may be underreported due to slow growth of the species [1].
Despite the above, the major threat posed by Wallemia spp. appears to be their allergenic potential, either through exposure by inhalation or, as shown by recent research, by the overgrowth of W. mellicola in the gastrointestinal tract. Wallemia spp. have long been associated with the development of farmer’s lung disease, a type of bronchial asthma or hypersensitivity pneumonitis (reviewed in [1]). A survey of air in animal and hay barns detected propagules of Wallemia spp. reaching up to 500 × 106 colony forming units (CFU)/m3, while only 20 to 500 CFU/m3 were found in residential buildings [5]. Immune sensitization to Wallemia spp. is frequently observed in asthmatic patients. Species of Wallemia were among the few fungi that increased the risk of asthma for inhabitants of homes damaged by water [20,21].
Wallemia spp. are often found in the human (and mice) gastrointestinal mycobiota. In mice the eradication of Candida spp. with antifungals leads to gastrointestinal overgrowth of W. mellicola, Aspergillus amsteoldami, and Epicoccum nigrum. While feeding healthy mice with these fungi did not lead to changes in their gut mycobiota, oral administration of W. mellicola after transient antibiotic therapy led to expansion of W. mellicola in the gut (a phenomenon not observed for either A. amstelodami or E. nigrum). This expansion in turn led to altered pulmonary immune responses to inhaled aeroallergens–without Wallemia present in the lungs [22,23].
The genome of W. mellicola (strain CBS 633.66, isolated from date honey and at the time classified as W. sebi) was published in 2012 [8]. The genome turned out to be unusually compact for a basidiomycete (9.8 Mbp) and contained a putative mating-type locus, even though sexual reproduction in W. mellicola has not been described to date.
To investigate the intraspecific relationships between strains of W. mellicola isolated from various indoor and outdoor environments in different parts of the world, we sequenced the whole genomes of 25 strains and analysed them using population and comparative genomic tools.
2. Materials and Methods
2.1. Culture, Medium, Growth Conditions and DNA Isolation
Twenty-five strains of W. mellicola (Table 1) were obtained from the Ex Culture Collection of the Department of Biology, Biotechnical Faculty, University of Ljubljana (Slovenia). They were cultivated and their DNA was isolated as described previously [24]. All strains used in this study are publicly available in the Ex Culture Collection under their EXF numbers (Table 1).

Table 1.
Strains sequenced in this study.
2.2. Genome Sequencing
The genome sequencing was performed using the platform BGISEQ-500, with 2 × 150 bp libraries, prepared as described previously [25]. Multiplexing of the samples was used, and after demultiplexing, the quality of the reads was investigated using FastQC. Trimming the reads for adaptors and quality (removal of bases with Q < 20) was performed with the ‘bbduk’ script (https://jgi.doe.gov/data-and-tools/bbtools/).
The sequencing reads, assembly and annotation data have been deposited in Genbank under BioProject PRJNA527769 and in CNGB Nucleotide Sequence Archive (CNSA) (https://db.cngb.org/cnsa/) of China National GeneBank DataBase (CNGBdb) with accession code CNP0000446.
2.3. Variant Calling
Sequencing reads were mapped to the reference W. mellicola genome of strain CBS 633.66 (GenBank AFQX00000000.1) [8] with ‘bwa mem’, using the default parameters. This was followed by sorting with Samtools 1.6 [26], and identification of duplicates with Picard 2.10.2. Variant calling was performed with Genome Analysis Toolkit 4.1 [27]. ‘Genome Analysis Toolkit (GATK) Best Practices’ were modified by using the ‘hard filtering’ and haploid ploidy.
2.4. Assembly and Annotation
IDBA-Hybrid 1.1.3 [28] was used to assemble the genomes. The process was guided by the W. mellicola CBS 633.66 reference genome [8]. The other parameters were: maximum k-value 120, seed kmer 20, minimum support 2, similarity for alignment 0.95, maximum allowed gap in the reference 100, minimum size of contigs 500.
Protein-coding genes were annotated with MAKER 2.31.8 [29]. The published predicted proteome of W. mellicola CBS 633.66 [8] and the fungal proteins of the Swissprot database (downloaded on 12.06.2018) were used as evidence. Semi-HMM-based Nucleic Acid Parser (SNAP) [30] was trained in three bootstrap iterations (W. mellicola CBS 633.66 proteins were used as evidence in the first iteration, W. mellicola and Swissprot database in the second and third), using protein-alignment-derived gene models following the workflow of Campbell et al. (2014). Augustus predictions with the Laccaria bicolor training parameters was also used [31].
BUSCO 3 software [32] in proteomic mode and with the Basidiomycota protein dataset [33] was used to investigate the genome assembly and gene prediction completeness. All of the parameters were used as the default values.
Genome Annotation Generator (GAG) 2.0.1 software [34] was used to prepare the files for submission to GenBank. All of the gene models with a coding region <150 bp or with introns <10 bp were removed.
