Integrative rDNAomics—Importance of the Oldest Repetitive Fraction of the Eukaryote Genome
Abstract
1. The Eukaryotic rDNAome
2. The Multifaceted Nucleolus
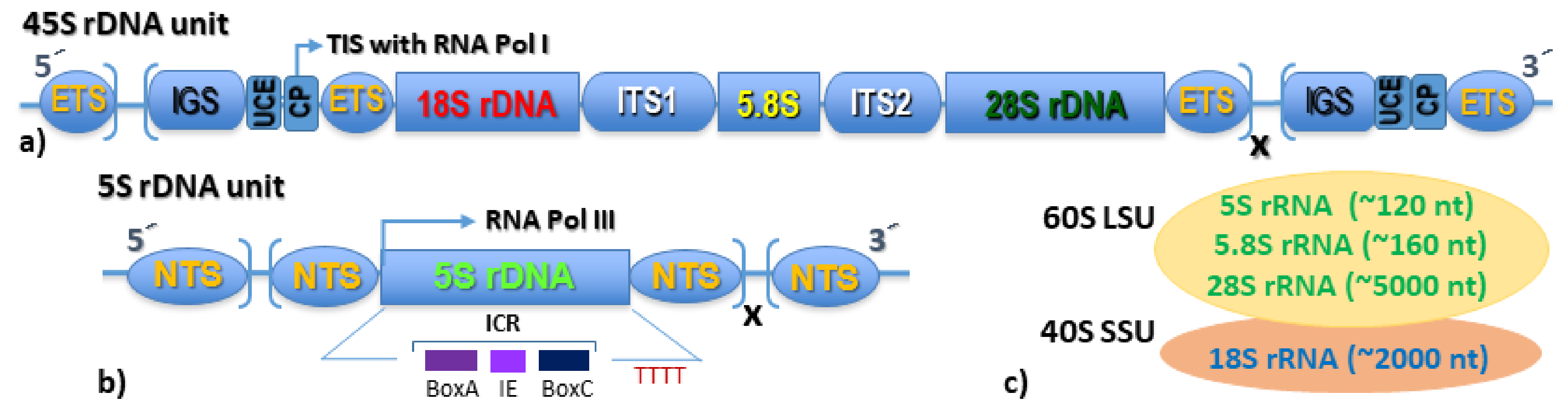
3. The Nucleolus Forming 45S rDNA
4. The Extra-Nucleolar 5S rDNA
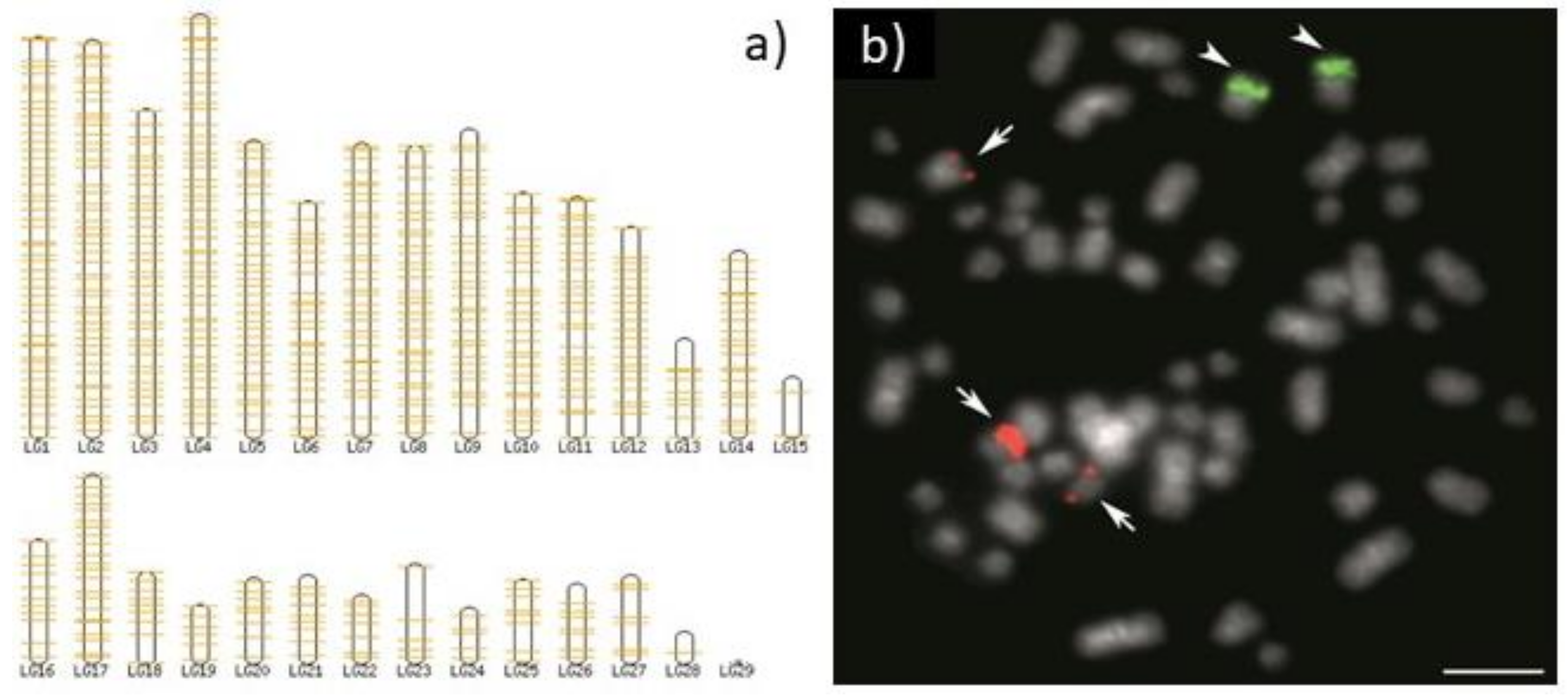
5. Copy Number Really Matters
6. Overview of Important Facts about rDNA
7. GC Content of rDNA
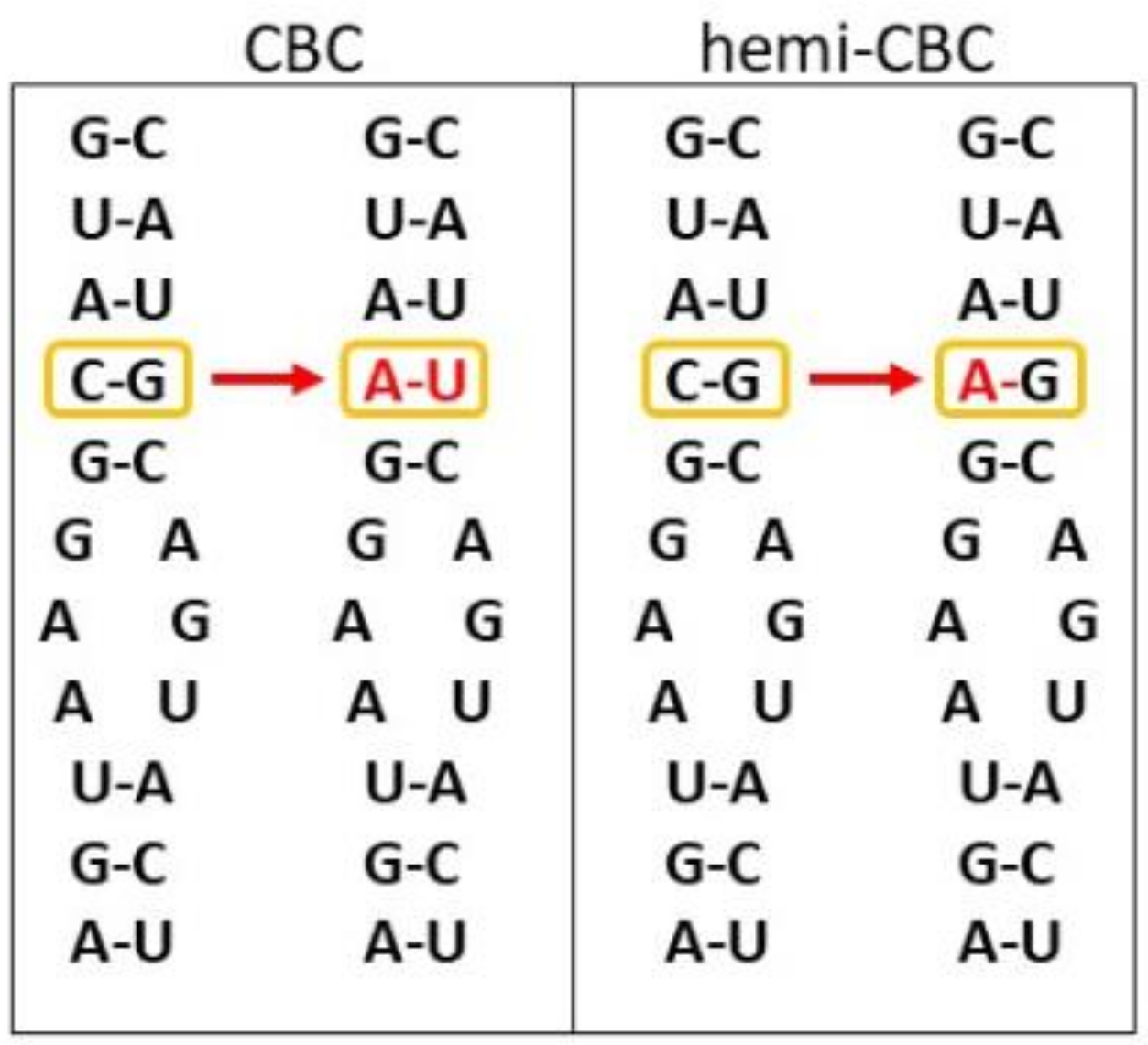
8. Phylogeny, Species Delimitation, and Secondary Structure of rRNAs—The Way How to Determine in Silico Whether Two Lineages Can Successfully Cross
9. Concluding Remarks
Supplementary Materials
Funding
Acknowledgments
Conflicts of Interest
References
- Bernhardt, H.S.; Tate, W.P. A Ribosome Without RNA. Front. Ecol. Evol. 2015, 3, 1–6. [Google Scholar] [CrossRef]
- Boisvert, F.-M.; van Koningsbruggen, S.; Navascués, J.; Lamond, A.I. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007, 8, 574–585. [Google Scholar] [CrossRef] [PubMed]
- Porokhovnik, L.; Gerton, J.L. Ribosomal DNA-connecting ribosome biogenesis and chromosome biology. Chromosome Res. Int. J. Mol. Supramol. Evol. Asp. Chromosome Biol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Grummt, I. The nucleolus—guardian of cellular homeostasis and genome integrity. Chromosoma 2013, 122, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Sochorová, J.; Garcia, S.; Gálvez, F.; Symonová, R.; Kovařík, A. Evolutionary trends in animal ribosomal DNA loci: introduction to a new online database. Chromosoma 2018, 127, 141–150. [Google Scholar] [CrossRef]
- James, S.A.; O’Kelly, M.J.T.; Carter, D.M.; Davey, R.P.; van Oudenaarden, A.; Roberts, I.N. Repetitive sequence variation and dynamics in the ribosomal DNA array of Saccharomyces cerevisiae as revealed by whole-genome resequencing. Genome Res. 2009, 19, 626–635. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T. Ribosomal RNA gene repeats, their stability and cellular senescence. Proc. Jpn. Acad. Ser. B 2014, 90, 119–129. [Google Scholar] [CrossRef]
- Russell, J.; Zomerdijk, J.C.B.M. RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem. Sci. 2005, 30, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Santoro, R.; Schmitz, K.-M.; Sandoval, J.; Grummt, I. Intergenic transcripts originating from a subclass of ribosomal DNA repeats silence ribosomal RNA genes in trans. EMBO Rep. 2010, 11, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Grimaldi, G.; Di Nocera, P.P. Multiple repeated units in Drosophila melanogaster ribosomal DNA spacer stimulate rRNA precursor transcription. Proc. Natl. Acad. Sci. USA 1988, 85, 5502–5506. [Google Scholar] [CrossRef]
- Mayer, C.; Neubert, M.; Grummt, I. The structure of NoRC-associated RNA is crucial for targeting the chromatin remodelling complex NoRC to the nucleolus. EMBO Rep. 2008, 9, 774–780. [Google Scholar] [CrossRef]
