The Biochemical Role of the Human NEIL1 and NEIL3 DNA Glycosylases on Model DNA Replication Forks
Abstract
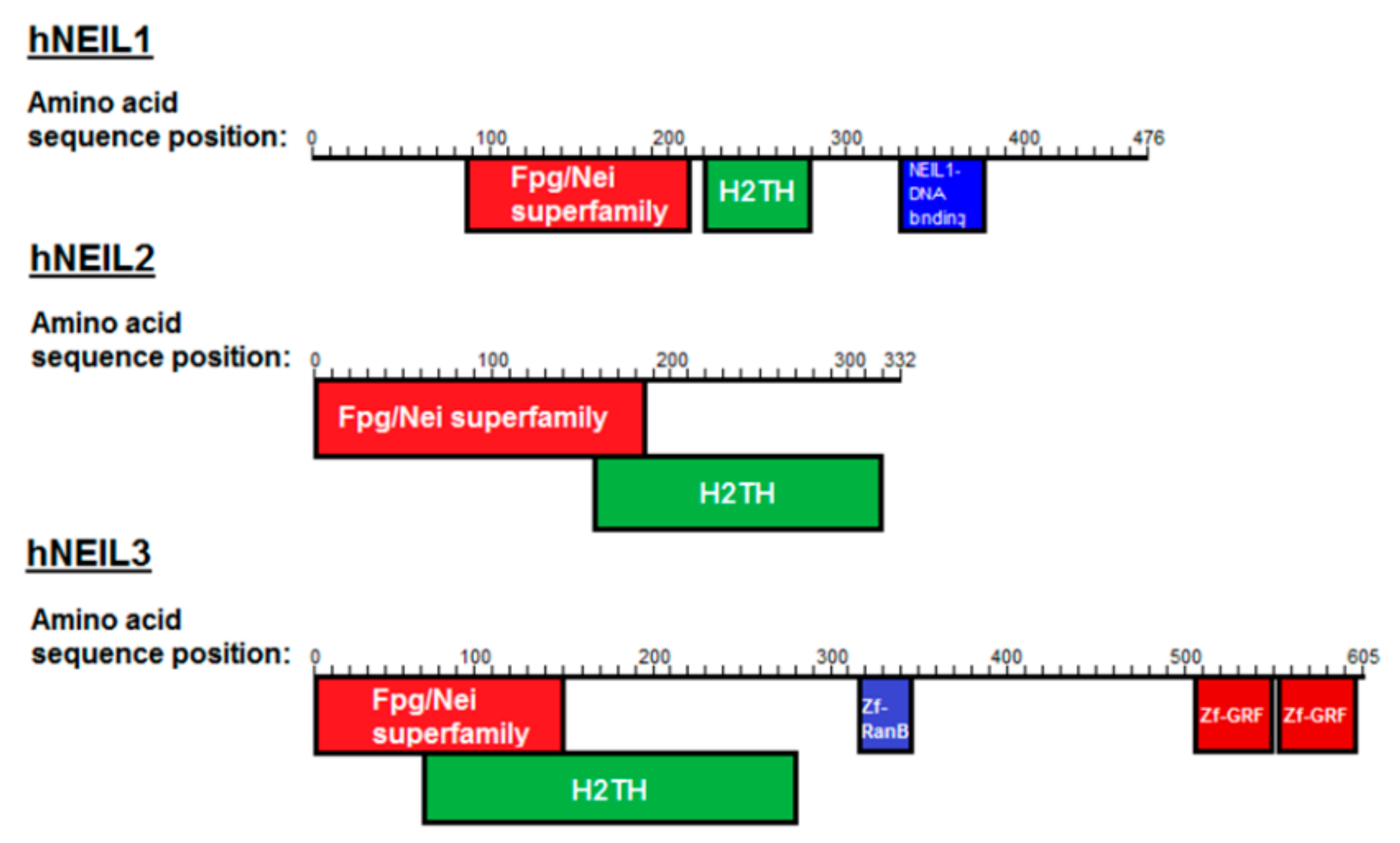
:1. Introduction
2. Materials and Methods
2.1. Materials
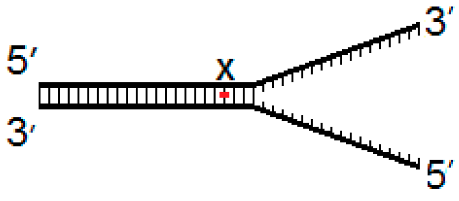
2.2. DNA substrates
2.3. Expression and Purification of hNEIL1 and hNEIL3
2.4. DNA Glycosylase Activity Assays
3. Results
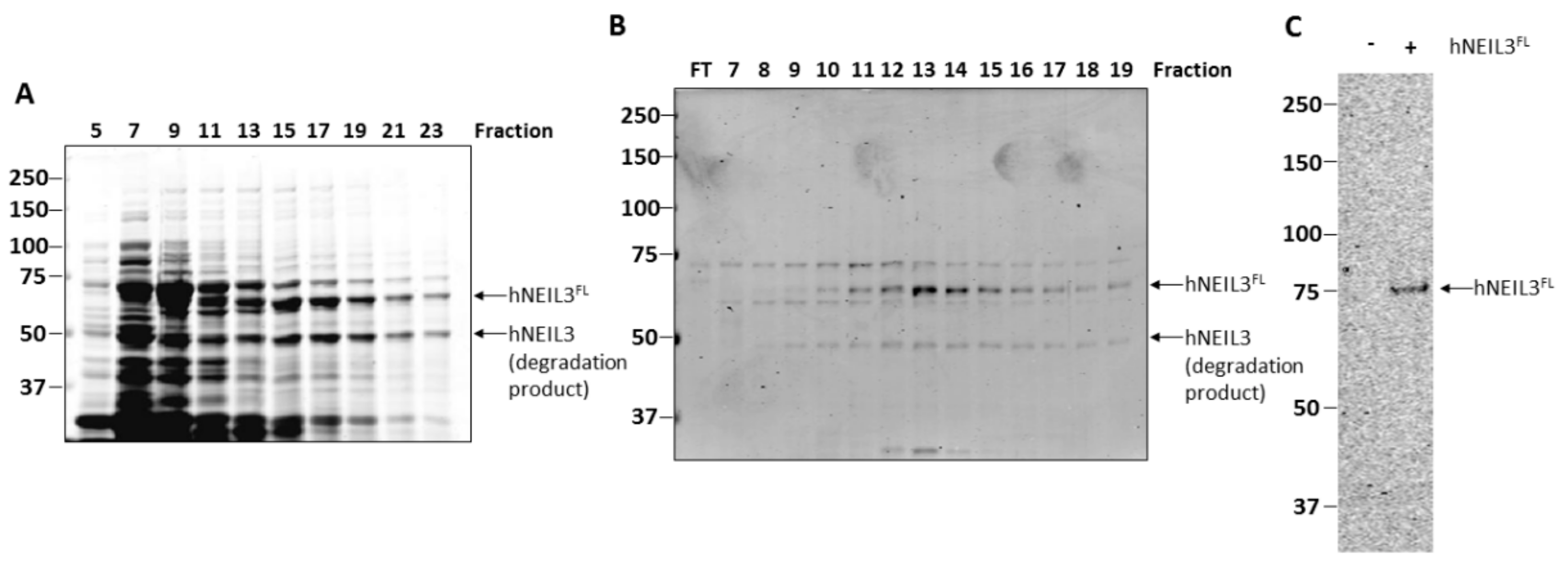
3.1. Purification of hNEIL3FL
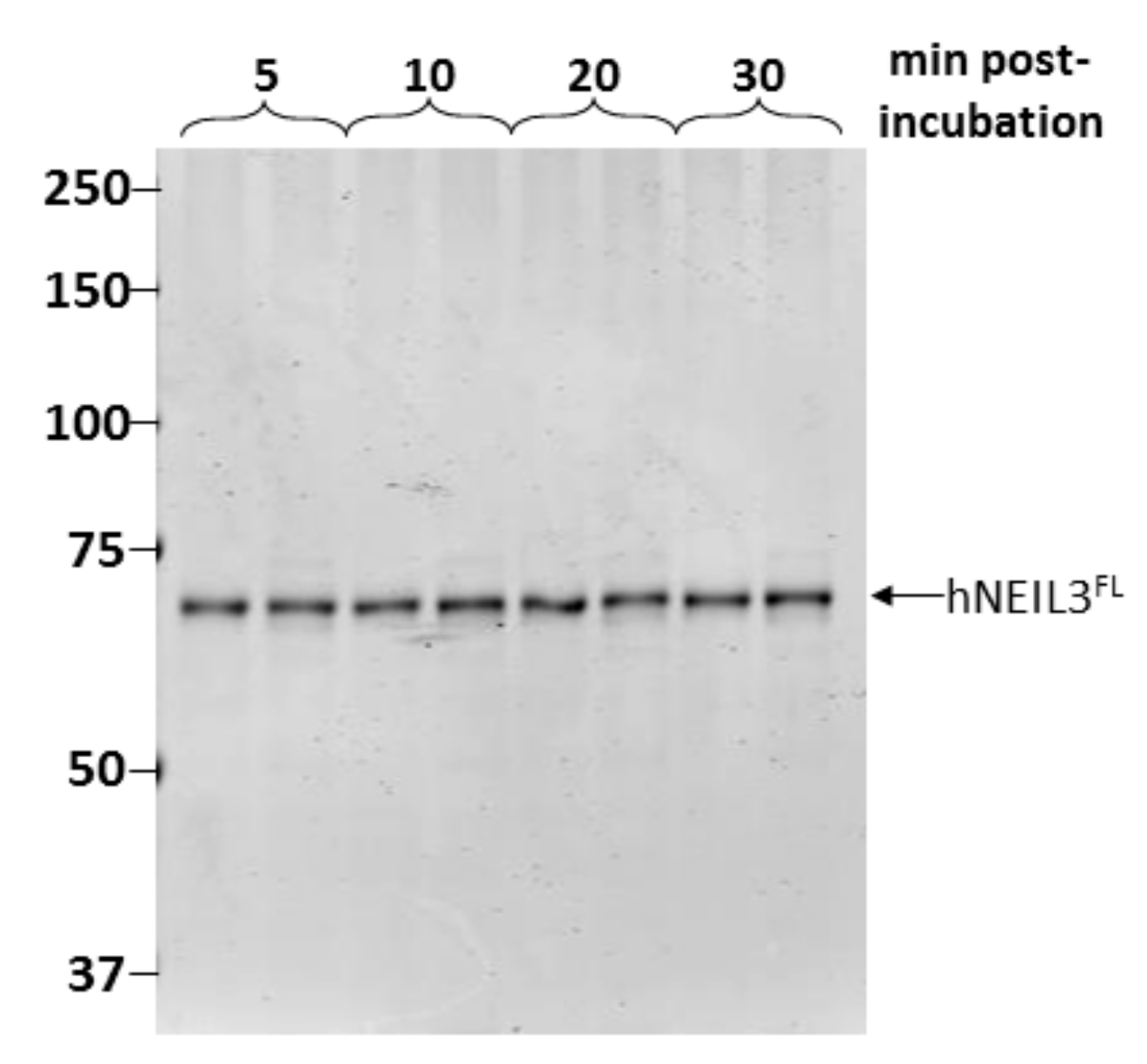
3.2. Assessment of Protein Stability of hNEIL3FL
3.3. Activity of hNEIL1 and hNEIL3FL on ssDNA and dsDNA Substrates
3.4. Activity of hNEIL1 and hNEIL3FL on Model DNA Replication Fork Substrates
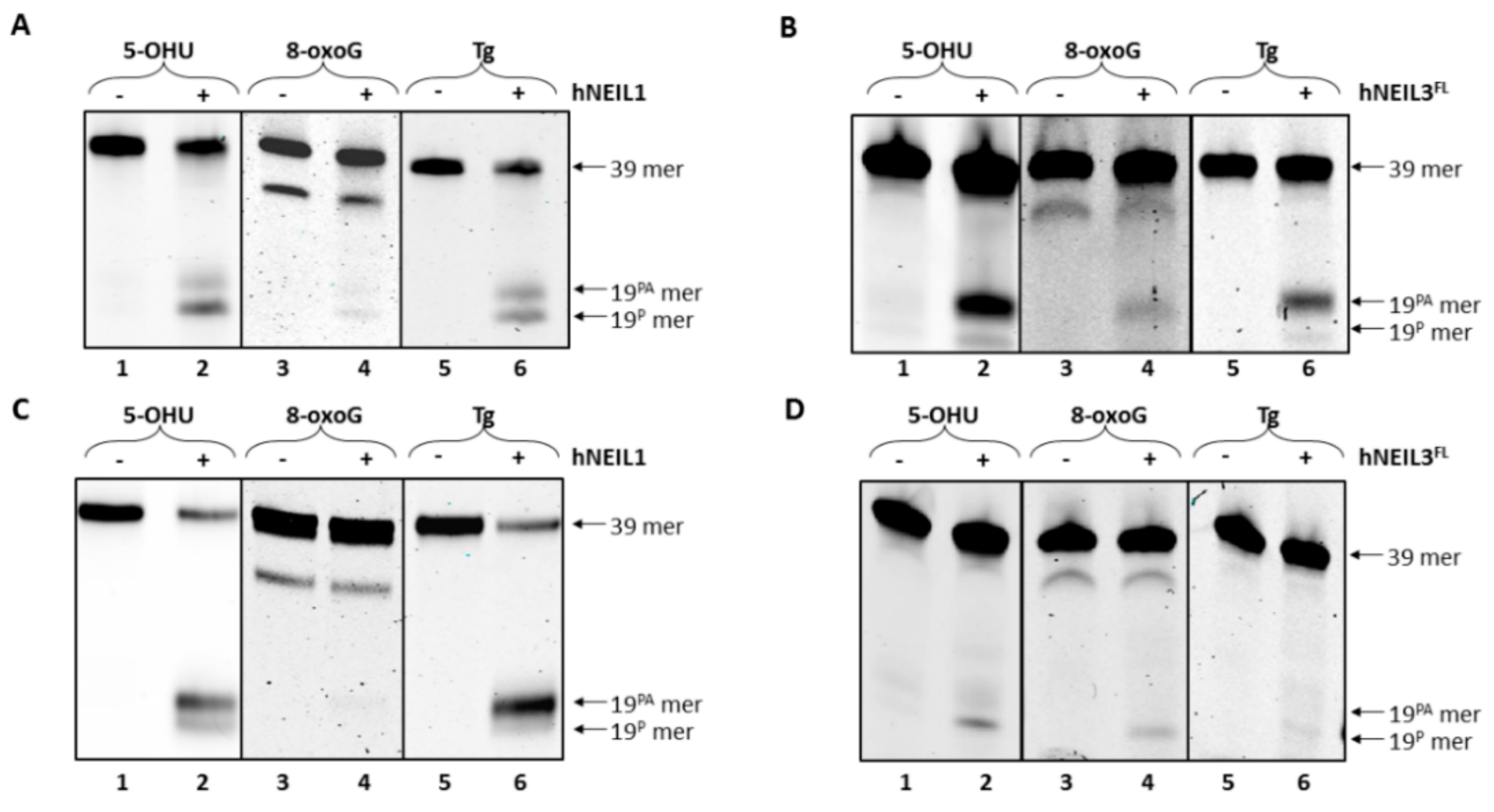
3.4.1. Activity of hNEIL1 on Model DNA Replication Fork Substrates