2.5. Variant-Based Analysis
Principal component analysis of the Single Nucleotide Polymorphism (SNP) data was performed with the ‘glPca’ function from the ‘adgenet’ package [35]. Linkage Disequilibrium (LD) was estimated on a dataset of biallelic SNP loci. For each pair of loci, the normalized coefficient of LD (D’) and the squared correlation coefficient (r2) were calculated using ‘vcftools’ [36]. To investigate LD decay, D’ and r2 of loci within 10,000 nucleotides from each other were plotted as a function of distance and a generalized additive model fitted curve was added using ‘ggplot2’ in R [37,38]. The LD decay range was determined as the interval outside which all of the arithmetic means of D’ or r2 were either higher (left interval border) or lower (right interval border) than half of the maximum observed D’ or r2 means.
2.6. Phylogenetic Analysis
Gene phylogenetic trees were constructed from the predicted coding sequences of complete and single-copy BUSCOs. Alignment was calculated with MAFFT 7.407 in ‘--auto’ mode [39]. Gblocks 0.91 was employed to optimize the alignment, with the options ‘-b3 = 10 -b4 = 3 -b5 = n′ [40]; if the resulting alignment length was > 200 nt and the mean number of different nucleotides between the sequence pairs was larger than 15 (as counted by the ‘infoalign’ tool included in EMBOSS 6.6.0.0 [41]), phylogeny was inferred from the alignment with PhyML 3.1 [42]. The nucleotide substitution model was Hasegawa-Kishino-Yano 85 [43], the proportion of invariable sites and the alpha parameter of the gamma distribution of substitution rate categories were estimated by PhyML. Trees were visualized in DensiTree 2.2.5 [44]. A majority rule consensus tree was constructed in R with the function ‘consensus.edges’ (package ‘phytools’), using the default parameters [38,45].
The phylogenetic network was reconstructed from the SNP data as described previously [24].
2.7. Core Genome, GO Enrichment
The core genome W. mellicola was estimated with the pipeline GET_HOMOLOGUES 3.0.8 [46] from the predicted proteomes of all here sequenced strains and the reference strain W. mellicola CBS 633.66 [8] as a consensus of COGtriangle and OrthoMCL algorithms using default parameters. Representative sequences of each protein cluster were annotated using the PANTHER HMM scoring tools 2.1 and the HMM library version 13.1 [47]. Statistically significant enrichment of GO-Slim Biological Process terms was investigated at www.pantherdb.org for the lists of core gene clusters (present in all 26 genomes) and soft core gene clusters (in at least 24 genomes) with a list of all gene clusters used as a reference. Fisher’s Exact test and the False Discovery Rate correction were used.
2.8. Mating-Type Loci
BLAST was used to search for mating genes in the assembled and annotated W. mellicola genomes and predicted proteomes, using homologues of putative mating genes identified in the reference genome [8] as queries. The functional annotations of the genes were assigned according to Padamsee et al. [8]. Putative mating loci and their flanking regions were visualized in R with ‘ggplot2’ [37,38]. The corresponding regions of the genomes were aligned and the alignments visualized using Mummer 3.23 [48].
3. Results
Wallemia mellicola has a worldwide distribution and, unlike most other species in the genus, it is frequently isolated. To investigate its population structure, 25 genomes of W. mellicola were sequenced and compared. Strains were selected to cover a variety of habitats (from hypersaline water and various low water activity food to house dust, air, soil, plants and soil) and isolation locations (14 countries), as listed in Table 1.
Genomes were sequenced at 318× average coverage and the minimum coverage was 194× in case of genome 5. Using the reference W. mellicola to guide the assembly process, the genomes were assembled into 202 to 422 contigs (average 239 ± 43 SD). The size of the genomes was small and similar between the strains (average 9.75 Mbp ± 0.05 Mbp SD). Nevertheless, the completeness of the genome was high, with 88.18% (±0.50% SD) complete basidiomycetous Benchmarking Universal Single-Copy Orthologues (BUSCOs) present in the predicted proteomes, most of them in a single copy, and only 5.89% (±0.38% SD) of BUSCOs missing entirely. Between 4327 and 4509 (average 4475 ± 37 SD) predicted genes covered 66.64% (±0.32% SD) of the assemblies. The average intron length of 64 and the average GC content of 39.95% were very similar between the individual genomes (Table 2, Supplementary Table S1).

Table 2.
Statistics for the sequenced Wallemia mellicola genomes.
All 25 genomes and the reference W. mellicola genome shared 2845 genes (the core genome, identified by the GET_HOMOLOGUES pipeline). The softcore genome (genes present in at least 24 of 26 genomes) contained additional 611 genes. When the genomes were classified with the PANTHER classification system, the following categories were identified as significantly overrepresented in the core genome: Molecular Function: nucleoside-triphosphatase activity, oxidoreductase activity, protein binding, transporter activity, transferase activity, nucleic acid binding; Biological Process: nucleic acid metabolic process, cellular localization, transport, cellular component organization, gene expression, regulation of biological process; Cellular Component: chromosomal part, endomembrane system, cytosol, plasma membrane, vacuole, protein-containing complex, nuclear lumen.
