Role of Autophagy-Related Gene atg22 in Developmental Process and Virulence of Fusarium oxysporum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Fungus and Culture Conditions
2.2. Generation of Deletion Mutants
2.3. Overexpression of Foatg22 Mutant Strains
2.4. Fungal Transformation
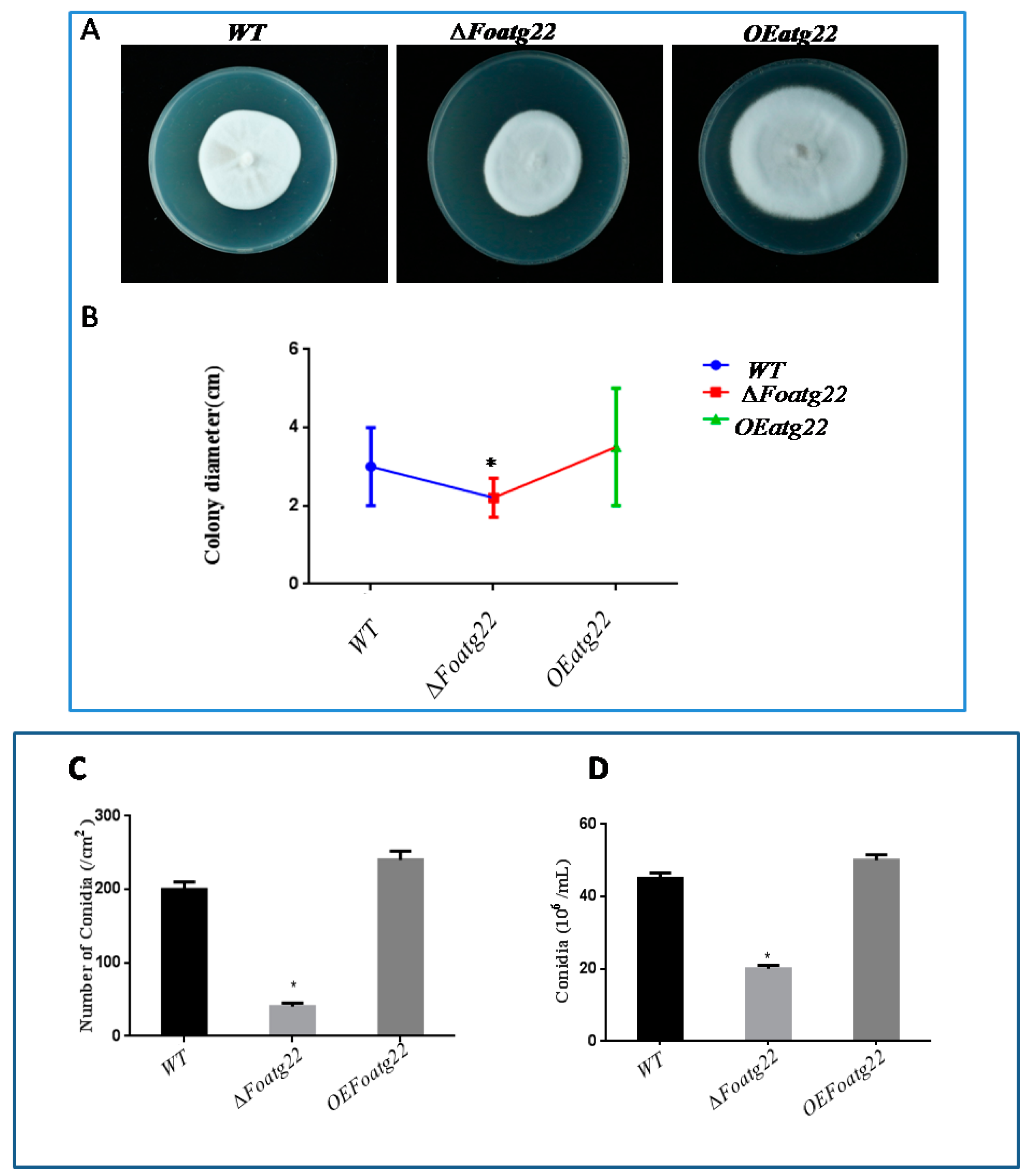
2.5. Evaluation of Radial Growth, Conidiation, Formation, and Germination
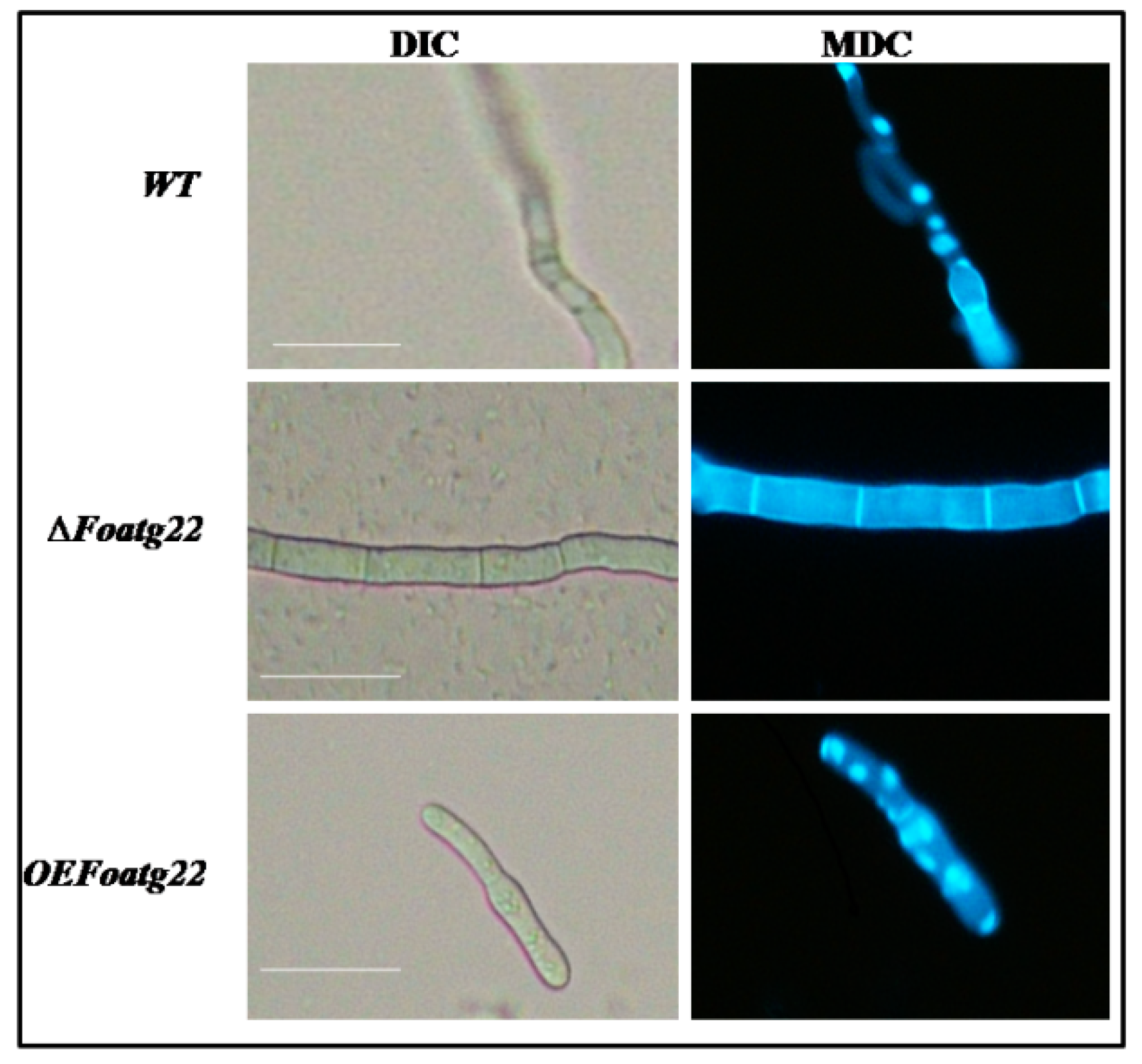
2.6. Analysis of Autophagy
2.7. Analysis of Gene Expression
2.8. Pathogenicity Test
2.9. Optical and Epifluorescence Microscopy
3. Results
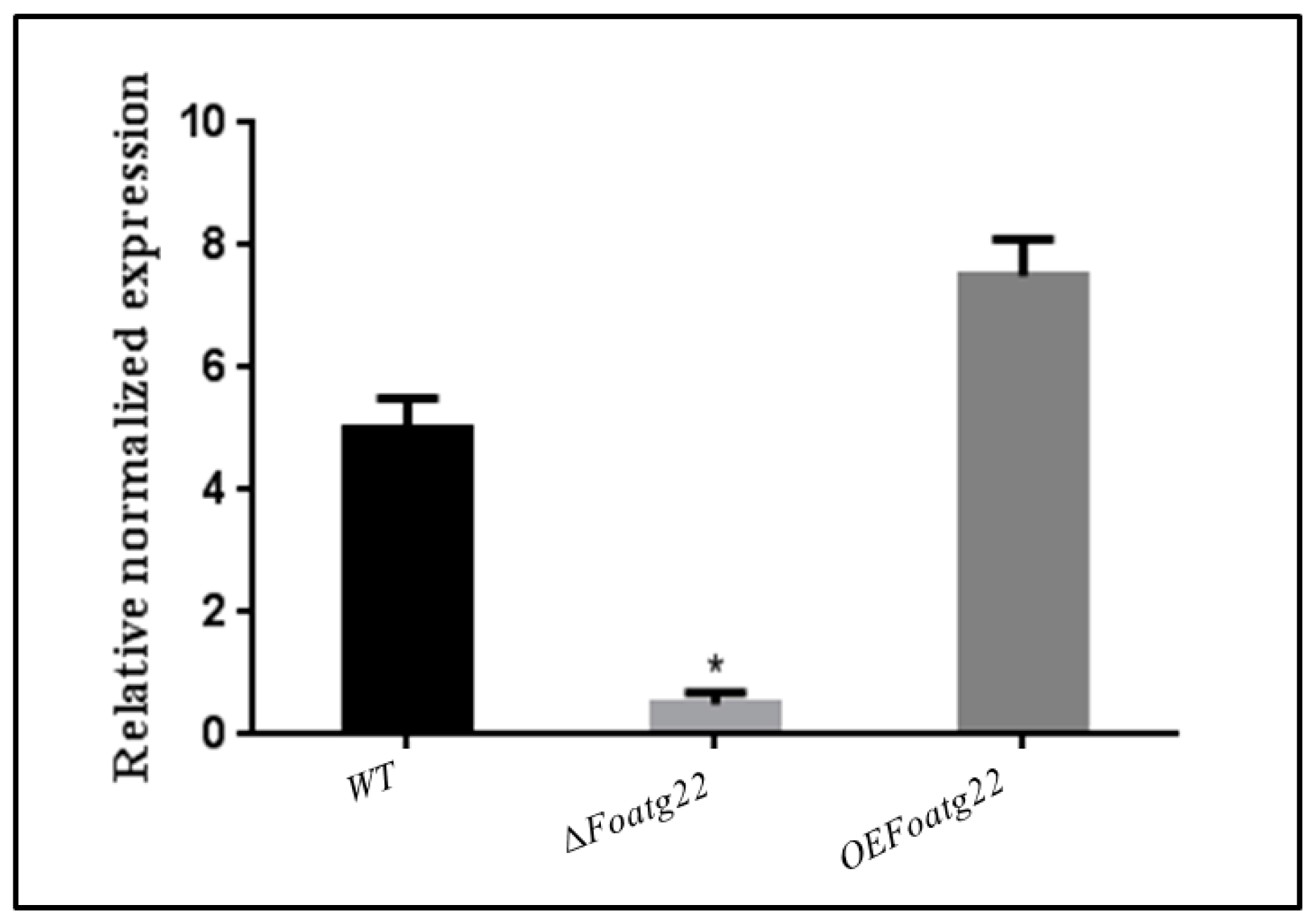
3.1. Deletion and Overexpression Mutants of Foatg22 in F. oxysporum
3.2. Role of ATG22 in Hyphal Formation
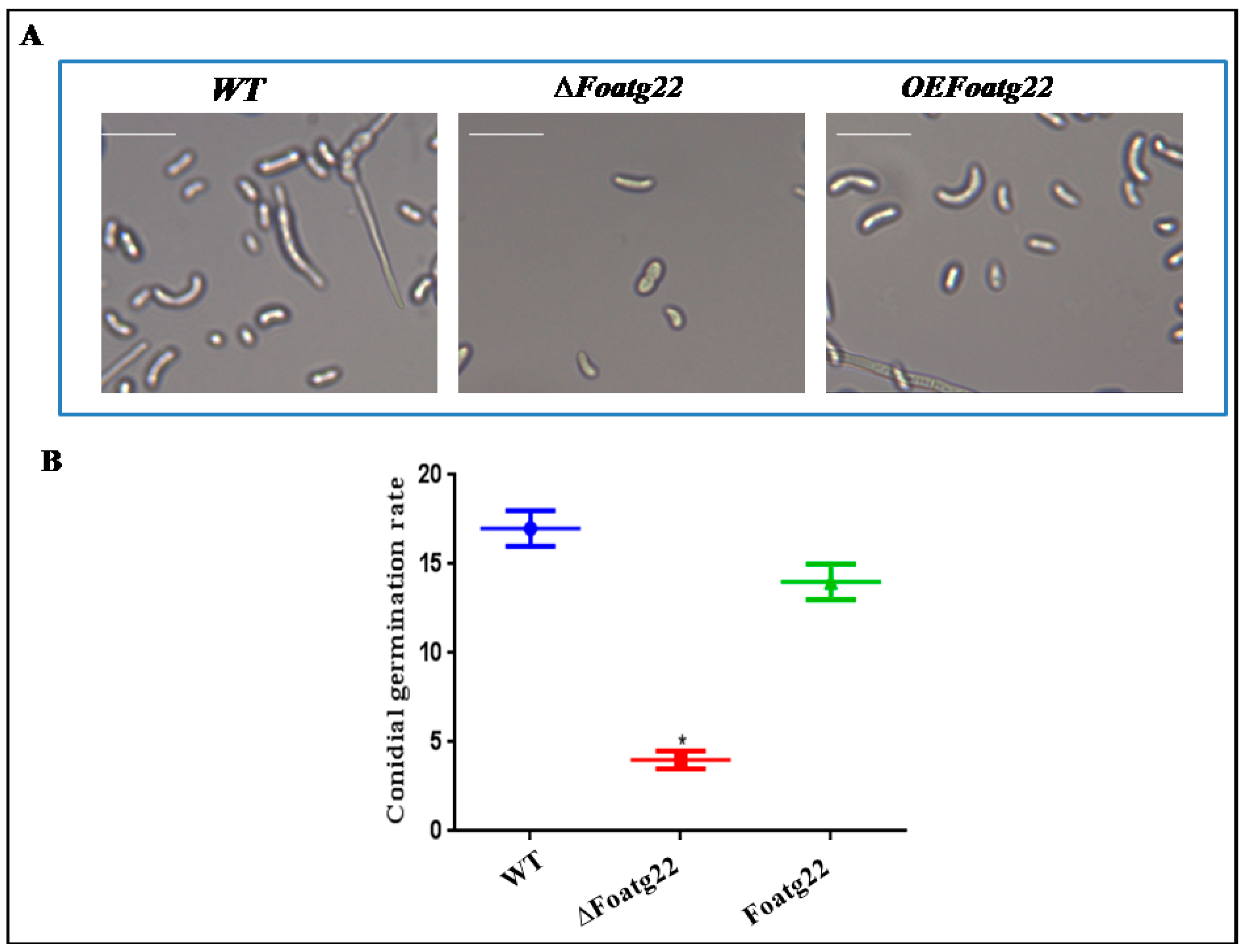
3.3. Autophagy Contributes toMycelial Growth and Conidiation
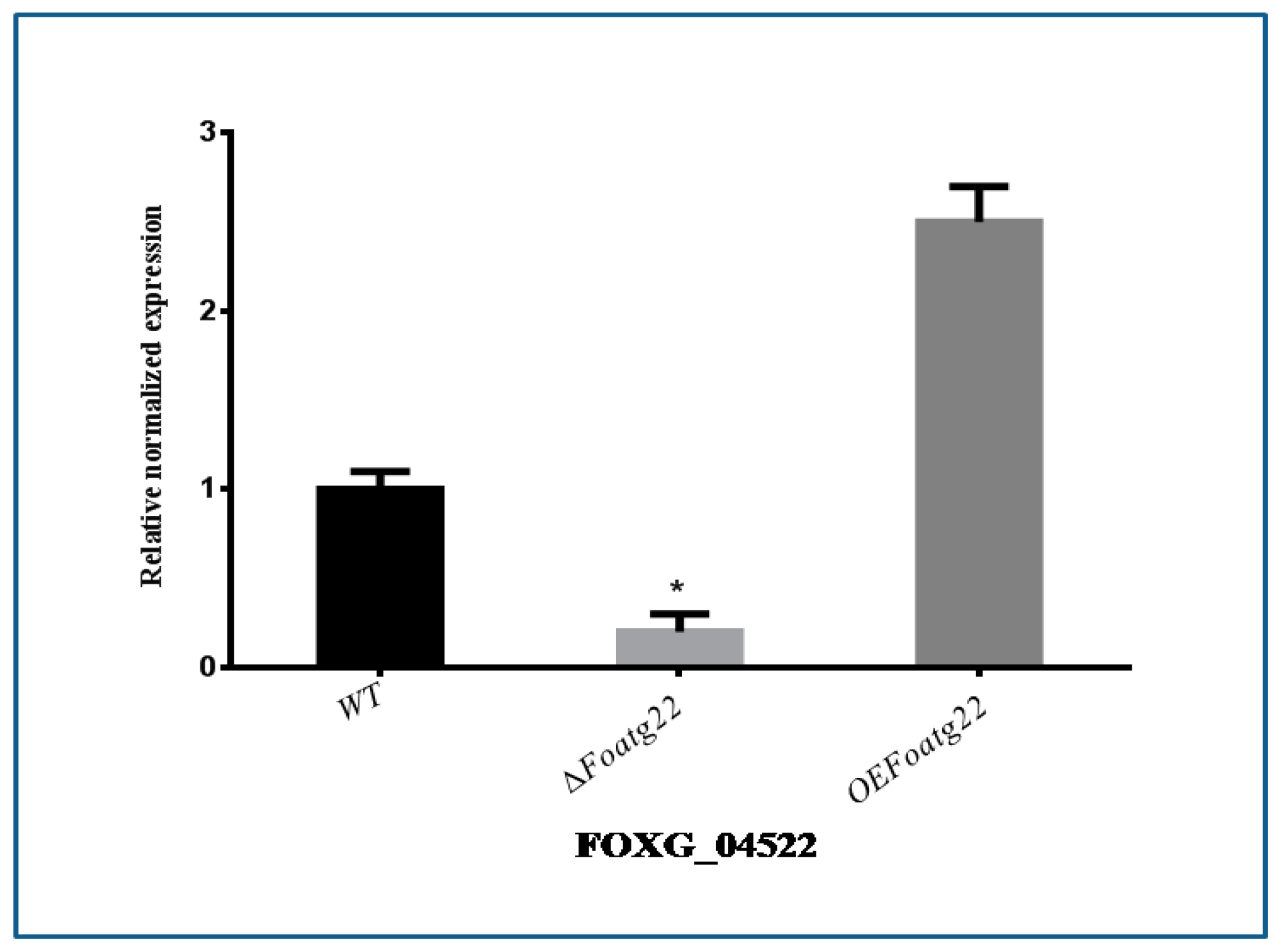
3.4. Expression of Vegetative Growth-Related Gene
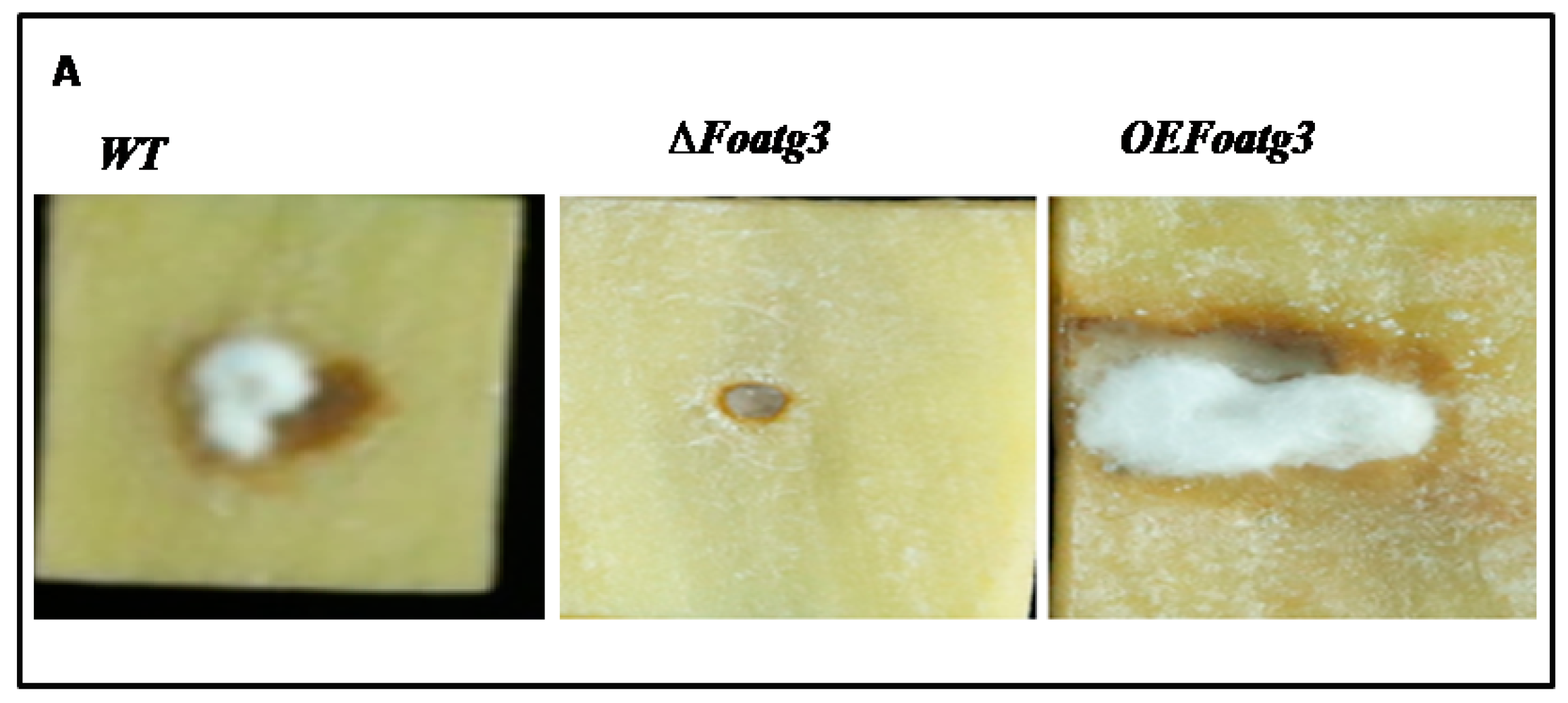
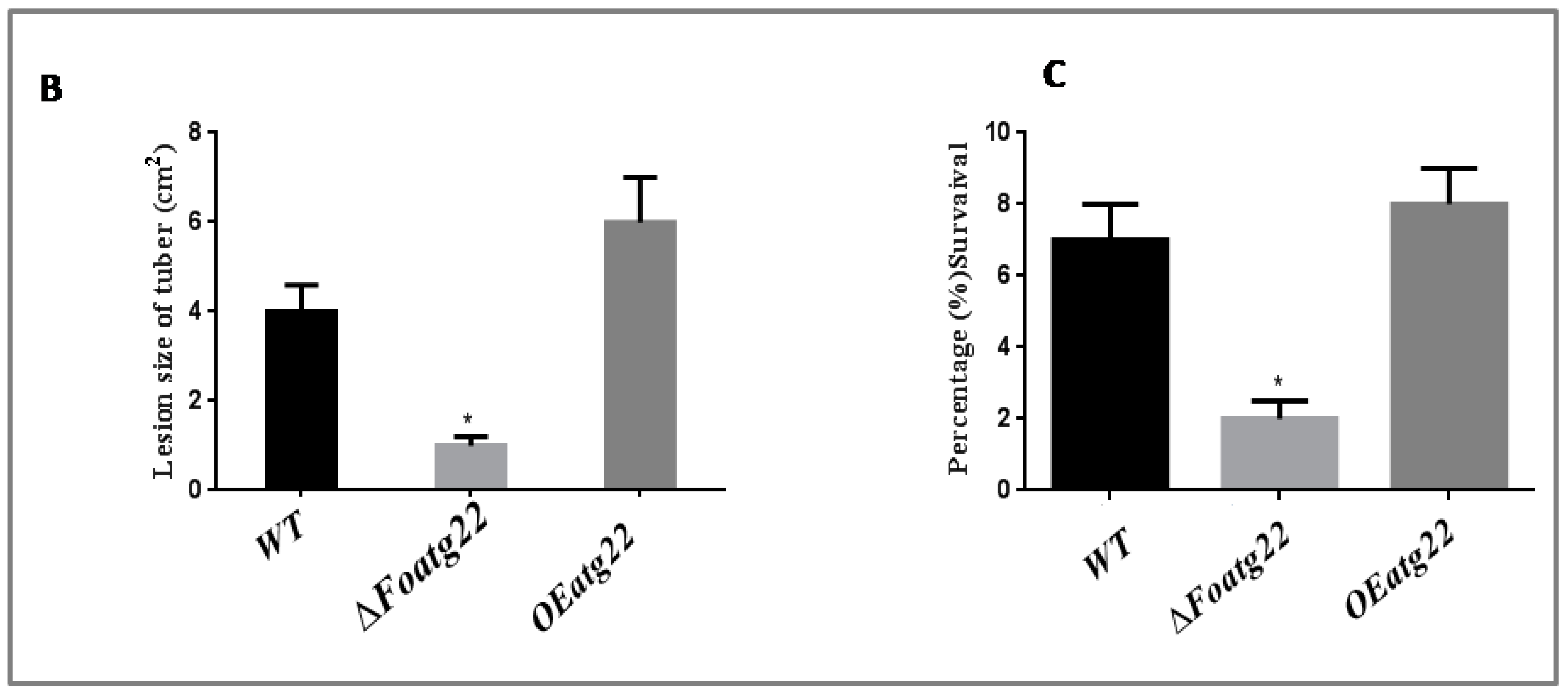
3.5. Plant Infection Assay
4. Discussion
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| ATG | Autophagy-related |
| WT | Wild type |
| OE | Overexpression |
| ∆ | Deletion mutant |
| MDC | Monodansylcadaverine |
| DIC | Differential interference contrast |
References
- El-Kassas, H.Y.; Khairy, H.M. A trial for biological control of a pathogenic fungus (Fusarium solani) by some marine microorganisms. Am. Eurasian. J. Agric. Environ. Sci. 2009, 3, 434–440. [Google Scholar]