- Németh, A.; Längst, G. Genome organization in and around the nucleolus. Trends Genet. TIG 2011, 27, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Howell, W.M.; Black, D.A. Controlled silver-staining of nucleolus organizer regions with a protective colloidal developer: A 1-step method. Experientia 1980, 36, 1014–1015. [Google Scholar] [CrossRef] [PubMed]
- Thiry, M.; Lafontaine, D.L.J. Birth of a nucleolus: The evolution of nucleolar compartments. Trends Cell Biol. 2005, 15, 194–199. [Google Scholar] [CrossRef]
- Thiry, M.; Lamaye, F.; Lafontaine, D.L.J. The nucleolus: When 2 became 3. Nucleus 2011, 2, 289–293. [Google Scholar] [CrossRef]
- Henras, A.K.; Plisson-Chastang, C.; O’Donohue, M.-F.; Chakraborty, A.; Gleizes, P.-E. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip. Rev. RNA 2015, 6, 225–242. [Google Scholar] [CrossRef] [PubMed]
- Garcia, S.; Garnatje, T.; Kovařík, A. Plant rDNA database: Ribosomal DNA loci information goes online. Chromosoma 2012, 121, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Sasaki, M. Ribosomal DNA stability is supported by many ‘buffer genes’—introduction to the Yeast rDNA Stability Database. FEMS Yeast Res. 2017, 17. [Google Scholar] [CrossRef]
- Pikaard, C.S. Nucleolar dominance: Uniparental gene silencing on a multi-megabase scale in genetic hybrids. Plant Mol. Biol. 2000, 43, 163–177. [Google Scholar] [CrossRef] [PubMed]
- Michalak, K.; Maciak, S.; Kim, Y.B.; Santopietro, G.; Oh, J.H.; Kang, L.; Garner, H.R.; Michalak, P. Nucleolar dominance and maternal control of 45S rDNA expression. Proc. R. Soc. B Biol. Sci. 2015, 282, 20152201. [Google Scholar] [CrossRef]
- Maciak, S.; Michalak, K.; Kale, S.D.; Michalak, P. Nucleolar Dominance and Repression of 45S Ribosomal RNA Genes in Hybrids between Xenopus borealis and X. muelleri (2n = 36). Cytogenet. Genome Res. 2016, 149, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Hu, F.; Luo, K.; Li, W.; Liu, S. Unique nucleolar dominance patterns in distant hybrid lineage derived from Megalobrama Amblycephala × Culter Alburnus. BMC Genet. 2016, 17. [Google Scholar] [CrossRef]
- Cao, L.; Qin, Q.; Xiao, Q.; Yin, H.; Wen, J.; Liu, Q.; Huang, X.; Huo, Y.; Tao, M.; Zhang, C.; et al. Nucleolar Dominance in a Tetraploidy Hybrid Lineage Derived From Carassius auratus red var. () × Megalobrama amblycephala (). Front. Genet. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Onishi, T.; Berglund, C.; Reeder, R.H. On the mechanism of nucleolar dominance in mouse-human somatic cell hybrids. Proc. Natl. Acad. Sci. USA 1984, 81, 484–487. [Google Scholar] [CrossRef] [PubMed]
- Greil, F.; Ahmad, K. Nucleolar Dominance of the Y Chromosome in Drosophila melanogaster. Genetics 2012, 191, 1119–1128. [Google Scholar] [CrossRef] [PubMed]
- Tautz, D.; Hancock, J.M.; Webb, D.A.; Tautz, C.; Dover, G.A. Complete sequences of the rRNA genes of Drosophila melanogaster. Mol. Biol. Evol. 1988, 5, 366–376. [Google Scholar] [PubMed]
- Symonová, R.; Majtánová, Z.; Arias-Rodriguez, L.; Mořkovský, L.; Kořínková, T.; Cavin, L.; Pokorná, M.J.; Doležálková, M.; Flajšhans, M.; Normandeau, E.; et al. Genome Compositional Organization in Gars Shows More Similarities to Mammals than to Other Ray-Finned Fish: CYTOGENOMICS OF GARS. J. Exp. Zoolog. B Mol. Dev. Evol. 2017, 328, 607–619. [Google Scholar] [CrossRef]