The density of single nucleotide polymorphisms (SNPs) when compared to the reference W. mellicola genome (Table 2, Supplementary Table S1) was very similar for all strains (0.41–0.60%). No strains with very high similarity to the reference genome were found. There was also little clustering of the strains in the principal component analysis (PCA) of SNP data (Figure 1). The first two axes explained 23.7% and 8.80% of the variation. A cluster of four strains isolated from food (three from Slovenia, one from United Kingdom) were observed and were most likely of clonal origin (Supplementary Table S2). Two pairs of strains from the same country clustered closely together (4 and 13, 6 and 16) but apart from that the strains from the same habitats or from the same geographic locations were spread relatively far from each other on the PCA plot.
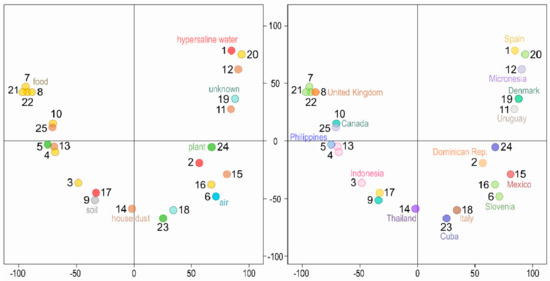
Figure 1.
Clustering of the Wallemia mellicola genomes. Principal component analysis of single nucleotide polymorphisms (SNP) data estimated by comparing 25 sequenced genomes to the reference genome. The genomes are represented by circles, the color of which corresponds to the habitat (left) or sampling location (right) of the sequenced strains. The first two axes explain 23.7% (x axis) and 8.80% (y axis) of variation.
Phylogenetic analysis of core BUSCOs (40 single-copy BUSCOs present in all sequenced genomes and with a minimum average of 15 different nucleotides between gene pairs) returned very different phylogenetic trees (Figure 2). There was little concordance between the tree topologies, resulting in a majority rule consensus tree with an extreme, star-like topology. The phylogenetic network analysis of the SNP data detected a fair amount of reticulation in the network.

Figure 2.
Phylogeny of W. mellicola strains. (A) Overlay of 40 core Benchmarking Universal Single-Copy Orthologue (BUSCO) gene trees estimated by PhyML 3.1 using the Hasegawa-Kishino-Yano 85 nucleotide substitution model and estimating the alpha parameter of the gamma distribution of the substitution rate categories and the proportion of invariable sites. (B) Majority rule consensus tree of 40 core gene trees described above. (C) Phylogenetic network reconstructed with the Neighbor-Net algorithm based on the dissimilarity distance matrix calculated from the SNP data.
A lack of concordance between phylogenetic histories of individual parts of the genome can be best explained by recombination shuffling these parts of the genome between individual organisms. A well-recognized estimator of the amount of recombination is the influence of the physical distance between the loci on the linkage disequilibrium (LD) between these loci. In non-recombining organisms the linkage between the loci should be absolute, no matter the distance between them, while in recombining populations the linkage between two loci is expected to decrease as a function of the distance between the two loci, approaching (but not necessarily reaching) equilibrium. In W. mellicola the LD decay, the average distance over which the LD falls to half of its maximum value, was 1291–3064 bp (intersect of the fitted curve was at 1990) when estimated with the squared correlation coefficient (r2; Figure 3) and 224–860 when estimated with the normalized coefficient of LD (D’; data not shown).
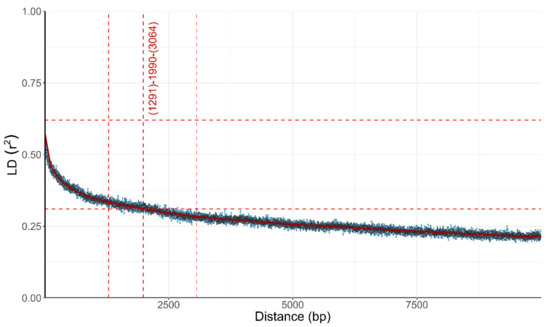
Figure 3.
Linkage disequilibrium (LD) decay in Wallemia mellicola estimated by calculation of the squared correlation coefficient (r2) between pairs of biallelic loci. r2 is plotted against the physical distance of the loci in the genome. Horizontal lines mark the maximum observed value and half of the maximum observed value. Vertical lines mark the interval of the physical distance delimited by the first point of the curve under half of the maximum r2 value (left vertical line), the last point above half of the maximum r2 value (right vertical line) and the point where the fitted curve intersects with half of the maximum r2 value (middle vertical line).
A putative mating-type locus of W. mellicola that was identified by Padamsee et al. [8] was also found in all 25 here sequenced genomes (Figure 4, Supplementary File S1). Two groups of strains (containing 12 and 13 strains each) were recognized by comparing the gene annotations in the locus - the two groups differed in some of the genes and in the orientation of the locus. No other chromosomal inversions were identified in the corresponding contig (Figure 4).
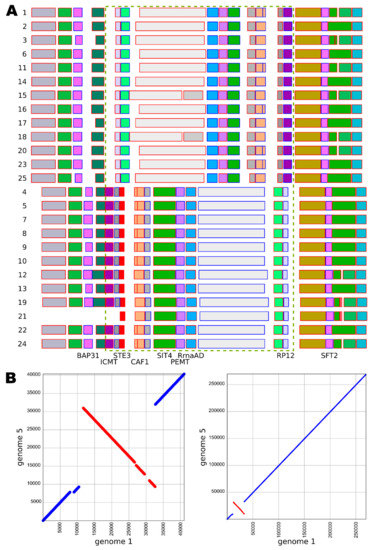
Figure 4.