- Wharton, P.K.W. Fusarium Dry Rot. Available online: http://www.Potatodiseases.Org/contact.html (accessed on 23 May 2007).
- Wharton, P.S.; Kirk, W.W.; Berry, D.; Tumbalam, P. Seed treatment application-timing options for control of Fusarium decay and sprout rot of cut seedpieces. Am. J. Potato Res. 2007, 3, 237–244. [Google Scholar] [CrossRef]
- Chełkowski, J. Toxinogenicity of Fusarium Species Causing Dry rot of Potato Tubers —Fusarium— Chapter 25; Elsevier B.V.: New York, NY, USA, 1989; pp. 435–440. [Google Scholar]
- Slininger, P.J.; Burkhead, K.D.; Schisler, D.A. Antifungal and sprout regulatory bioactivities of phenylacetic acid, indole-3-acetic acid, and tyrosol isolated from the potato dry rot suppressive bacterium Enterobacter cloacae S11 : T : 07. J. Ind. Microbiol. Biotechnol. 2004, 11, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Chełkowski, J. Chapter 25 — Toxinogenicity of fusarium species causing dry rot of potato tubers. In Fusarium; Chełkowski, J., Ed.; Elsevier: Amsterdam, The Netherlands, 1989; Volume 2, pp. 435–440. [Google Scholar]
- Secor, G.A.; Gudmestad, N.C. Managing fungal diseases of potato. Can. J. Plant Pathol. 1999, 3, 213–221. [Google Scholar] [CrossRef]
- Shattock, R. Compendium of Potato Diseases, Second Edition. W.R. Stevenson. Plant Pathol. 2002, 4. [Google Scholar] [CrossRef]
- Secor, G.; Salas, B. Fusarium dry rot and Fusarium wilt. Compend. Potato Dis. 2001, 23–25. [Google Scholar]
