A Four-Pseudogene Classifier Identified by Machine Learning Serves as a Novel Prognostic Marker for Survival of Osteosarcoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition
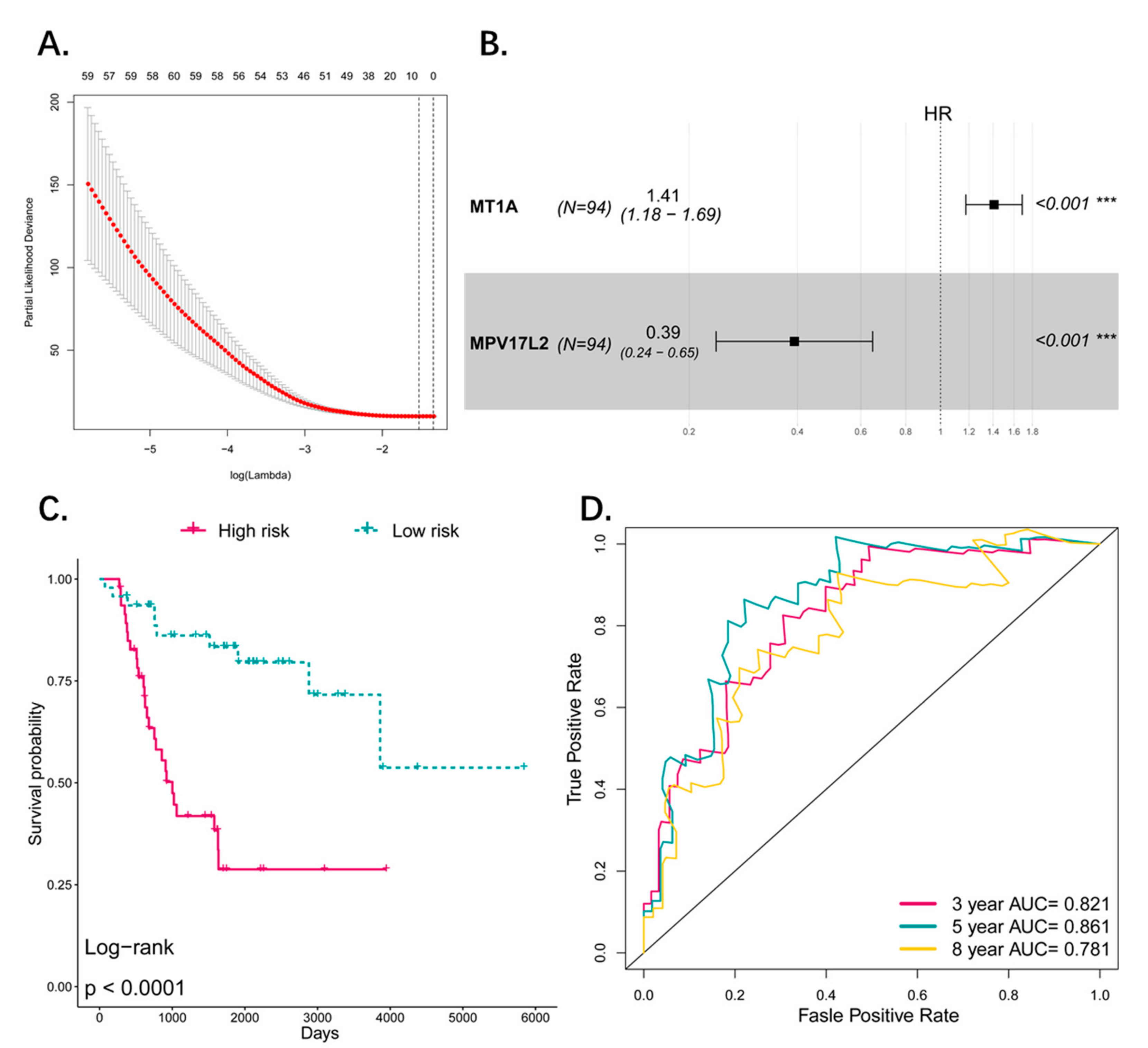
2.2. Construction of Prognostic Signature
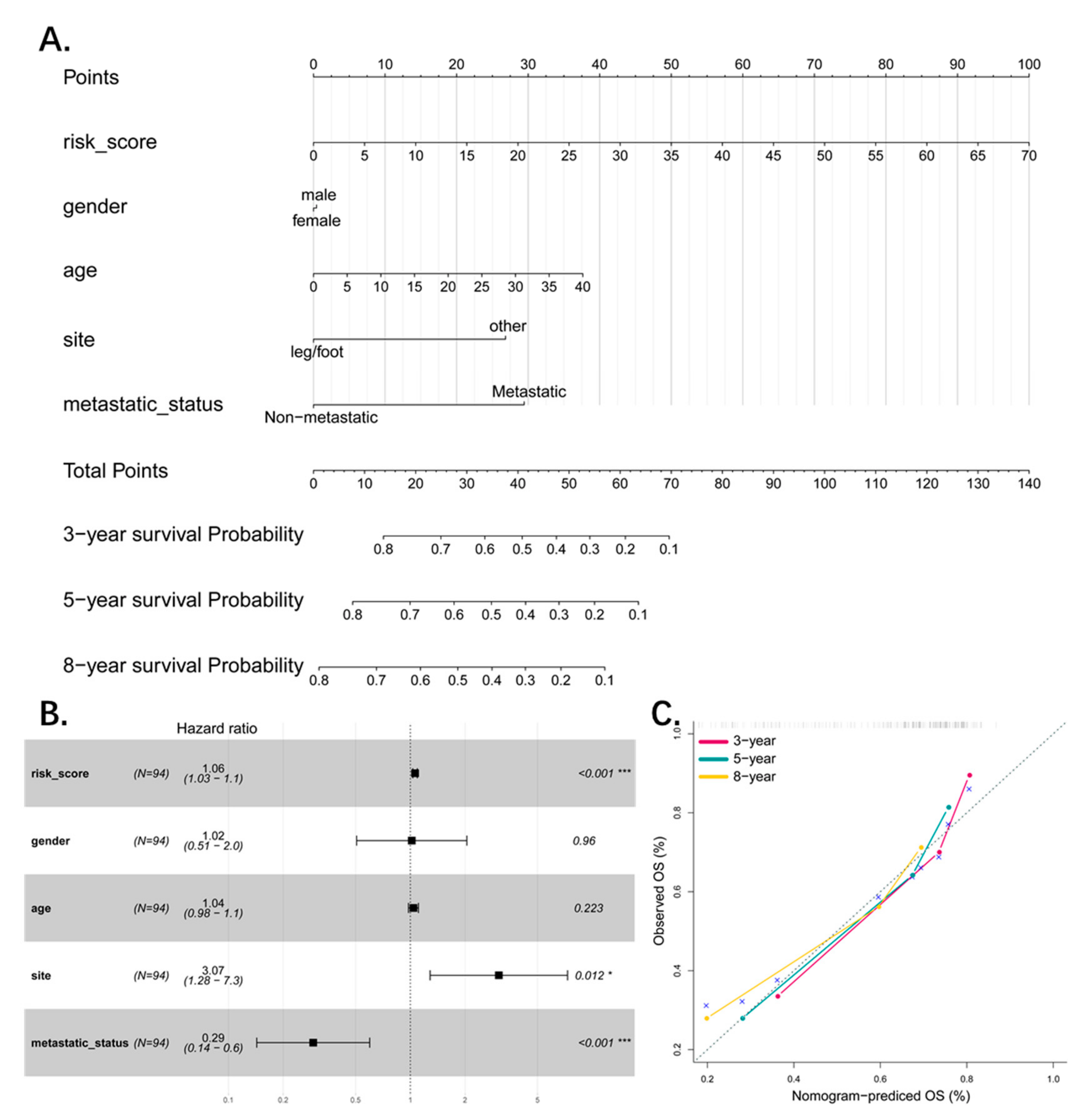
2.3. Nomogram and Calibration
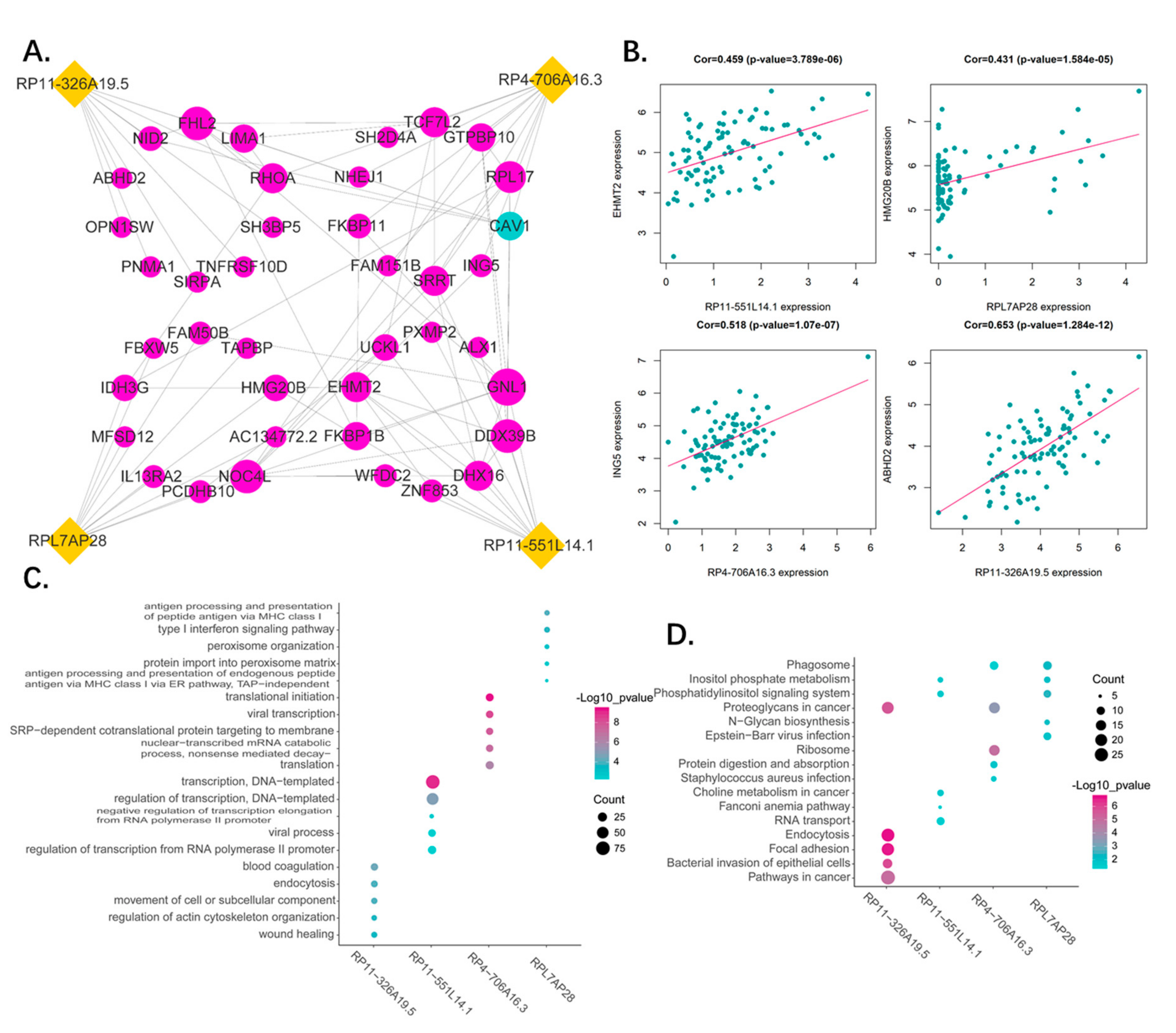
2.4. Correlation Analysis of the Four Pseudogenes and Annotation of Their Function
3. Results
3.1. Clinical Characteristics of the Patients
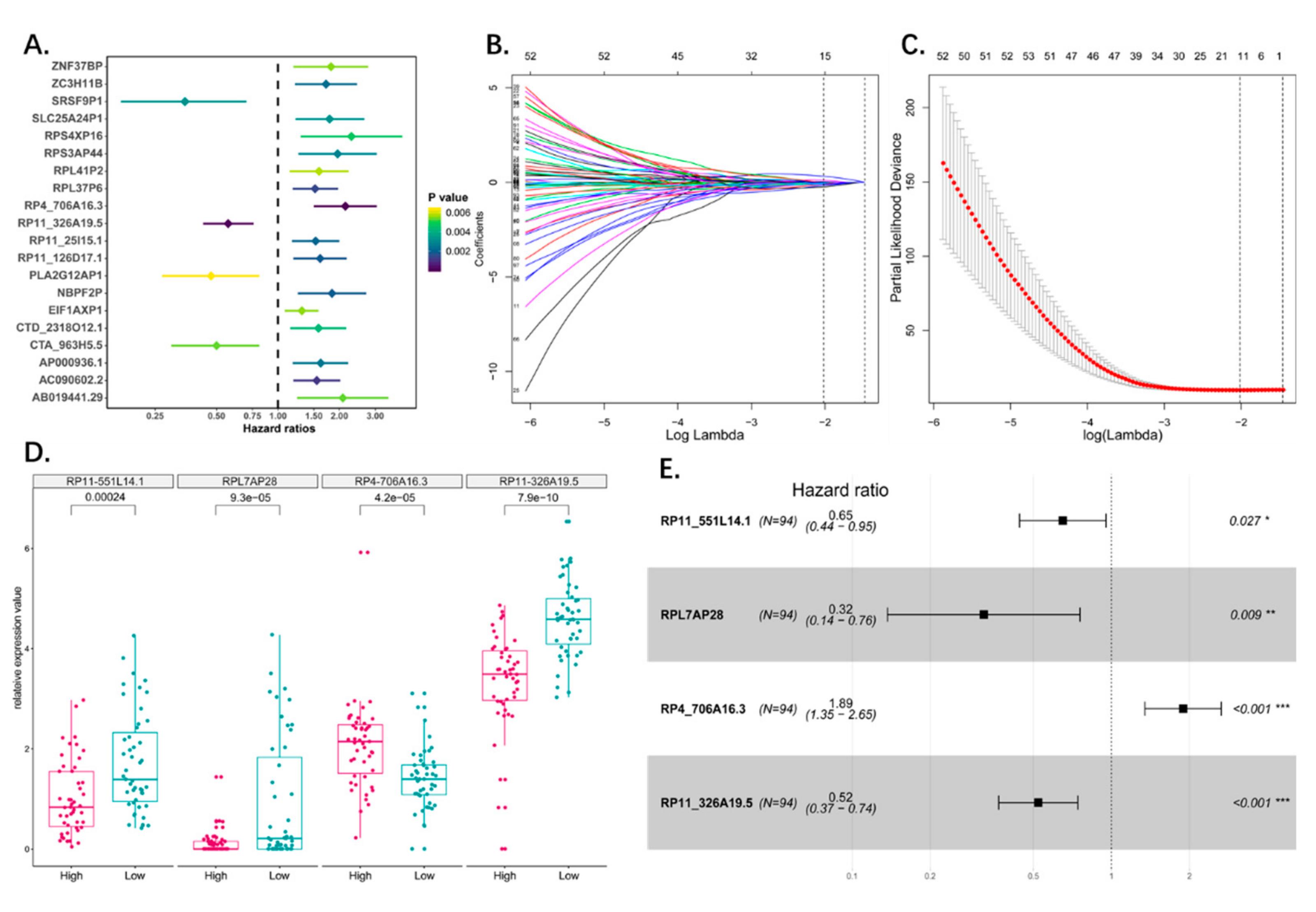
3.2. Identification of Osteosarcoma Survival-Related Pseudogenes
3.3. Construction of the Prognostic Pseudogene Signature for Osteosarcoma
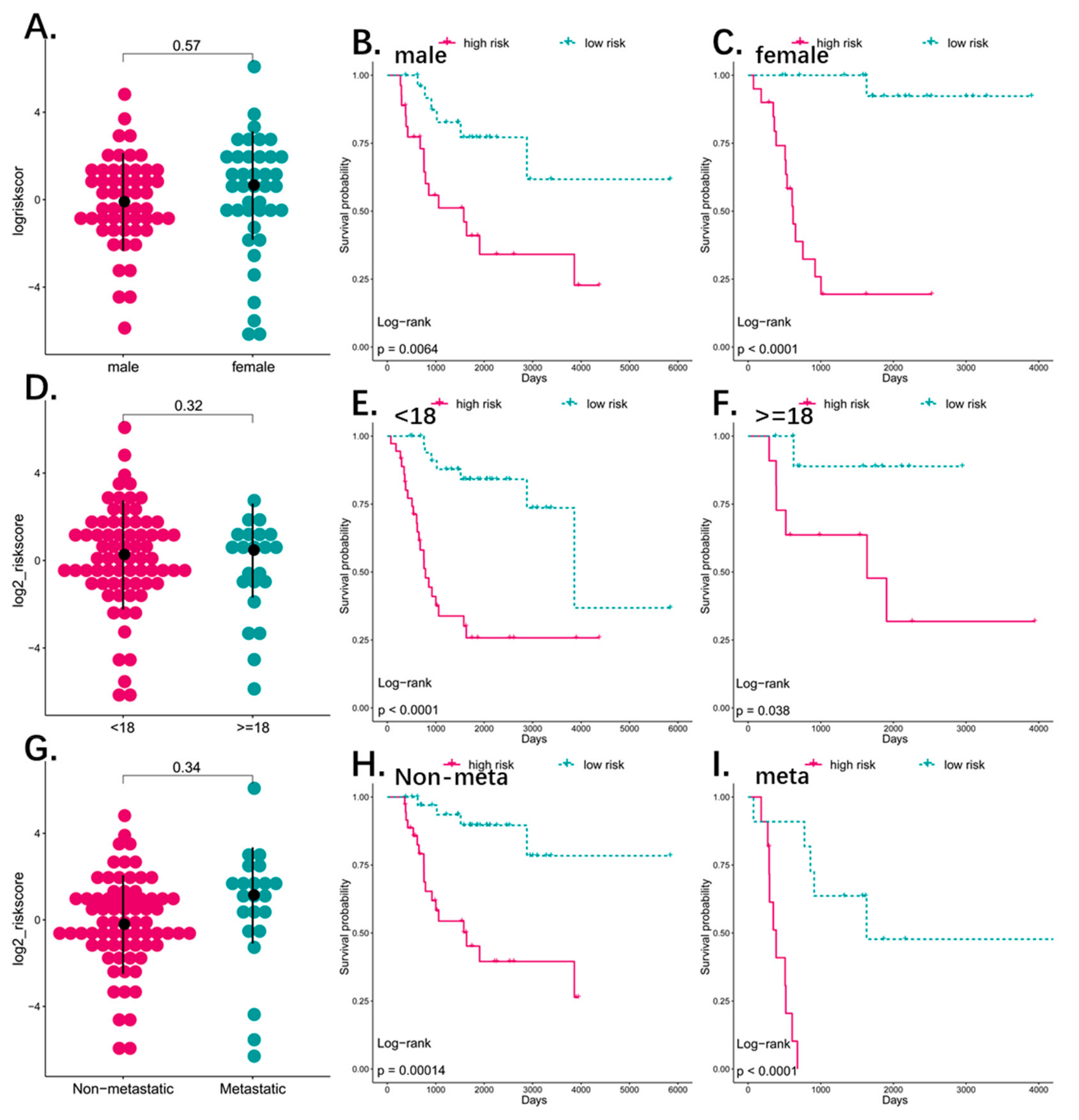
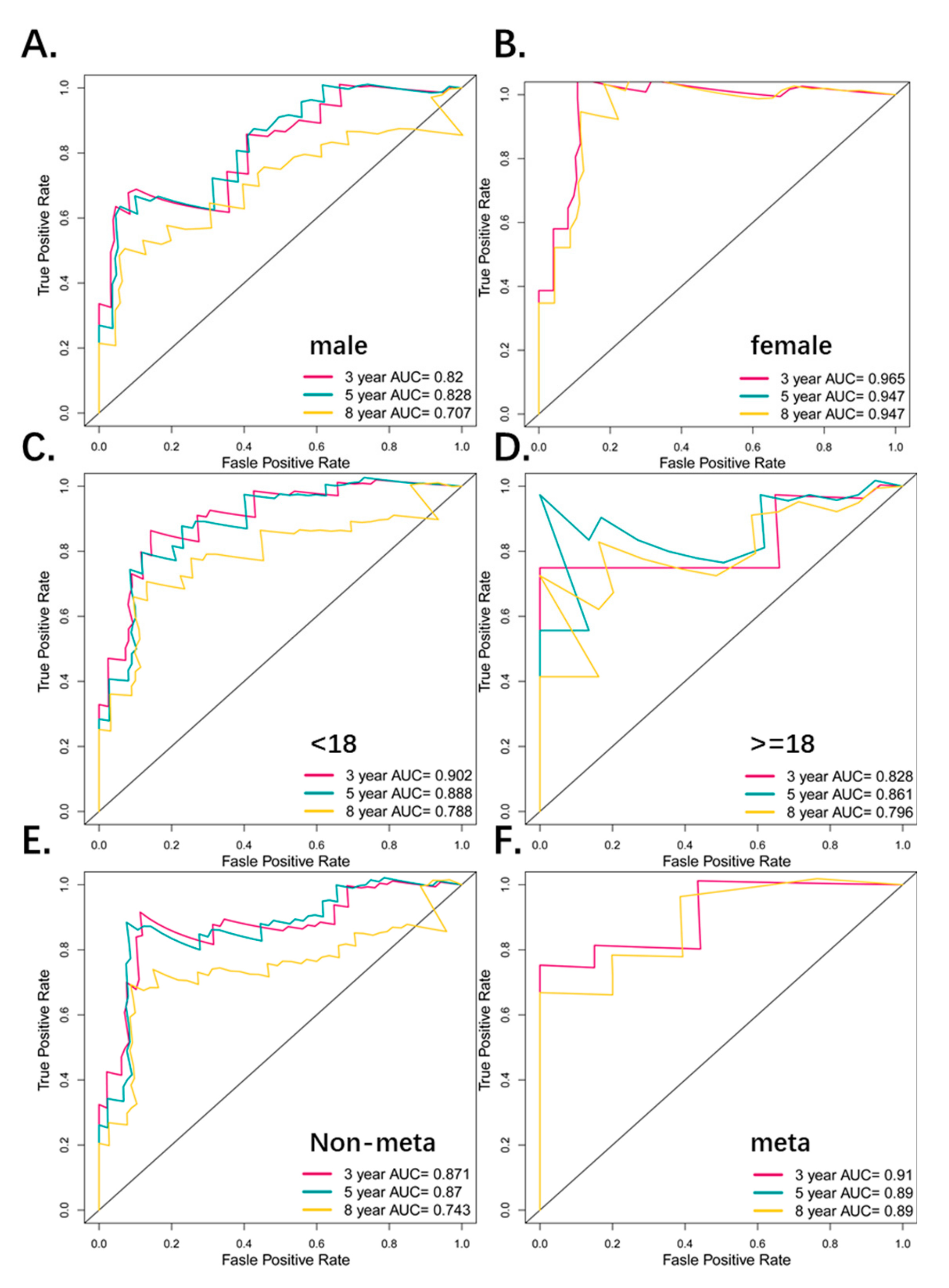
3.4. Predictive Value of the Four-Pseudogene Signature
3.5. The Four-Pseudogene Signature Is an Independent Prognostic Predictor of Osteosarcoma
3.6. Combined Pseudogene and Clinical Risk Score Is an Independent Predictor of Survival
3.7. Functional Analysis of the Predictive Pseudogenes
3.8. Comparison and Combination of Gene-Signature and Pseudogene-Signature
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AUC | Area under curve |
| BP | Biological process |
| DAVID | Database for Annotation, Visualization, and Integrated Discovery |
| GO | Gene ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| K–M | Kaplan–Meier |
| OS | Overall survival |
| PCGs | Protein coding genes |
| ROC | Receiver operating characteristic |
References
- Simpson, S.; Dunning, M.D.; de Brot, S.; Grau-Roma, L.; Mongan, N.P.; Rutland, C.S. Comparative review of human and canine osteosarcoma: Morphology, epidemiology, prognosis, treatment and genetics. Acta Vet. Scand. 2017, 59, 71. [Google Scholar] [CrossRef]
- Moore, D.D.; Luu, H.H. Osteosarcoma. Cancer Treat. Res. 2014, 162, 65–92. [Google Scholar] [CrossRef]
- Shankar, G.M.; Clarke, M.J.; Ailon, T.; Rhines, L.D.; Patel, S.R.; Sahgal, A.; Laufer, I.; Chou, D.; Bilsky, M.H.; Sciubba, D.M.; et al. The role of revision surgery and adjuvant therapy following subtotal resection of osteosarcoma of the spine: A systematic review with meta-analysis. J. Neurosurg. Spine 2017, 27, 97–104. [Google Scholar] [CrossRef]
- Chen, Y.; Gokavarapu, S.; Shen, Q.; Liu, F.; Cao, W.; Ling, Y.; Ji, T. Chemotherapy in head and neck osteosarcoma: Adjuvant chemotherapy improves overall survival. Oral Oncol. 2017, 73, 124–131. [Google Scholar] [CrossRef]