- Castro, S.I.; Hleap, J.S.; Cárdenas, H.; Blouin, C. Molecular organization of the 5S rDNA gene type II in elasmobranchs. RNA Biol. 2016, 13, 391–399. [Google Scholar] [CrossRef][Green Version]
- Szymanski, M.; Barciszewska, M.Z.; Erdmann, V.A.; Barciszewski, J. 5S Ribosomal RNA Database. Nucleic Acids Res. 2002, 30, 176–178. [Google Scholar] [CrossRef] [PubMed]
- Dinman, J.D. 5S rRNA: Structure and Function from Head to Toe. Int. J. Biomed. Sci. IJBS 2005, 1, 2–7. [Google Scholar]
- Locati, M.D.; Pagano, J.F.B.; Ensink, W.A.; van Olst, M.; van Leeuwen, S.; Nehrdich, U.; Zhu, K.; Spaink, H.P.; Girard, G.; Rauwerda, H.; et al. Linking maternal and somatic 5S rRNA types with different sequence-specific non-LTR retrotransposons. RNA 2017, 23, 446–456. [Google Scholar] [CrossRef]
- Peterson, R.C.; Doering, J.L.; Brown, D.D. Characterization of two xenopus somatic 5S DNAs and one minor oocyte-specific 5S DNA. Cell 1980, 20, 131–141. [Google Scholar] [CrossRef]
- Tomaszewski, R.; Jerzmanowski, A. The AT-rich flanks of the oocyte-type 5S RNA gene of Xenopus laevis act as a strong local signal for histone H1-mediated chromatin reorganization in vitro. Nucleic Acids Res. 1997, 25, 458–466. [Google Scholar] [CrossRef] [PubMed]
- Wolffe, A.P.; Brown, D.D. Developmental regulation of two 5S ribosomal RNA genes. Science 1988, 241, 1626–1632. [Google Scholar] [CrossRef]
- Martins, C.; Galetti, P.M. Two 5S rDNA arrays in neotropical fish species: is it a general rule for fishes? Genetica 2001, 111, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Fish Cytogenetics; Pisano, E., Ed.; Science Publishers: Enfield, NH, USA, 2007; ISBN 978-1-57808-330-5. [Google Scholar]
- Dimarco, E.; Cascone, E.; Bellavia, D.; Caradonna, F. Functional variants of 5S rRNA in the ribosomes of common sea urchin Paracentrotus lividus. Gene 2012, 508, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Vierna, J.; Wehner, S.; Höner zu Siederdissen, C.; Martínez-Lage, A.; Marz, M. Systematic analysis and evolution of 5S ribosomal DNA in metazoans. Heredity 2013, 111, 410–421. [Google Scholar] [CrossRef] [PubMed]
- Conrad, B.; Antonarakis, S.E. Gene Duplication: A Drive for Phenotypic Diversity and Cause of Human Disease. Annu. Rev. Genomics Hum. Genet. 2007, 8, 17–35. [Google Scholar] [CrossRef]
- The Wellcome Trust Case Control Consortium; Conrad, D.F.; Pinto, D.; Redon, R.; Feuk, L.; Gokcumen, O.; Zhang, Y.; Aerts, J.; Andrews, T.D.; Barnes, C.; et al. Origins and functional impact of copy number variation in the human genome. Nature 2010, 464, 704–712. [Google Scholar] [CrossRef]
- Rice, A.M.; McLysaght, A. Dosage sensitivity is a major determinant of human copy number variant pathogenicity. Nat. Commun. 2017, 8, 14366. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, J.G.; Branco, A.T.; Yu, S.; Lemos, B. Ribosomal DNA copy number is coupled with gene expression variation and mitochondrial abundance in humans. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef]