- Santhanam, P.; van Esse, H.P.; Albert, I.; Faino, L.; Nurnberger, T.; Thomma, B.P.H.J. Evidence for functional diversification within a fungal nep1-like protein family. Mol. Plant Microbe Interact. 2013, 3, 278–286. [Google Scholar] [CrossRef]
- Corral-Ramos, C.; Roca, M.G.; Di Pietro, A.; Roncero, M.I.G.; Ruiz-Roldan, C. Autophagy contributes to regulation of nuclear dynamics during vegetative growth and hyphal fusion in Fusarium oxysporum. Autophagy 2015, 1, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.L.; Li, M.; Suo, F.; Liu, X.M.; Shen, E.Z.; Yang, B.; Dong, M.Q.; He, W.Z.; Du, L.L. Global analysis of fission yeast mating genes reveals new autophagy factors. PLoS Genet. 2013, 8, 244–258. [Google Scholar]
- Chardwiriyapreecha, S.; Mukaiyama, H.; Sekito, T.; Iwaki, T.; Takegawa, K.; Kakinuma, Y. Avt5p is required for vacuolar uptake of amino acids in the fission yeast Schizosaccharomyces pombe. FEBS Lett. 2010, 11, 2339–2345. [Google Scholar] [CrossRef] [PubMed]
- Suriapranata, I.; Epple, U.D.; Bernreuther, D.; Bredschneider, M.; Sovarasteanu, K.; Thumm, M. The breakdown of autophagic vesicles inside the vacuole depends on Aut4p. J. Cell Sci. 2000, 22, 4025–4033. [Google Scholar]
- Veneault-Fourrey, C.; Barooah, M.; Egan, M.; Wakley, G.; Talbot, N.J. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science 2006, 5773, 580–583. [Google Scholar] [CrossRef]
- Asakura, M.; Ninomiya, S.; Oku, M.; Okuno, T.; Sakai, Y.; Takano, Y. Atg26-mediated pexophagy is required for host invasion by the plant pathogenic fungus Colletotrichum orbiculare. Autophagy 2009, 6. [Google Scholar]