- Kansara, M.; Teng, M.W.; Smyth, M.J.; Thomas, D.M. Translational biology of osteosarcoma. Nat. Rev. Cancer 2014, 14, 722–735. [Google Scholar] [CrossRef] [PubMed]
- Ballman, K.V. Biomarker: Predictive or Prognostic? J. Clin. Oncol. 2015, 33, 3968–3971. [Google Scholar] [CrossRef]
- Nonaka, T.; Wong, D.T.W. Liquid Biopsy in Head and Neck Cancer: Promises and Challenges. J. Dent. Res. 2018, 97, 701–708. [Google Scholar] [CrossRef] [PubMed]
- Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef] [PubMed]
- Pei, B.; Sisu, C.; Frankish, A.; Howald, C.; Habegger, L.; Mu, X.J.; Harte, R.; Balasubramanian, S.; Tanzer, A.; Diekhans, M.; et al. The GENCODE pseudogene resource. Genome Biol. 2012, 13, R51. [Google Scholar] [CrossRef]
- Watanabe, T.; Totoki, Y.; Toyoda, A.; Kaneda, M.; Kuramochi-Miyagawa, S.; Obata, Y.; Chiba, H.; Kohara, Y.; Kono, T.; Nakano, T.; et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 2008, 453, 539–543. [Google Scholar] [CrossRef] [PubMed]
- Abdollahzadeh, R.; Daraei, A.; Mansoori, Y.; Sepahvand, M.; Amoli, M.M.; Tavakkoly-Bazzaz, J. Competing endogenous RNA (ceRNA) cross talk and language in ceRNA regulatory networks: A new look at hallmarks of breast cancer. J. Cell. Physiol. 2019, 234, 10080–10100. [Google Scholar] [CrossRef]
- Zhou, B.S.; Beidler, D.R.; Cheng, Y.C. Identification of antisense RNA transcripts from a human DNA topoisomerase I pseudogene. Cancer Res. 1992, 52, 4280–4285. [Google Scholar] [PubMed]
- Poliseno, L.; Salmena, L.; Zhang, J.; Carver, B.; Haveman, W.J.; Pandolfi, P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010, 465, 1033–1038. [Google Scholar] [CrossRef] [PubMed]
- Pink, R.C.; Wicks, K.; Caley, D.P.; Punch, E.K.; Jacobs, L.; Carter, D.R. Pseudogenes: Pseudo-functional or key regulators in health and disease? RNA 2011, 17, 792–798. [Google Scholar] [CrossRef]
- Liu, C.; Guo, T.; Xu, G.; Sakai, A.; Ren, S.; Fukusumi, T.; Ando, M.; Sadat, S.; Saito, Y.; Khan, Z.; et al. Characterization of Alternative Splicing Events in HPV-Negative Head and Neck Squamous Cell Carcinoma Identifies an Oncogenic DOCK5 Variant. Clin. Cancer Res. 2018, 24, 5123–5132. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Yang, J.; Zhang, Y.; Zhou, J.; Li, Y.; Hao, X. Integrated analysis of pseudogene RP11-564D11.3 expression and its potential roles in hepatocellular carcinoma. Epigenomics 2019, 11, 267–280. [Google Scholar] [CrossRef] [PubMed]