- Long, E.O.; Dawid, I.B. Repeated genes in eukaryotes. Annu. Rev. Biochem. 1980, 49, 727–764. [Google Scholar] [CrossRef] [PubMed]
- Oakes, M.; Siddiqi, I.; Vu, L.; Aris, J.; Nomura, M. Transcription factor UAF, expansion and contraction of ribosomal DNA (rDNA) repeats, and RNA polymerase switch in transcription of yeast rDNA. Mol. Cell. Biol. 1999, 19, 8559–8569. [Google Scholar] [CrossRef]
- Jack, C.V.; Cruz, C.; Hull, R.M.; Keller, M.A.; Ralser, M.; Houseley, J. Regulation of ribosomal DNA amplification by the TOR pathway. Proc. Natl. Acad. Sci. 2015, 112, 9674–9679. [Google Scholar] [CrossRef] [PubMed]
- Chestkov, I.V.; Jestkova, E.M.; Ershova, E.S.; Golimbet, V.E.; Lezheiko, T.V.; Kolesina, N.Y.; Porokhovnik, L.N.; Lyapunova, N.A.; Izhevskaya, V.L.; Kutsev, S.I.; et al. Abundance of ribosomal RNA gene copies in the genomes of schizophrenia patients. Schizophr. Res. 2018, 197, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, J.G.; Branco, A.T.; Godinho, S.A.; Yu, S.; Lemos, B. Concerted copy number variation balances ribosomal DNA dosage in human and mouse genomes. Proc. Natl. Acad. Sci. USA 2015, 112, 2485–2490. [Google Scholar] [CrossRef]
- Porokhovnik, L.N.; Lyapunova, N.A. Dosage effects of human ribosomal genes (rDNA) in health and disease. Chromosome Res. 2019, 27, 5–17. [Google Scholar] [CrossRef]
- Wang, M.; Lemos, B. Ribosomal DNA copy number amplification and loss in human cancers is linked to tumor genetic context, nucleolus activity, and proliferation. PLoS Genet. 2017, 13, e1006994. [Google Scholar] [CrossRef]
- Xu, B.; Li, H.; Perry, J.M.; Singh, V.P.; Unruh, J.; Yu, Z.; Zakari, M.; McDowell, W.; Li, L.; Gerton, J.L. Ribosomal DNA copy number loss and sequence variation in cancer. PLoS Genet. 2017, 13, e1006771. [Google Scholar] [CrossRef] [PubMed]
- Eagle, S.H.; Crease, T.J. Copy number variation of ribosomal DNA and Pokey transposons in natural populations of Daphnia. Mob. DNA 2012, 3, 4. [Google Scholar] [CrossRef] [PubMed]
- Locati, M.D.; Pagano, J.F.B.; Girard, G.; Ensink, W.A.; van Olst, M.; van Leeuwen, S.; Nehrdich, U.; Spaink, H.P.; Rauwerda, H.; Jonker, M.J.; et al. Expression of distinct maternal and somatic 5.8S, 18S, and 28S rRNA types during zebrafish development. RNA 2017, 23, 1188–1199. [Google Scholar] [CrossRef] [PubMed]
- Dion-Cote, A.-M.; Symonova, R.; Rab, P.; Bernatchez, L. Reproductive isolation in a nascent species pair is associated with aneuploidy in hybrid offspring. Proc. R. Soc. B Biol. Sci. 2015, 282, 20142862. [Google Scholar] [CrossRef] [PubMed]
- Dion-Côté, A.-M.; Symonová, R.; Lamaze, F.C.; Pelikánová, Š.; Ráb, P.; Bernatchez, L. Standing chromosomal variation in Lake Whitefish species pairs: The role of historical contingency and relevance for speciation. Mol. Ecol. 2017, 26, 178–192. [Google Scholar] [CrossRef]