- Dong, B.; Liu, X.H.; Lu, J.P.; Zhang, F.S.; Gao, H.M.; Wang, H.K.; Lin, F.C. MgAtg9 trafficking in Magnaportheoryzae. Autophagy 2009, 7, 946–953. [Google Scholar] [CrossRef]
- Lu, J.P.; Liu, X.H.; Feng, X.X.; Min, H.; Lin, F.C. An autophagy gene, MgATG5, is required for cell differentiation and pathogenesis in Magnaportheoryzae. Curr. Genet. 2009, 4, 461–473. [Google Scholar] [CrossRef]
- Nadal, M.; Gold, S.E. The autophagy genes atg8 and atg1 affect morphogenesis and pathogenicity in Ustilago maydis. Mol. Plant Pathol. 2010, 4, 463–478. [Google Scholar] [CrossRef] [PubMed]
- Lv, W.; Wang, C.; Yang, N.; Que, Y.; Talbot, N.J.; Wang, Z. Genome-wide functional analysis reveals that autophagy is necessary for growth, sporulation, deoxynivalenol production and virulence in Fusarium graminearum. Sci. Rep. 2017, 1, 110–125. [Google Scholar] [CrossRef]
- Liu, X.H.; Lu, J.P.; Zhang, L.; Dong, B.; Min, H.; Lin, F.C. Involvement of a magnaporthegrisea serine/threonine kinase gene, mgatg1, in appressorium turgor and pathogenesis. Eukaryot. Cell 2007, 6, 997–1005. [Google Scholar] [CrossRef]
- Liu, T.B.; Liu, X.H.; Lu, J.P.; Zhang, L.; Min, H.; Lin, F.C. The cysteine protease MoAtg4 interacts with MoAtg8 and is required for differentiation and pathogenesis in Magnaportheoryzae. Autophagy 2010, 1, 74–85. [Google Scholar] [CrossRef]