- Ganapathi, M.K.; Jones, W.D.; Sehouli, J.; Michener, C.M.; Braicu, I.E.; Norris, E.J.; Biscotti, C.V.; Vaziri, S.A.; Ganapathi, R.N. Expression profile of COL2A1 and the pseudogene SLC6A10P predicts tumor recurrence in high-grade serous ovarian cancer. Int. J. Cancer 2016, 138, 679–688. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Ma, J.; Wang, A.; Wang, W.; Luo, S.; Liu, Y.; Ye, X. A support vector machine and a random forest classifier indicates a 15-miRNA set related to osteosarcoma recurrence. OncoTargets Ther. 2018, 11, 253–269. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Feng, H.; Li, Z.; Li, D.; Liu, S.; Huang, H.; Li, M. Application of weighted gene co-expression network analysis to identify key modules and hub genes in oral squamous cell carcinoma tumorigenesis. OncoTargets Ther. 2018, 11, 6001–6021. [Google Scholar] [CrossRef]
- Mao, X.; Qin, X.; Li, L.; Zhou, J.; Zhou, M.; Li, X.; Xu, Y.; Yuan, L.; Liu, Q.N.; Xing, H. A 15-long non-coding RNA signature to improve prognosis prediction of cervical squamous cell carcinoma. Gynecol. Oncol. 2018, 149, 181–187. [Google Scholar] [CrossRef]
- Zhang, X.; Feng, H.; Li, D.; Liu, S.; Amizuka, N.; Li, M. Identification of differentially expressed genes induced by aberrant methylation in oral squamous cell carcinomas using integrated bioinformatic analysis. Int. J. Mol. Sci. 2018, 19, 1698. [Google Scholar] [CrossRef] [PubMed]
- Xing, L.; Zhang, X.; Tong, D. Systematic Profile Analysis of Prognostic Alternative Messenger RNA Splicing Signatures and Splicing Factors in Head and Neck Squamous Cell Carcinoma. DNA Cell Biol. 2019. [Google Scholar] [CrossRef]
- Xiao-Jie, L.; Ai-Mei, G.; Li-Juan, J.; Jiang, X. Pseudogene in cancer: Real functions and promising signature. J. Med. Genet. 2015, 52, 17–24. [Google Scholar] [CrossRef]
- Vera Alvarez, R.; Pongor, L.S.; Marino-Ramirez, L.; Landsman, D. TPMCalculator: One-step software to quantify mRNA abundance of genomic features. Bioinformatics 2018, 1–3. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Sauerbrei, W.; Royston, P.; Binder, H. Selection of important variables and determination of functional form for continuous predictors in multivariable model building. Stat. Med. 2007, 26, 5512–5528. [Google Scholar] [CrossRef] [PubMed]
- Tibshirani, R. The lasso method for variable selection in the Cox model. Stat. Med. 1997, 16, 385–395. [Google Scholar] [CrossRef]
- Zhang, J.X.; Song, W.; Chen, Z.H.; Wei, J.H.; Liao, Y.J.; Lei, J.; Hu, M.; Chen, G.Z.; Liao, B.; Lu, J.; et al. Prognostic and predictive value of a microRNA signature in stage II colon cancer: A microRNA expression analysis. Lancet Oncol. 2013, 14, 1295–1306. [Google Scholar] [CrossRef]