Putative mating-type loci in different strains of Wallemia mellicola. (A) Annotated mating-type loci and their flanking regions with putative gene functions assigned according to Padamsee et al. [8]. The chromosomal inversion is marked with a dashed green rectangle. Genome numbers are marked on the left. The blue or red outline of the rectangles representing the genes shows the gene orientation: left to right (blue) or right to left (red). (B) Alignment of the contigs containing the putative mating-type loci from the genomes 1 (x axis) and 5 (y axis) at different magnifications.
4. Discussion
Some extremotolerant fungal species are found in a large number of diverse environments, presumably using their stress tolerance to endure the often challenging conditions in their chosen habitats [49,50,51]. Although an infrequently mentioned example of such species, Wallemia mellicola, a species described upon a taxonomic revision of Wallemia sebi, can be isolated from habitats as different as air and house dust, hypersaline water of solar salterns, soil, salted, food preserved with low water activity, plant surface and pollen, straw and seeds [1] and is even a common part of the human gastrointestinal mycobiota [23]. To compare W. mellicola isolates from a variety of habitats and locations, twenty-five strains (Table 1) were genome sequenced and compared to investigate the possible existence of local subpopulations or cryptic specialization towards specific habitats and to check for evidence of recombination within the species. Such result would not be unusual—in recent years population genomics enabled the discovery of several cases of cryptic diversification or specialization in fungal species, which had previously appeared homogenous due to the lower phylogenetic resolution of standard taxonomic markers [52,53,54,55].
The sequenced genomes showed high similarity to the reference W. mellicola genome, which was reported to be very compact and with a large gene density [8]. The genomes of Wallemia spp. thus remain among the smallest in Basidiomycota [56,57]. The twenty-five genomes sequenced here were on average 9.75 Mbp large (compared to 9.8 Mbp of the reference genome) and 4475 protein coding genes were annotated per genome—with very little variation between the genomes (Table 2, Supplementary Table S1). This was less than 5284 genes annotated in the reference genome, which might be a consequence of different approaches to the genome assembly (de novo for the reference genome and reference-guided from short reads here) and/or to the gene annotation. At the same time 94% of the Benchmarking Universal Single-Copy Orthologues (BUSCOs) were found in the genome on average, indicating that the genomes were assembled to a high degree of completeness.
The variation between genomes (0.52% average SNP density compared to the reference genome (Table 2, Supplementary Table S1)) was comparable to variation observed in other fungal species, for example 0.55% in wild strains of Saccharomyces cerevisiae [58], 0.41% in Neurospora crassa [59] or up to 0.66% in Candida glabrata [60]. Only one clonal cluster of nearly identical genomes was observed on the PCA plot of SNP data, representing strains 7, 8, 21 and 22, which differed by less than 500 SNPs per genome pair (Supplementary Table S2). All four strains were isolated from food, three from chocolate in Slovenia, one from bread in UK. The next most similar pair of genomes (4 and 13, both isolated in Indonesia) differed by 17223 SNPs. However, despite this there was no general trend of higher similarity based on geographical proximity—although only a rough estimate, the average number of SNPs between the isolates isolated in different countries and the average difference between the genomes from the same country differed by only 2.4%. Similarly, the PCA did not uncover any clustering of strains based on either their habitat or geographical origin (Figure 1). This indicates that adaptation of localized populations or cryptic specialization for specific (types of) habitats is not widespread in W. mellicola. Considering the many habitats W. mellicola is able to inhabit, such generalization is not self-evident. To name just a few examples, using genomic data, it was discovered that the mushroom Suillus brevipes shows strong population differentiation and environmental adaptation [61], the pathogen Cryptococcus neoformans diversified into lineages with different pathogenic potential [62], the arbuscular mycorrhizal fungus Rhizophagus irregularis diverged into four main genetic groups, which were not related to the geographical origin of the strains [63], and the yeast Metschnikowia reukaufii diverged into lineages, which were again not related to geographical origin, but were shown to be metabolically distinct [53].