- Symonová, R.; Howell, W. Vertebrate Genome Evolution in the Light of Fish Cytogenomics and rDNAomics. Genes 2018, 9, 96. [Google Scholar] [CrossRef]
- Symonová, R.; Ocalewicz, K.; Kirtiklis, L.; Delmastro, G.B.; Pelikánová, Š.; Garcia, S.; Kovařík, A. Higher-order organisation of extremely amplified, potentially functional and massively methylated 5S rDNA in European pikes (Esox sp.). BMC Genomics 2017, 18. [Google Scholar] [CrossRef]
- Martins, C.; Wasko, A.P.; Oliveira, C.; Porto-Foresti, F.; Parise-Maltempi, P.P.; Wright, J.M.; Foresti, F. Dynamics of 5S rDNA in the tilapia (Oreochromis niloticus) genome: Repeat units, inverted sequences, pseudogenes and chromosome loci. Cytogenet. Genome Res. 2002, 98, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, R.T.; Cabral-de-Mello, D.C.; Valente, G.T.; Venere, P.C.; Martins, C. Evolutionary dynamics of rRNA gene clusters in cichlid fish. BMC Evol. Biol. 2012, 12, 198. [Google Scholar] [CrossRef]
- Lu, K.L.; Nelson, J.O.; Watase, G.J.; Warsinger-Pepe, N.; Yamashita, Y.M. Transgenerational dynamics of rDNA copy number in Drosophila male germline stem cells. eLife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Dong, J.; Liu, X.; Massana, R. Extremely High Copy Numbers and Polymorphisms of the rDNA Operon Estimated from Single Cell Analysis of Oligotrich and Peritrich Ciliates. Protist 2013, 164, 369–379. [Google Scholar] [CrossRef]
- Prescott, D.M. The DNA of ciliated protozoa. Microbiol. Rev. 1994, 58, 233–267. [Google Scholar] [CrossRef] [PubMed]
- Heyse, G.; Jönsson, F.; Chang, W.-J.; Lipps, H.J. RNA-dependent control of gene amplification. Proc. Natl. Acad. Sci. USA 2010, 107, 22134–22139. [Google Scholar] [CrossRef]
- Pan, W.-C.; Orias, E.; Flacks, M.; Blackburn, E.H. Allele-specific, selective amplification of a ribosomal RNA gene in tetrahymena thermophila. Cell 1982, 28, 595–604. [Google Scholar] [CrossRef]
- Symonová, R.; Majtánová, Z.; Sember, A.; Staaks, G.B.; Bohlen, J.; Freyhof, J.; Rábová, M.; Ráb, P. Genome differentiation in a species pair of coregonine fishes: An extremely rapid speciation driven by stress-activated retrotransposons mediating extensive ribosomal DNA multiplications. BMC Evol. Biol. 2013, 13, 42. [Google Scholar] [CrossRef]
- Szathmáry, E.; Smith, J.M. The major evolutionary transitions. Nature 1995, 374, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Parks, M.M.; Kurylo, C.M.; Dass, R.A.; Bojmar, L.; Lyden, D.; Vincent, C.T.; Blanchard, S.C. Variant ribosomal RNA alleles are conserved and exhibit tissue-specific expression. Sci. Adv. 2018, 4, eaao0665. [Google Scholar] [CrossRef]
- Bughio, F.; Maggert, K.A. The peculiar genetics of the ribosomal DNA blurs the boundaries of transgenerational epigenetic inheritance. Chromosome Res. Int. J. Mol. Supramol. Evol. Asp. Chromosome Biol. 2018. [Google Scholar] [CrossRef]
- Torres-Machorro, A.L.; Hernández, R.; Cevallos, A.M.; López-Villaseñor, I. Ribosomal RNA genes in eukaryotic microorganisms: Witnesses of phylogeny? FEMS Microbiol. Rev. 2010, 34, 59–86. [Google Scholar] [CrossRef]