- Huang, Y.Q.; Liang, C.H.; He, L.; Tian, J.; Liang, C.S.; Chen, X.; Ma, Z.L.; Liu, Z.Y. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J. Clin. Oncol. 2016, 34, 2157–2164. [Google Scholar] [CrossRef]
- Mao, Y.; Dong, L.; Zheng, Y.; Dong, J.; Li, X. Prediction of recurrence in cervical cancer using a nine-lncRNA signature. Front. Genet. 2019, 10, 284. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Xie, H.; Wang, H.; Han, C.; Yang, J.; Lin, Z.; Li, Y.; He, Q.; Wang, R.; Cui, Y.; et al. MRI-based radiomics signature for the preoperative prediction of extracapsular extension of prostate cancer. J. Magn. Reson. Imaging 2019. [Google Scholar] [CrossRef]
- Tian, X.P.; Huang, W.J.; Huang, H.Q.; Liu, Y.H.; Wang, L.; Zhang, X.; Lin, T.Y.; Rao, H.L.; Li, M.; Liu, F.; et al. Prognostic and predictive value of a microRNA signature in adults with T-cell lymphoblastic lymphoma. Leukemia 2019. [Google Scholar] [CrossRef]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Zhang, X.; Feng, H.; Du, J.; Sun, J.; Li, D.; Hasegawa, T.; Amizuka, N.; Li, M. Aspirin promotes apoptosis and inhibits proliferation by blocking G0/G1 into S phase in rheumatoid arthritis fibroblast-like synoviocytes via downregulation of JAK/STAT3 and NF-kappaB signaling pathway. Int. J. Mol. Med. 2018, 42, 3135–3148. [Google Scholar] [CrossRef]
- Zhang, X.; Feng, H.; Li, Z.; Guo, J.; Li, M. Aspirin is Involved in the Cell Cycle Arrest, Apoptosis, Cell Migration, and Invasion of Oral Squamous Cell Carcinoma. Int. J. Mol. Sci. 2018, 19, 2029. [Google Scholar] [CrossRef]
- Luetke, A.; Meyers, P.A.; Lewis, I.; Juergens, H. Osteosarcoma treatment—Where do we stand? A state of the art review. Cancer Treat. Rev. 2014, 40, 523–532. [Google Scholar] [CrossRef]
- Han, L.; Yuan, Y.; Zheng, S.; Yang, Y.; Li, J.; Edgerton, M.E.; Diao, L.; Xu, Y.; Verhaak, R.G.W.; Liang, H. The Pan-Cancer analysis of pseudogene expression reveals biologically and clinically relevant tumour subtypes. Nat. Commun. 2014, 5, 3963. [Google Scholar] [CrossRef]
- De Martino, M.; Forzati, F.; Arra, C.; Fusco, A.; Esposito, F. HMGA1-pseudogenes and cancer. Oncotarget 2016, 7, 28724–28735. [Google Scholar] [CrossRef]
- Muller, C.I.; Miller, C.W.; Hofmann, W.K.; Gross, M.E.; Walsh, C.S.; Kawamata, N.; Luong, Q.T.; Koeffler, H.P. Rare mutations of the PIK3CA gene in malignancies of the hematopoietic system as well as endometrium, ovary, prostate and osteosarcomas, and discovery of a PIK3CA pseudogene. Leuk. Res. 2007, 31, 27–32. [Google Scholar] [CrossRef]