3.4.2. Activity of hNEIL3FL on Model DNA Replication Fork Substrates
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lindahl, T. Instability and decay of the primary structure of DNA. Nature 1993, 362, 709–715. [Google Scholar] [CrossRef]
- Maynard, S.; Schurman, S.H.; Harboe, C.; de Souza-Pinto, N.C.; Bohr, V.A. Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis 2009, 30, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Wallace, S.S.; Murphy, D.L.; Sweasy, J.B. Base excision repair and cancer. Cancer Lett. 2012, 327, 73–89. [Google Scholar] [CrossRef] [PubMed]
- Kelley, M.R.; Logsdon, D.; Fishel, M.L. Targeting DNA repair pathways for cancer treatment: What’s new? Future Oncol. 2014, 10, 1215–1237. [Google Scholar] [CrossRef] [PubMed]
- Parsons, J.L.; Dianov, G.L. Co-ordination of base excision repair and genome stability. DNA Repair (Amst.) 2013, 12, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Carter, R.J.; Parsons, J.L. Base Excision Repair, a Pathway Regulated by Posttranslational Modifications. Mol. Cell Biol. 2016, 36, 1426–1437. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, A.L.; Schar, P. DNA glycosylases: in DNA repair and beyond. Chromosoma 2012, 121, 1–20. [Google Scholar] [CrossRef]
- Regnell, C.E.; Hildrestrand, G.A.; Sejersted, Y.; Medin, T.; Moldestad, O.; Rolseth, V.; Krokeide, S.Z.; Suganthan, R.; Luna, L.; Bjørås, M.; et al. Hippocampal adult neurogenesis is maintained by Neil3-dependent repair of oxidative DNA lesions in neural progenitor cells. Cell Rep. 2012, 2, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Wallace, S.S. Base