- Drouin, G.; Tsang, C. 5S rRNA Gene Arrangements in Protists: A Case of Nonadaptive Evolution. J. Mol. Evol. 2012, 74, 342–351. [Google Scholar] [CrossRef][Green Version]
- Mallatt, J.; Chittenden, K.D. The GC content of LSU rRNA evolves across topological and functional regions of the ribosome in all three domains of life. Mol. Phylogenet. Evol. 2014, 72, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, S.; Ganley, A.R.D. The conservation landscape of the human ribosomal RNA gene repeats. PLOS ONE 2018, 13, e0207531. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T. A new role of the rDNA and nucleolus in the nucleus—rDNA instability maintains genome integrity. BioEssays 2008, 30, 267–272. [Google Scholar] [CrossRef]
- Escobar, J.S.; Glémin, S.; Galtier, N. GC-Biased Gene Conversion Impacts Ribosomal DNA Evolution in Vertebrates, Angiosperms, and Other Eukaryotes. Mol. Biol. Evol. 2011, 28, 2561–2575. [Google Scholar] [CrossRef]
- Li, B.; Kremling, K.A.G.; Wu, P.; Bukowski, R.; Romay, M.C.; Xie, E.; Buckler, E.S.; Chen, M. Coregulation of ribosomal RNA with hundreds of genes contributes to phenotypic variation. Genome Res. 2018, 28, 1555–1565. [Google Scholar] [CrossRef]
- Ganley, A.R.D.; Kobayashi, T. Highly efficient concerted evolution in the ribosomal DNA repeats: Total rDNA repeat variation revealed by whole-genome shotgun sequence data. Genome Res. 2007, 17, 184–191. [Google Scholar] [CrossRef]
- Stults, D.M.; Killen, M.W.; Pierce, H.H.; Pierce, A.J. Genomic architecture and inheritance of human ribosomal RNA gene clusters. Genome Res. 2007, 18, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Stults, D.M.; Killen, M.W.; Williamson, E.P.; Hourigan, J.S.; Vargas, H.D.; Arnold, S.M.; Moscow, J.A.; Pierce, A.J. Human rRNA Gene Clusters Are Recombinational Hotspots in Cancer. Cancer Res. 2009, 69, 9096–9104. [Google Scholar] [CrossRef]
- Paredes, S.; Branco, A.T.; Hartl, D.L.; Maggert, K.A.; Lemos, B. Ribosomal DNA deletions modulate genome-wide gene expression: “rDNA-sensitive” genes and natural variation. PLoS Genet. 2011, 7, e1001376. [Google Scholar] [CrossRef]
- Paredes, S.; Maggert, K.A. Ribosomal DNA contributes to global chromatin regulation. Proc. Natl. Acad. Sci. USA 2009, 106, 17829–17834. [Google Scholar] [CrossRef]
- Prokopowich, C.D.; Gregory, T.R.; Crease, T.J. The correlation between rDNA copy number and genome size in eukaryotes. Genome 2003, 46, 48–50. [Google Scholar] [CrossRef] [PubMed]
- Banditt, M.; Koller, T.; Sogo, J.M. Transcriptional activity and chromatin structure of enhancer-deleted rRNA genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999, 19, 4953–4960. [Google Scholar] [CrossRef] [PubMed]
- Pederson, T. The Nucleolus. Cold Spring Harb. Perspect. Biol. 2011, 3, a000638. [Google Scholar] [CrossRef]