- Yan, B.; Wubuli, A.; Liu, Y.; Wang, X. Long non-coding RNA phosphatase and tensin homolog pseudogene 1 suppresses osteosarcoma cell growth via the phosphoinositide 3-kinase/protein kinase B signaling pathway. Exp. Ther. Med. 2018, 15, 4829–4837. [Google Scholar] [CrossRef]
- Li, Q.; Li, H.; Zhang, L.; Zhang, C.; Yan, W.; Wang, C. Identification of novel long non-coding RNA biomarkers for prognosis prediction of papillary thyroid cancer. Oncotarget 2017, 8, 46136–46144. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, Y.; Sun, N.; Lu, Z.; Sun, S.; Huang, J.; Chen, Z.; He, J. Prognostic alternative mRNA splicing signature in non-small cell lung cancer. Cancer Lett. 2017, 393, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Guglas, K.; Bogaczynska, M.; Kolenda, T.; Rys, M.; Teresiak, A.; Blizniak, R.; Lasinska, I.; Mackiewicz, J.; Lamperska, K. lncRNA in HNSCC: Challenges and potential. Contemp. Oncol. (Pozn) 2017, 21, 259–266. [Google Scholar] [CrossRef] [PubMed]



| Alive (n = 57) | Dead (n = 37) | Total (n = 94) | |
|---|---|---|---|
| Gender | |||
| FEMALE | 25 | 15 | 40 |
| MALE | 32 | 22 | 54 |
| Age | |||
| Mean (SD) | 15.19 (5.75) | 14.62 (4.40) | 14.97 (5.27) |
| Median (MIN, MAX) | 15 (3, 39) | 14 (5, 32) | 14.5 (3, 39) |
| Site | |||
| Leg/foot | 53 | 30 | 83 |
| other | 4 | 7 | 11 |
| Metastatic status | |||
| Metastatic | 7 | 15 | 22 |
| Non-metastatic | 50 | 22 | 72 |
| Pseudogene | HR | CI95 | p-Value | Pseudogene | HR | CI95 | p-Value |
|---|---|---|---|---|---|---|---|
| RP4_706A16.3 | 2.14 | 1.5–3.05 | 2.60 × 10−5 | RP11_271C24.3 | 1.65 | 1.07–2.55 | 0.023498 |
| RP11_326A19.5 | 0.57 | 0.43–0.76 | 0.000127 | NCF1C | 0.61 | 0.4–0.94 | 0.023523 |
| AC090602.2 | 1.55 | 1.19–2.02 | 0.001094 | RP11_551L14.1 | 0.64 | 0.43–0.94 | 0.023633 |
| RPL37P6 | 1.52 | 1.18–1.97 | 0.001415 | RPL23AP53 | 1.54 | 1.06–2.23 | 0.023792 |
| RP11_126D17.1 | 1.61 | 1.19–2.17 | 0.00183 | ZNF815P | 0.53 | 0.31–0.92 | 0.024488 |
| NBPF2P | 1.84 | 1.25–2.71 | 0.001885 | AC002075.4 | 1.41 | 1.04–1.91 | 0.025616 |
| RP11_25I15.1 | 1.53 | 1.17–2 | 0.002111 | RPL7P47 | 1.73 | 1.07–2.8 | 0.025737 |
| ZC3H11B | 1.72 | 1.21–2.44 | 0.002279 | CTC_451P13.1 | 1.49 | 1.05–2.11 | 0.026269 |
| AP000936.1 | 1.62 | 1.18–2.21 | 0.002517 | RP11_36C20.1 | 1.81 | 1.07–3.07 | 0.026794 |
| RPS3AP44 | 1.96 | 1.26–3.05 | 0.00281 | RP11_62J1.3 | 0.54 | 0.31–0.93 | 0.026951 |
| SRSF9P1 | 0.35 | 0.17–0.7 | 0.00321 | RPL7P9 | 1.47 | 1.04–2.06 | 0.027115 |