- Kershaw, M.J.; Talbot, N.J. Genome-wide functional analysis reveals that infection-associated fungal autophagy is necessary for rice blast disease. Proc. Natl. Acad. Sci. USA 2009, 37, 15967–15972. [Google Scholar] [CrossRef]
- Rauyaree, P.; Ospina-Giraldo, M.D.; Kang, S.; Bhat, R.G.; Subbarao, K.V.; Grant, S.J.; Dobinson, K.F. Mutations in VMK1, a mitogen-activated protein kinase gene, affect microsclerotia formation and pathogenicity in Verticillium dahliae. Curr. Genet. 2005, 2, 109–116. [Google Scholar] [CrossRef]
- Tzima, A.; Paplomatas, E.J.; Rauyaree, P.; Kang, S. Roles of the catalytic subunit of cAMP-dependent protein kinase A in virulence and development of the soilborne plant pathogen Verticillium dahliae. Fungal Genet. Biol. 2010, 5, 406–415. [Google Scholar] [CrossRef]
- Tzima, A.K.; Paplomatas, E.J.; Rauyaree, P.; Ospina-Giraldo, M.D.; Kang, S. VdSNF1, the Sucrose nonfermenting protein kinase gene of Verticillium dahliae, is required for virulence and expression of genes involved in cell-wall degradation. Mol. Plant Microbe Interact. 2011, 1, 129–142. [Google Scholar] [CrossRef] [PubMed]
- Tzima, A.K.; Paplomatas, E.J.; Tsitsigiannis, D.I.; Kang, S. The G protein β subunit controls virulence and multiple growth- and development-related traits in Verticillium dahliae. Fungal Genet. Biol. 2012, 4, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Szewczyk, E.; Nayak, T.; Oakley, C.E.; Edgerton, H.; Xiong, Y.; Taheri-Talesh, N.; Osmani, S.A.; Oakley, B.R. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 2006, 6, 3111–3120. [Google Scholar] [CrossRef]