- Yu, S.; Lemos, B. A Portrait of Ribosomal DNA Contacts with Hi-C Reveals 5S and 45S rDNA Anchoring Points in the Folded Human Genome. Genome Biol. Evol. 2016, 8, 3545–3558. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Lemos, B. The long-range interaction map of ribosomal DNA arrays. PLoS Genet. 2018, 14, e1007258. [Google Scholar] [CrossRef] [PubMed]
- Bierhoff, H.; Postepska-Igielska, A.; Grummt, I. Noisy silence: Non-coding RNA and heterochromatin formation at repetitive elements. Epigenetics 2014, 9, 53–61. [Google Scholar] [CrossRef] [PubMed]
- McStay, B.; Grummt, I. The Epigenetics of rRNA Genes: From Molecular to Chromosome Biology. Annu. Rev. Cell Dev. Biol. 2008, 24, 131–157. [Google Scholar] [CrossRef]
- Schöfer, C.; Weipoltshammer, K. Nucleolus and chromatin. Histochem. Cell Biol. 2018, 150, 209–225. [Google Scholar] [CrossRef] [PubMed]
- Eickbush, T.H.; Eickbush, D.G. Finely Orchestrated Movements: Evolution of the Ribosomal RNA Genes. Genetics 2007, 175, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Robicheau, B.M.; Susko, E.; Harrigan, A.M.; Snyder, M. Ribosomal RNA Genes Contribute to the Formation of Pseudogenes and Junk DNA in the Human Genome. Genome Biol. Evol. 2017, 9, 380–397. [Google Scholar] [CrossRef]
- Bernardi, G. Structural and Evolutionary Genomics: Natural Selection in Genome Evolution; Elsevier: Amsterdam, The Netherlands, 2005; ISBN 978-0-08-046187-8. [Google Scholar]
- Galtier, N.; Piganeau, G.; Mouchiroud, D.; Duret, L. GC-content evolution in mammalian genomes: The biased gene conversion hypothesis. Genetics 2001, 159, 907–911. [Google Scholar]
- Varriale, A.; Torelli, G.; Bernardi, G. Compositional properties and thermal adaptation of 18S rRNA in vertebrates. RNA 2008, 14, 1492–1500. [Google Scholar] [CrossRef][Green Version]
- Wang, H.-C.; Xia, X.; Hickey, D. Thermal adaptation of the small subunit ribosomal RNA gene: A comparative study. J. Mol. Evol. 2006, 63, 120–126. [Google Scholar] [CrossRef]
- Parker, M.S.; Balasubramaniam, A.; Sallee, F.R.; Parker, S.L. The Expansion Segments of 28S Ribosomal RNA Extensively Match Human Messenger RNAs. Front. Genet. 2018, 9. [Google Scholar] [CrossRef]
- Parker, M.S.; Sallee, F.R.; Park, E.A.; Parker, S.L. Homoiterons and expansion in ribosomal RNAs. FEBS Open Bio 2015, 5, 864–876. [Google Scholar] [CrossRef]
- Demeshkina, N.; Repkova, M.; Ven’Yaminova, A.; Graifer, D.; Karpova, G. Nucleotides of 18S rRNA surrounding mRNA codons at the human ribosomal A, P, and E sites: A crosslinking study with mRNA analogs carrying an aryl azide group at either the uracil or the guanine residue. RNA 2000, 6, 1727–1736. [Google Scholar] [CrossRef]
- Barendt, P.A.; Shah, N.A.; Barendt, G.A.; Kothari, P.A.; Sarkar, C.A. Evidence for Context-Dependent Complementarity of Non-Shine-Dalgarno Ribosome Binding Sites to Escherichia coli rRNA. ACS Chem. Biol. 2013, 8, 958–966. [Google Scholar] [CrossRef]
- Forsdyke, D.R. Evolutionary Bioinformatics, 3rd ed.; Springer: Cham, Switzerland, 2016; ISBN 978-3-319-28755-3. [Google Scholar]