| SLC25A24P1 | 1.79 | 1.22–2.65 | 0.003255 | RP11_10G12.1 | 1.45 | 1.04–2.01 | 0.027132 |
| CTD_2318O12.1 | 1.58 | 1.15–2.16 | 0.004281 | EEF1A1P11 | 1.44 | 1.04–1.99 | 0.027742 |
| RPS4XP16 | 2.29 | 1.29–4.07 | 0.00472 | RPL13P12 | 0.79 | 0.64–0.98 | 0.028947 |
| AB019441.29 | 2.08 | 1.24–3.48 | 0.005308 | RP11_155G14.5 | 1.47 | 1.04–2.08 | 0.029335 |
| CTA_963H5.5 | 0.5 | 0.3–0.81 | 0.005308 | RPL13AP7 | 1.57 | 1.04–2.37 | 0.030481 |
| ZNF37BP | 1.82 | 1.19–2.77 | 0.0055 | RP11_488L18.4 | 1.47 | 1.04–2.08 | 0.030529 |
| EIF1AXP1 | 1.31 | 1.08–1.58 | 0.005551 | TMSB4XP8 | 0.75 | 0.57–0.97 | 0.031485 |
| RPL41P2 | 1.59 | 1.14–2.22 | 0.005779 | FCF1P2 | 0.6 | 0.38–0.96 | 0.031956 |
| PLA2G12AP1 | 0.47 | 0.27–0.81 | 0.006369 | CSPG4P13 | 1.35 | 1.03–1.78 | 0.03249 |
| RP11_23J18.1 | 0.44 | 0.24–0.79 | 0.006523 | RPL41P1 | 1.09 | 1.01–1.17 | 0.0327 |
| RP11_535M15.2 | 1.83 | 1.18–2.83 | 0.006615 | RPL7P23 | 1.75 | 1.05–2.93 | 0.032774 |
| PTGES3P1 | 0.5 | 0.3–0.83 | 0.006904 | RP11_175B9.3 | 1.32 | 1.02–1.69 | 0.033142 |
| RP11_16F15.2 | 1.79 | 1.17–2.72 | 0.006917 | RP11_1166P10.1 | 1.55 | 1.03–2.31 | 0.033608 |
| RPL10P3 | 1.44 | 1.1–1.87 | 0.007168 | SRP68P3 | 1.28 | 1.02–1.6 | 0.034297 |
| RP11_197B12.1 | 1.41 | 1.1–1.81 | 0.007359 | RP11_51F16.9 | 0.57 | 0.34–0.96 | 0.034541 |
| MTND4P12 | 1.3 | 1.07–1.57 | 0.007377 | FAM86C2P | 2.07 | 1.05–4.08 | 0.034789 |
| RP11_120B7.1 | 1.81 | 1.16–2.82 | 0.008388 | AC073850.6 | 1.41 | 1.02–1.94 | 0.035167 |
| AC005077.14 | 1.42 | 1.09–1.83 | 0.008434 | HTR7P1 | 0.41 | 0.18–0.94 | 0.0355 |
| RP11_255H23.5 | 1.78 | 1.16–2.73 | 0.008551 | CALM2P2 | 1.41 | 1.02–1.95 | 0.035662 |
| RP11_494O16.3 | 0.55 | 0.35–0.86 | 0.008747 | RP11_587D21.1 | 1.34 | 1.02–1.76 | 0.035695 |
| RP1_95L4.4 | 0.46 | 0.25–0.82 | 0.009153 | DSTNP2 | 0.6 | 0.37–0.97 | 0.038699 |
| AC010468.1 | 1.83 | 1.16–2.9 | 0.009748 | AC144530.1 | 1.69 | 1.03–2.78 | 0.039254 |
| MST1L | 1.66 | 1.13–2.44 | 0.009936 | AC004967.7 | 1.45 | 1.02–2.07 | 0.039533 |
| AC079781.5 | 1.39 | 1.08–1.8 | 0.010811 | NCF1B | 0.68 | 0.46–0.98 | 0.039578 |
| HSP90AB2P | 1.87 | 1.15–3.04 | 0.011429 | RP11_381E24.1 | 1.64 | 1.02–2.63 | 0.039904 |
| AC009474.2 | 1.45 | 1.09–1.93 | 0.01172 | RPS3AP6 | 1.46 | 1.02–2.08 | 0.040027 |
| CTB_75G16.1 | 1.76 | 1.13–2.74 | 0.012065 | RPL35P2 | 0.59 | 0.36–0.98 | 0.040168 |
| TMSB10P2 | 0.64 | 0.45–0.91 | 0.012102 | RPL4P4 | 2.04 | 1.03–4.04 | 0.040672 |
| RP3_342P20.2 | 1.88 | 1.15–3.08 | 0.012513 | HLA_H | 0.77 | 0.6–0.99 | 0.040894 |