- Maruthachalam, K.; Klosterman, S.J.; Kang, S.; Hayes, R.J.; Subbarao, K.V. Identification of pathogenicity-related genes in the vascular wilt fungus Verticillium dahliae by Agrobacterium tumefaciens-mediated t-dna insertional mutagenesis. Mol. Biotechnol. 2011, 3, 209–221. [Google Scholar] [CrossRef]
- Di Pietro, A.; Roncero, M.I.G. Cloning, expression, and role in pathogenicity of pg1 encoding the major extracellular endopolygalacturonase of the vascular wilt pathogen Fusarium oxysporum. Mol. Plant Microbe Interact. 1998, 2, 91–98. [Google Scholar] [CrossRef]
- Biederbick, A.; Kern, H.F.; Elsasser, H.P. Monodansylcadaverine (Mdc) is a specific in-vivo marker for autophagic vacuoles. Eur. J. Cell Biol. 1995, 1, 3–14. [Google Scholar]
- Bartoszewska, M.; Kiel, J.A.K.W. The role of macroautophagy in development of filamentous fungi. Antioxid. Redox Sign. 2011, 11, 2271–2287. [Google Scholar] [CrossRef]
- Josefsen, L.; Droce, A.; Sondergaard, T.E.; Sorensen, J.L.; Bormann, J.; Schafer, W.; Giese, H.; Olsson, S. Autophagy provides nutrients for nonassimilating fungal structures and is necessary for plant colonization but not for infection in the necrotrophic plant pathogen Fusarium graminearum. Autophagy 2012, 3, 326–337. [Google Scholar] [CrossRef] [PubMed]
- Kikuma, T.; Ohneda, M.; Arioka, M.; Kitamoto, K. Functional analysis of the ATG8 homologue Aoatg8 and role of autophagy in differentiation and germination in Aspergillus oryzae. Eukaryot. Cell 2006, 8, 1328–1336. [Google Scholar] [CrossRef] [PubMed]