The general absence of clear clonal clusters (apart from the above-mentioned exception) raised a question about signs of recombination within W. mellicola genomes. This would be in line with the proposal made by Padamsee et al. [8] that W. mellicola is capable of sexual reproduction and their description of a putative mating locus in its genome. The availability of the genome sequences of W. mellicola for the first time enabled us to check for evidence of recombination in the species. If an organism is recombining (either by meiotic recombination or through other mechanisms), different parts of its genome are expected to have different phylogenetic histories. Two main approaches are used to check this in practice. The first is to reconstruct the phylogenies of individual genes and compare them. If, on the one hand, the gene trees share the same topology, they fulfil the “strong phylogenetic signal” criterion for clonality [64]. If, on the other hand, there is a lack of concordance between the phylogenies, this can be interpreted as a good indication of recombination. In the case of W. mellicola the latter scenario was observed and the differences between the gene trees were so numerous that the majority rule tree had an extreme star-shaped topology (Figure 2). The second approach for investigating recombination on the genome level is to look at the inheritance of loci on the same DNA molecule. In clonal organisms two loci will always be inherited together—they will be linked and in maximum linkage disequilibrium (LD). In sexual organisms two loci can segregate randomly and even if they are on the same DNA molecule, the linkage between them can be broken by chromosomal crossover—which is more likely to happen the further apart the loci are located. Such decrease of LD with distance, and particularly the distance over which half of the maximum observed LD is reached (a value known as LD decay distance), is a good measure of the amount of recombination in the population [65]. In the sequenced strains of W. mellicola the LD decay distance calculated from biallelic SNPs was around 1990 bp (Figure 3). This value is higher than observed in many other fungi [55,58,66], but much lower than in highly clonal species, where LD decay distances can be larger by two orders of magnitude [66]. This indicates that W. mellicola is at least occasionally recombining, but also extensively combining this with clonal reproduction—an observation in line with the identification of four clones in the sequenced dataset.
The indications for recombination in W. mellicola appear to support its proposed capability for sexual reproduction [8]. Indeed, the putative mating-type locus has been found in all 25 here sequenced genomes. Furthermore, the locus was found to exist in two variants, which share several regions of high similarity, but differ in their orientations relative to the rest of the contigs on which they are located, and which contain no other chromosomal inversions (Figure 4). These two variants possibly represent two different mating types of W. mellicola—a hypothesis to be tested in the future.
5. Conclusions
Wallemia mellicola is an extremotolerant basidiomycetous fungus from the sub-phylum Wallemiomycotina, with a distinct phylogenetic position and capable of inhabiting a wide variety of habitats. Sequencing and analysis of twenty-five W. mellicola genomes showed that the strains form no clusters based on the habitat or geographical location from which they were isolated. The sequenced strains appear to represent a relatively homogenous and widespread population with only one clonal lineage detected in the dataset. The lack of concordance between individual gene phylogenies and the decay of linkage disequilibrium indicated that W. mellicola is at least occasionally recombining. The mechanism of recombination could be sexual reproduction—two versions of a putative mating-type locus have been found in all sequenced genomes, each present in approximately half of the strains.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/10/6/427/s1, Table S1: Statistics of the sequenced W. mellicola genomes, Table S2: Number of different SNP loci between pairs of W. mellicola genomes, File S1: Sequence of the putative mating-type locus and flanking genomic regions.
Author Contributions
Conceptualization, N.G.-C., Z.S. and C.G.; methodology, C.G., X.S. and J.Z.; investigation, X.S., J.Z. and C.G.; resources, Z.S. and N.G.-C.; data curation, C.G.; writing—original draft preparation, C.G.; writing—review and editing, C.G., N.G.-C., X.S. and Z.S.; visualization, C.G.; supervision, Z.S. and N.G.-C.; software, F.C.; funding acquisition, Y.H., N.G.-C. and Z.S.; Z.S. and N.G.C. contributed equally to this work as leading authors.