| HLA_U | 0.6 | 0.41–0.9 | 0.012586 | RPS15AP12 | 1.76 | 1.02–3.04 | 0.041171 |
| RPL7P32 | 1.69 | 1.12–2.55 | 0.01278 | AC141586.5 | 1.57 | 1.02–2.44 | 0.041404 |
| RP11_302I18.1 | 1.96 | 1.15–3.32 | 0.013077 | EEF1A1P12 | 1.4 | 1.01–1.93 | 0.041698 |
| RP11_360D2.2 | 1.39 | 1.07–1.81 | 0.013836 | ANKRD36BP2 | 1.39 | 1.01–1.9 | 0.042281 |
| EEF1A1P1 | 1.63 | 1.1–2.41 | 0.014059 | U47924.6 | 0.67 | 0.45–0.99 | 0.042954 |
| RP11_20O24.1 | 1.49 | 1.08–2.05 | 0.014879 | RP11_372E1.1 | 1.72 | 1.02–2.89 | 0.042994 |
| RPS20P22 | 1.44 | 1.07–1.95 | 0.015777 | RSL24D1P6 | 0.56 | 0.32–0.98 | 0.043269 |
| RP11_583F2.6 | 1.5 | 1.08–2.08 | 0.016158 | RPS26P31 | 1.49 | 1.01–2.21 | 0.044184 |
| SMG1P1 | 1.57 | 1.09–2.26 | 0.016217 | GGNBP1 | 1.51 | 1.01–2.24 | 0.044241 |
| RP11_592N21.1 | 1.55 | 1.08–2.22 | 0.016333 | FAM195CP | 0.67 | 0.45–0.99 | 0.044479 |
| SPATA20P1 | 0.7 | 0.53–0.94 | 0.016488 | EEF1A1P9 | 1.32 | 1.01–1.74 | 0.044928 |
| RP11_501C14.7 | 0.55 | 0.34–0.9 | 0.016961 | CES5AP1 | 1.25 | 1–1.55 | 0.045574 |
| RPL13AP6 | 1.75 | 1.1–2.78 | 0.017922 | RP11_553P9.1 | 1.61 | 1.01–2.57 | 0.045933 |
| RPS10L | 1.65 | 1.09–2.49 | 0.018544 | RP11_43F13.1 | 1.53 | 1.01–2.34 | 0.046476 |
| CSAG4 | 0.65 | 0.45–0.93 | 0.019462 | CTD_2192J16.15 | 1.56 | 1.01–2.42 | 0.046513 |
| TOB2P1 | 1.56 | 1.07–2.27 | 0.019856 | CH507_42P11.2 | 0.67 | 0.45–0.99 | 0.046872 |
| RP11_344N17.15 | 0.6 | 0.39–0.92 | 0.020105 | ADCY10P1 | 1.5 | 1.01–2.25 | 0.047079 |
| MLLT10P1 | 0.66 | 0.47–0.94 | 0.020725 | RP11_761N21.2 | 1.33 | 1–1.76 | 0.04724 |
| ESPNP | 1.29 | 1.04–1.61 | 0.021376 | RP11_736N17.9 | 1.29 | 1–1.66 | 0.047554 |
| RPL7AP28 | 0.43 | 0.21–0.89 | 0.021683 | RP11_504P24.3 | 1.43 | 1–2.04 | 0.047587 |
| RP11_241F15.10 | 0.64 | 0.44–0.94 | 0.022711 | RPS26P5 | 1.35 | 1–1.81 | 0.04809 |
| AC011737.2 | 1.61 | 1.07–2.42 | 0.023174 | REXO1L1P | 1.45 | 1–2.1 | 0.048596 |
| RPS11P5 | 1.45 | 1–2.09 | 0.048755 |
| Characteristics | Hazard Ratio | CI95 | p-Value | Significance |
|---|---|---|---|---|
| RP11_551L14.1 | 0.63 | 0.4–0.99 | 0.044 | * |
| RPL7AP28 | 0.42 | 0.2–0.89 | 0.023 | * |
| SRSF9P1 | 0.68 | 0.3–1.55 | 0.36 | |
| ZC3H11B | 1.29 | 0.85–1.96 | 0.235 | |
| RP4_706A16.3 | 1.55 | 1–2.41 | 0.049 | * |
| AC079781.5 | 1.16 | 0.84–1.6 | 0.372 | |
| NBPF2P | 1.56 | 0.86–2.82 | 0.14 | |
| TMSB10P2 | 0.88 | 0.55–1.41 | 0.589 | |
| ZNF815P | 0.62 | 0.3–1.27 | 0.193 | |
| SLC25A24P1 | 1.35 | 0.8–2.3 | 0.264 | |
| CTA_963H5.5 | 0.71 | 0.37–1.37 | 0.311 | |
| RP11_241F15.10 | 0.64 | 0.4–1.01 | 0.057 | |