- Duan, Z.B.; Chen, Y.X.; Huang, W.; Shang, Y.F.; Chen, P.L.; Wang, C.S. Linkage of autophagy to fungal development, lipid storage and virulence in Metarhiziumrobertsii. Autophagy 2013, 4, 538–549. [Google Scholar] [CrossRef]
- Nitsche, B.M.; Burggraaf-van Welzen, A.M.; Lamers, G.; Meyer, V.; Ram, A.F.J. Autophagy promotes survival in aging submerged cultures of the filamentous fungus Aspergillus niger. Appl. Microbiol. Biotechnol. 2013, 18, 8205–8218. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Roldan, M.C.; Kohli, M.; Roncero, M.I.G.; Philippsen, P.; Di Pietro, A.; Espeso, E.A. Nuclear Dynamics during Germination, Conidiation, and Hyphal Fusion of Fusarium oxysporum. Eukaryot. Cell 2010, 8, 1216–1224. [Google Scholar] [CrossRef] [PubMed]
- Richie, D.L.; Fuller, K.K.; Fortwendel, J.; Miley, M.D.; McCarthy, J.W.; Feldmesser, M.; Rhodes, J.C.; Askew, D.S. Unexpected link between metal ion deficiency and autophagy in Aspergillus fumigatus. Eukaryot. Cell 2007, 12, 2437–2447. [Google Scholar] [CrossRef]
- Shoji, J.Y.; Arioka, M.; Kitamoto, K. Possible involvement of pleiomorphic vacuolar networks in nutrient recycling in filamentous fungi. Autophagy 2006, 3, 226–227. [Google Scholar] [CrossRef]
- Luo, X.M.; Mao, H.Q.; Wei, Y.M.; Cai, J.; Xie, C.J.; Sui, A.P.; Yang, X.Y.; Dong, J.Y. The fungal-specific transcription factor Vdpf influences conidia production, melanized microsclerotia formation and pathogenicity in Verticillium dahliae. Mol. Plant Pathol. 2016, 9, 1364–1381. [Google Scholar]