Funding
The authors acknowledge the financial support from the Slovenian Research Agency to the Infrastructural Centre Mycosmo (MRIC UL), to the programs P1-0170 and P1-0198, and to the Postdoctoral Project Z7-7436 J. Zajc. The authors would like to thank Chris Berrie for language editing assistance.
Acknowledgments
The authors would like to thank Yuchong Tang for his invaluable help in project management, organization of scientific visits and facilitation of collaboration between the project partners. We thank Anja Černoša and Teja Lavrin for their valuable assistance in preparation of DNA samples. The authors would also like to thank Keith A. Seifert for providing the strains EXF-6156 (UAMH 2651), EXF-6157 (UAMH 2757) and EXF-6158 (UAMH 6689); and Jens Frisvad for the strain EXF-1443 (IBT 19078).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zajc, J.; Gunde-Cimerman, N. The Genus Wallemia—From Contamination of Food to Health Threat. Microorganisms 2018, 6, 46. [Google Scholar] [CrossRef] [PubMed]
- Johan-Olsen, O. Om Sop på Klipfisk den Såkaldte Mid; Dybwad: Christiania, Norway, 1887. [Google Scholar]
- Zalar, P.; Sybren de Hoog, G.; Schroers, H.-J.; Frank, J.M.; Gunde-Cimerman, N. Taxonomy and phylogeny of the xerophilic genus Wallemia (Wallemiomycetes and Wallemiales, cl. et ord. nov.). Antonie Van Leeuwenhoek 2005, 87, 311–328. [Google Scholar] [CrossRef] [PubMed]
- Jančič, S.; Nguyen, H.D.T.; Frisvad, J.C.; Zalar, P.; Schroers, H.-J.; Seifert, K.A.; Gunde-Cimerman, N. A Taxonomic Revision of the Wallemia sebi Species Complex. PLoS ONE 2015, 10, e0125933. [Google Scholar] [CrossRef] [PubMed]
- Jančič, S.; Zalar, P.; Kocev, D.; Schroers, H.J.; Džeroski, S.; Gunde-Cimerman, N. Halophily reloaded: New insights into the extremophilic life-style of Wallemia with the description of Wallemia hederae sp. nov. Fungal Divers. 2016, 76, 97–118. [Google Scholar] [CrossRef]
- Díaz-Valderrama, J.R.; Nguyen, H.D.T.; Aime, M.C. Wallemia peruviensis sp. nov., a new xerophilic fungus from an agricultural setting in South America. Extremophiles 2017, 21, 1017–1025. [Google Scholar] [CrossRef]
- Matheny, P.B.; Gossmann, J.A.; Zalar, P.; Kumar, T.K.A.; Hibbett, D.S. Resolving the phylogenetic position of the Wallemiomycetes: An enigmatic major lineage of Basidiomycota. Can. J. Bot. 2006, 84, 1794–1805. [Google Scholar] [CrossRef]
- Padamsee, M.; Kumar, T.K.A.; Riley, R.; Binder, M.; Boyd, A.; Calvo, A.M.; Furukawa, K.; Hesse, C.; Hohmann, S.; James, T.Y.; et al. The genome of the xerotolerant mold Wallemia sebi reveals adaptations to osmotic stress and suggests cryptic sexual reproduction. Fungal Genet. Biol. 2012, 49, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Zajc, J.; Liu, Y.; Dai, W.; Yang, Z.; Hu, J.; Gostinčar, C.; Gunde-Cimerman, N. Genome and transcriptome sequencing of the halophilic fungus Wallemia ichthyophaga: Haloadaptations present and absent. BMC Genom. 2013, 14, 617. [Google Scholar] [CrossRef]
- Zhao, R.L.; Li, G.J.; Sánchez-Ramírez, S.; Stata, M.; Yang, Z.L.; Wu, G.; Dai, Y.C.; He, S.H.; Cui, B.K.; Zhou, J.L.; et al. A six-gene phylogenetic overview of Basidiomycota and allied phyla with estimated divergence times of higher taxa and a phyloproteomics perspective. Fungal Divers. 2017, 84, 43–74. [Google Scholar] [CrossRef]
- Pitt, J.I.; Hocking, A.D. Fungi and Food Spoilage, 2nd ed.; Aspen Publishers, Inc.: Gaithersburg, MD, USA, 1999; ISBN 978-0-387-92206-5. [Google Scholar]
- Samson, R.A.; Hoekstra, E.S.; Frisvad, J.C.; Filtenborg, O. Introduction to Food- and Airborne Fungi, 6th ed.; Centraalbureau voor Schimmelcultures: Baarn, The Netherlands, 2002. [Google Scholar]
- Takahashi, T. Airborne fungal colony-forming units in outdoor and indoor environments in Yokohama, Japan. Mycopathologia 1997, 139, 23–33. [Google Scholar] [CrossRef]
- Fröhlich-Nowoisky, J.; Pickersgill, D.A.; Despres, V.R.; Poschl, U. High diversity of fungi in air particulate matter. Proc. Natl. Acad. Sci. USA 2009, 106, 12814–12819. [Google Scholar] [CrossRef] [PubMed]
- Hubert, J.; Nesvorna, M.; Kopecky, J.; Erban, T.; Klimov, P. Population and Culture Age Influence the Microbiome Profiles of House Dust Mites. Microb. Ecol. 2019, 77, 1048–1066. [Google Scholar] [CrossRef] [PubMed]
- Zajc, J.; Kogej, T.; Ramos, J.; Galinski, E.A.; Gunde-Cimerman, N. The osmoadaptation strategy of the most halophilic fungus Wallemia ichthyophaga, growing optimally at salinities above 15% NaCl. Appl. Environ. Microbiol. 2014, 80, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Jančič, S.; Frisvad, J.C.; Kocev, D.; Gostinčar, C.; Džeroski, S.; Gunde-Cimerman, N. Production of secondary metabolites in extreme environments: Food- and airborne Wallemia spp. produce toxic metabolites at hypersaline conditions. PLoS ONE 2016, 11, e0169116. [Google Scholar] [CrossRef] [PubMed]