| RP11_23J18.1 | 0.91 | 0.46–1.78 | 0.773 | |
| RP11_326A19.5 | 0.51 | 0.32–0.8 | 0.004 | ** |
| RP11_344N17.15 | 0.72 | 0.44–1.18 | 0.193 |
| High Risk (47) | Low Risk (47) | Total (94) | |
|---|---|---|---|
| Gender | |||
| Female | 23 | 17 | 40 |
| Male | 24 | 30 | 54 |
| Age | |||
| Mean (SD) | 14.8 (5.27) | 15.13 (5.36) | 14.97 (5.29) |
| Median (min,max) | 14 (5, 39) | 15 (3, 32) | 14.5 (3, 39) |
| Metastatic status | |||
| Metastatic | 15 | 7 | 22 |
| Non-metastatic | 32 | 40 | 72 |
| Site | |||
| Leg/foot | 39 | 44 | 83 |
| Other | 8 | 3 | 11 |
| Risk score | |||
| Mean (SD) | 5.55 (10.15) | 0.47 (0.32) | 3.01 (7.58) |
| Median (min,max) | 2.53 (1.23, 66.4) | 0.5 (0.01, 1.21) | 1.23 (0.01, 66.4) |
| Status | |||
| Alive | 28 | 38 | 66 |
| Dead | 19 | 9 | 28 |
| Total PG | Studied PG | Survival-Related PG | Signature-PG |
|---|---|---|---|
| 14,126 | 1333 | 125 | 4 |
| 3Y-AUC | 5Y-AUC | 8Y-AUC | KM-P-value |
| Whole patients | |||
| 0.885 | 0.878 | 0.796 | <0.0001 |
| Gender subgroup (male) | |||
| 0.82 | 0.828 | 0.707 | 0.0064 |
| Gender subgroup (female) | |||
| 0.965 | 0.947 | 0.947 | <0.0001 |
| Age subgroup (<18) | |||
| 0.902 | 0.888 | 0.788 | <0.0001 |
| Age subgroup (≥18) | |||
| 0.828 | 0.861 | 0.796 | 0.038 |
| Metastatic subgroup (non-meta) | |||
| 0.871 | 0.87 | 0.743 | 0.00014 |
| Metastatic subgroup (meta) | |||
| 0.91 | 0.89 | 0.89 | <0.0001 |
| Gene signature | |||
| 0.821 | 0.861 | 0.781 | <0.0001 |
| Combined signature | |||
| 0.937 | 0.956 | 0.871 | <0.0001 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, F.; Xing, L.; Zhang, X.; Zhang, X. A Four-Pseudogene Classifier Identified by Machine Learning Serves as a Novel Prognostic Marker for Survival of Osteosarcoma. Genes 2019, 10, 414. https://doi.org/10.3390/genes10060414
Liu F, Xing L, Zhang X, Zhang X. A Four-Pseudogene Classifier Identified by Machine Learning Serves as a Novel Prognostic Marker for Survival of Osteosarcoma. Genes. 2019; 10(6):414. https://doi.org/10.3390/genes10060414
Chicago/Turabian StyleLiu, Feng, Lu Xing, Xiaoqian Zhang, and Xiaoqi Zhang. 2019. "A Four-Pseudogene Classifier Identified by Machine Learning Serves as a Novel Prognostic Marker for Survival of Osteosarcoma" Genes 10, no. 6: 414. https://doi.org/10.3390/genes10060414
APA StyleLiu, F., Xing, L., Zhang, X., & Zhang, X. (2019). A Four-Pseudogene Classifier Identified by Machine Learning Serves as a Novel Prognostic Marker for Survival of Osteosarcoma. Genes, 10(6), 414. https://doi.org/10.3390/genes10060414