- Zhou, L.; Zhao, J.; Guo, W.Z.; Zhang, T.Z. Functional Analysis of Autophagy Genes via Agrobacterium-Mediated Transformation in the Vascular Wilt Fungus Verticillium dahliae. J. Genet. Genom. 2013, 8, 421–431. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khalid, A.R.; Zhang, S.; Luo, X.; Mehmood, K.; Rahim, J.; Shaheen, H.; Dong, P.; Qiu, D.; Ren, M. Role of Autophagy-Related Gene atg22 in Developmental Process and Virulence of Fusarium oxysporum. Genes 2019, 10, 365. https://doi.org/10.3390/genes10050365
Khalid AR, Zhang S, Luo X, Mehmood K, Rahim J, Shaheen H, Dong P, Qiu D, Ren M. Role of Autophagy-Related Gene atg22 in Developmental Process and Virulence of Fusarium oxysporum. Genes. 2019; 10(5):365. https://doi.org/10.3390/genes10050365
Chicago/Turabian StyleKhalid, A. Rehman, Shumin Zhang, Xiumei Luo, Khalid Mehmood, Junaid Rahim, Hamayun Shaheen, Pan Dong, Dan Qiu, and Maozhi Ren. 2019. "Role of Autophagy-Related Gene atg22 in Developmental Process and Virulence of Fusarium oxysporum" Genes 10, no. 5: 365. https://doi.org/10.3390/genes10050365
APA StyleKhalid, A. R., Zhang, S., Luo, X., Mehmood, K., Rahim, J., Shaheen, H., Dong, P., Qiu, D., & Ren, M. (2019). Role of Autophagy-Related Gene atg22 in Developmental Process and Virulence of Fusarium oxysporum. Genes, 10(5), 365. https://doi.org/10.3390/genes10050365