- Moss, M.O. Recent studies of mycotoxins. J. Appl. Microbiol. 1998, 84, 62–76. [Google Scholar] [CrossRef]
- Guarro, J.; Gugnani, H.C.; Sood, N.; Batra, R.; Mayayo, E.; Gene, J.; Kakkar, S. Subcutaneous phaeohyphomycosis caused by Wallemia sebi in an immunocompetent host. J. Clin. Microbiol. 2008, 46, 1129–1131. [Google Scholar] [CrossRef]
- Sakamoto, T.; Urisu, A.; Yamada, M.; Matsuda, Y.; Tanaka, K.; Torii, S. Studies on the Osmophilic Fungus Wallemia sebi as an Allergen Evaluated by Skin Prick Test and Radioallergosorbent Test. Int. Arch. Allergy Immunol. 1989, 90, 368–372. [Google Scholar] [CrossRef]
- Vesper, S.J.; McKinstry, C.; Yang, C.; Haugland, R.A.; Kercsmar, C.M.; Yike, I.; Schluchter, M.D.; Kirchner, H.L.; Sobolewski, J.; Allan, T.M.; et al. Specific molds associated with asthma in water-damaged homes. J. Occup. Environ. Med. 2006, 48, 852–858. [Google Scholar] [CrossRef]
- Wheeler, M.L.; Limon, J.J.; Bar, A.S.; Leal, C.A.; Gargus, M.; Tang, J.; Brown, J.; Funari, V.A.; Wang, H.L.; Crother, T.R.; et al. Immunological Consequences of Intestinal Fungal Dysbiosis. Cell Host Microbe 2016, 19, 865–873. [Google Scholar] [CrossRef]
- Skalski, J.H.; Limon, J.J.; Sharma, P.; Gargus, M.D.; Nguyen, C.; Tang, J.; Coelho, A.L.; Hogaboam, C.M.; Crother, T.R.; Underhill, D.M. Expansion of commensal fungus Wallemia mellicola in the gastrointestinal mycobiota enhances the severity of allergic airway disease in mice. PLoS Pathog. 2018, 14, e1007260. [Google Scholar] [CrossRef]
- Gostinčar, C.; Stajich, J.E.; Zupančič, J.; Zalar, P.; Gunde-Cimerman, N. Genomic evidence for intraspecific hybridization in a clonal and extremely halotolerant yeast. BMC Genom. 2018, 19, 364. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Zhong, H.; Lin, Y.; Chen, B.; Han, M.; Ren, H.; Lu, H.; Luber, J.M.; Xia, M.; Li, W.; et al. Assessment of the cPAS-based BGISEQ-500 platform for metagenomic sequencing. Gigascience 2018, 7, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Alkan, C.; Coe, B.P.; Eichler, E.E. GATK toolkit. Nat. Rev. Genet. 2011, 12, 363–376. [Google Scholar] [CrossRef]
- Peng, Y.; Leung, H.C.M.; Yiu, S.M.; Chin, F.Y.L. IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012, 28, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.S.; Holt, C.; Moore, B.; Yandell, M. Genome Annotation and Curation Using MAKER and MAKER-P. Curr. Protoc. Bioinform. 2014, 48, 1–39. [Google Scholar] [CrossRef]
- Korf, I. Gene finding in novel genomes. BMC Bioinform. 2004, 5, 59. [Google Scholar] [CrossRef] [PubMed]
- Stanke, M.; Morgenstern, B. AUGUSTUS: A web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005, 33, 465–467. [Google Scholar] [CrossRef] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Kriventseva, E.V.; Tegenfeldt, F.; Petty, T.J.; Waterhouse, R.M.; Simão, F.A.; Pozdnyakov, I.A.; Ioannidis, P.; Zdobnov, E.M. OrthoDB v8: Update of the hierarchical catalog of orthologs and the underlying free software. Nucleic Acids Res. 2015, 43, 250–256. [Google Scholar] [CrossRef]
- Geib, S.M.; Hall, B.; Derego, T.; Bremer, F.T.; Cannoles, K.; Sim, S.B. Genome Annotation Generator: A simple tool for generating and correcting WGS annotation tables for NCBI submission. Gigascience 2018, 7, 4. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T.; Ahmed, I. adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 2011, 27, 3070–3071. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2; Springer New York: New York, NY, USA, 2009; ISBN 978-0-387-98140-6. [Google Scholar]
- R Development Core Team R: A Language and Environment for Statistical Computing. 2019. Available online: ftp://ftp.uvigo.es/CRAN/web/packages/dplR/vignettes/intro-dplR.pdf (accessed on 23 March 2019).
- Katoh, K.; Toh, H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 2008, 9, 286–298. [Google Scholar] [CrossRef] [PubMed]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Hasegawa, M.; Kishino, H.; Yano, T. aki Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985, 22, 160–174. [Google Scholar] [CrossRef]
- Bouckaert, R.R. DensiTree: Making sense of sets of phylogenetic trees. Bioinformatics 2010, 26, 1372–1373. [Google Scholar] [CrossRef]
- Revell, L.J. phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 2012, 3, 217–223. [Google Scholar] [CrossRef]
- Vinuesa, P.; Contreras-Moreira, B. Robust identification of orthologues and paralogues for microbial pan-genomics using GET_HOMOLOGUES: A case study of pIncA/C plasmids. Methods Mol. Biol. 2015, 1231, 203–232. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.D. PANTHER: A Library of Protein Families and Subfamilies Indexed by Function. Genome Res. 2003, 13, 2129–2141. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004, 5, 2. [Google Scholar] [CrossRef] [PubMed]