- Coleman, A.W. ITS2 is a double-edged tool for eukaryote evolutionary comparisons. Trends Genet. 2003, 19, 370–375. [Google Scholar] [CrossRef]
- Coleman, A.W. Pan-eukaryote ITS2 homologies revealed by RNA secondary structure. Nucleic Acids Res. 2007, 35, 3322–3329. [Google Scholar] [CrossRef]
- Coleman, A.W. Nuclear rRNA transcript processing versus internal transcribed spacer secondary structure. Trends Genet. 2015, 31, 157–163. [Google Scholar] [CrossRef]
- Ruhl, M.W.; Wolf, M.; Jenkins, T.M. Compensatory base changes illuminate morphologically difficult taxonomy. Mol. Phylogenet. Evol. 2010, 54, 664–669. [Google Scholar] [CrossRef] [PubMed]
- Muller, T.; Philippi, N.; Dandekar, T.; Schultz, J.; Wolf, M. Distinguishing species. RNA 2007, 13, 1469–1472. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Shi, L.; Li, D.; Sun, Y.; Niu, Y.; Chen, Z.; Luo, H.; Pang, X.; Sun, Z.; Liu, C.; et al. Extensive Pyrosequencing Reveals Frequent Intra-Genomic Variations of Internal Transcribed Spacer Regions of Nuclear Ribosomal DNA. PLoS ONE 2012, 7, e43971. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.; Wolf, M. ITS2 sequence–structure analysis in phylogenetics: A how-to manual for molecular systematics. Mol. Phylogenet. Evol. 2009, 52, 520–523. [Google Scholar] [CrossRef] [PubMed]
- Ruhl, W.M. Compensatory Base Changes Illuminate Morphologically Difficult Taxonomy. Master Thesis, University of Georgia in Athens, Athens, Greece, 2009. [Google Scholar]
- Nakhoul, H.; Ke, J.; Zhou, X.; Liao, W.; Zeng, S.X.; Lu, H. Ribosomopathies: mechanisms of disease. Clin. Med. Insights Blood Disord. 2014, 7, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Hetman, M. Role of the nucleolus in human diseases. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2014, 1842, 757. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Quin, J.E.; Devlin, J.R.; Cameron, D.; Hannan, K.M.; Pearson, R.B.; Hannan, R.D. Targeting the nucleolus for cancer intervention. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2014, 1842, 802–816. [Google Scholar] [CrossRef] [PubMed]
- Woods, S.J.; Hannan, K.M.; Pearson, R.B.; Hannan, R.D. The nucleolus as a fundamental regulator of the p53 response and a new target for cancer therapy. Biochim. Biophys. Acta BBA Gene Regul. Mech. 2015, 1849, 821–829. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Symonová, R. Integrative rDNAomics—Importance of the Oldest Repetitive Fraction of the Eukaryote Genome. Genes 2019, 10, 345. https://doi.org/10.3390/genes10050345
Symonová R. Integrative rDNAomics—Importance of the Oldest Repetitive Fraction of the Eukaryote Genome. Genes. 2019; 10(5):345. https://doi.org/10.3390/genes10050345
Chicago/Turabian StyleSymonová, Radka. 2019. "Integrative rDNAomics—Importance of the Oldest Repetitive Fraction of the Eukaryote Genome" Genes 10, no. 5: 345. https://doi.org/10.3390/genes10050345
APA StyleSymonová, R. (2019). Integrative rDNAomics—Importance of the Oldest Repetitive Fraction of the Eukaryote Genome. Genes, 10(5), 345. https://doi.org/10.3390/genes10050345