- Gostinčar, C.; Grube, M.; De Hoog, S.; Zalar, P.; Gunde-Cimerman, N. Extremotolerance in fungi: Evolution on the edge. FEMS Microbiol. Ecol. 2010, 71, 2–11. [Google Scholar] [CrossRef]
- Gostinčar, C.; Grube, M.; Gunde-Cimerman, N. Evolution of fungal pathogens in domestic environments? Fungal Biol. 2011, 115, 1008–1018. [Google Scholar] [CrossRef] [PubMed]
- Gostinčar, C.; Gunde-Cimerman, N.; Grube, M. 10 Polyextremotolerance as the fungal answer to changing environments. In Microbial Evolution under Extreme Conditions; Bakermans, C., Ed.; DE GRUYTER: Berlin/München,Germany; Boston, MA, USA, 2015; pp. 185–208. ISBN 9783110335064. [Google Scholar]
- Silva, D.N.; Várzea, V.; Paulo, O.S.; Batista, D. Population genomic footprints of host adaptation, introgression and recombination in coffee leaf rust. Mol. Plant Pathol. 2018, 19, 1742–1753. [Google Scholar] [CrossRef] [PubMed]
- Dhami, M.K.; Hartwig, T.; Letten, A.D.; Banf, M.; Fukami, T. Genomic diversity of a nectar yeast clusters into metabolically, but not geographically, distinct lineages. Mol. Ecol. 2018, 27, 2067–2076. [Google Scholar] [CrossRef] [PubMed]
- Branco, S.; Gladieux, P.; Ellison, C.E.; Kuo, A.; LaButti, K.; Lipzen, A.; Grigoriev, I.V.; Liao, H.L.; Vilgalys, R.; Peay, K.G.; et al. Genetic isolation between two recently diverged populations of a symbiotic fungus. Mol. Ecol. 2015, 24, 2747–2758. [Google Scholar] [CrossRef]
- Ellison, C.E.; Hall, C.; Kowbel, D.; Welch, J.; Brem, R.B.; Glass, N.L.; Taylor, J.W. Population genomics and local adaptation in wild isolates of a model microbial eukaryote. Proc. Natl. Acad. Sci. USA 2011, 108, 2831–2836. [Google Scholar] [CrossRef]
- Gregory, T.R.; Nicol, J.A.; Tamm, H.; Kullman, B.; Kullman, K.; Leitch, I.J.; Murray, B.G.; Kapraun, D.F.; Greilhuber, J.; Bennett, M.D. Eukaryotic genome size databases. Nucleic Acids Res. 2007, 35, D332–D338. [Google Scholar] [CrossRef]
- Mohanta, T.K.; Bae, H. The diversity of fungal genome. Biol. Proced. Online 2015, 17, 8. [Google Scholar] [CrossRef] [PubMed]
- Peter, J.; De Chiara, M.; Friedrich, A.; Yue, J.-X.; Pflieger, D.; Bergström, A.; Sigwalt, A.; Barre, B.; Freel, K.; Llored, A.; et al. Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 2018, 556, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Pomraning, K.R.; Smith, K.M.; Freitag, M. Bulk Segregant Analysis Followed by High-Throughput Sequencing Reveals the Neurospora Cell Cycle Gene, ndc-1, To Be Allelic with the Gene for Ornithine Decarboxylase, spe-1. Eukaryot. Cell 2011, 10, 724–733. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Carreté, L.; Ksiezopolska, E.; Pegueroles, C.; Gómez-Molero, E.; Saus, E.; Iraola-Guzmán, S.; Loska, D.; Bader, O.; Fairhead, C.; Gabaldón, T. Patterns of Genomic Variation in the Opportunistic Pathogen Candida glabrata Suggest the Existence of Mating and a Secondary Association with Humans. Curr. Biol. 2018, 28, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Branco, S.; Bi, K.; Liao, H.-L.; Gladieux, P.; Badouin, H.; Ellison, C.E.; Nguyen, N.H.; Vilgalys, R.; Peay, K.G.; Taylor, J.W.; et al. Continental-level population differentiation and environmental adaptation in the mushroom Suillus brevipes. Mol. Ecol. 2017, 26, 2063–2076. [Google Scholar] [CrossRef] [PubMed]
- Desjardins, C.A.; Giamberardino, C.; Sykes, S.M.; Yu, C.-H.; Tenor, J.L.; Chen, Y.; Yang, T.; Jones, A.M.; Sun, S.; Haverkamp, M.R.; et al. Population genomics and the evolution of virulence in the fungal pathogen Cryptococcus neoformans. Genome Res. 2017, 27, 1207–1219. [Google Scholar] [CrossRef] [PubMed]
- Savary, R.; Masclaux, F.G.; Wyss, T.; Droh, G.; Cruz Corella, J.; Machado, A.P.; Morton, J.B.; Sanders, I.R. A population genomics approach shows widespread geographical distribution of cryptic genomic forms of the symbiotic fungus Rhizophagus irregularis. ISME J. 2018, 12, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Tibayrenc, M.; Ayala, F.J. Reproductive clonality of pathogens: A perspective on pathogenic viruses, bacteria, fungi, and parasitic protozoa. Proc. Natl. Acad. Sci. USA 2012, 109, 3305–3313. [Google Scholar] [CrossRef]
- Taylor, J.W.; Hann-Soden, C.; Branco, S.; Sylvain, I.; Ellison, C.E. Clonal reproduction in fungi. Proc. Natl. Acad. Sci. USA 2015, 112, 8901–8908. [Google Scholar] [CrossRef]
- Nieuwenhuis, B.P.S.; James, T.Y. The frequency of sex in fungi. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 20150540. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).