excision repair: a critical player in many games. DNA Repair (Amst.) 2014, 19, 14–26. [Google Scholar] [CrossRef] [PubMed]
- Massaad, M.J.; Zhou, J.; Tsuchimoto, D.; Chou, J.; Jabara, H.; Janssen, E.; Glauzy, S.; Olson, B.G.; Morbach, H.; Ohsumi, T.K.; et al. Deficiency of base excision repair enzyme NEIL3 drives increased predisposition to autoimmunity. J. Clin. Invest. 2016, 126, 4219–4236. [Google Scholar] [CrossRef] [PubMed]
- Robson, C.N.; Hickson, I.D. Isolation of cDNA clones encoding a human apurinic/apyrimidinic endonuclease that corrects DNA repair and mutagenesis defects in E. coli xth (exonuclease III) mutants. Nucleic Acids Res. 1991, 19, 5519–5523. [Google Scholar] [CrossRef] [PubMed]
- Demple, B.; Herman, T.; Chen, D.S. Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proc. Natl. Acad. Sci. USA 1991, 88, 11450–11454. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, Y.; Kim, K. Excision of deoxyribose phosphate residues by DNA polymerase b during DNA repair. Science 1995, 269, 699–702. [Google Scholar] [CrossRef]
- Sobol, R.W.; Horton, J.K.; Kühn, R.; Gu, H.; Singhal, R.K.; Prasad, R.; Rajewsky, K.; Wilson, S.H. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature 1996, 379, 183–186. [Google Scholar] [CrossRef]
- Cappelli, E.; Taylor, R.; Cevasco, M.; Abbondandolo, A.; Caldecott, K.; Frosina, G. Involvement of XRCC1 and DNA ligase III gene products in DNA base excision repair. J. Biol. Chem. 1997, 272, 23970–23975. [Google Scholar] [CrossRef]
- Nash, R.A.; Caldecott, K.W.; Barnes, D.E.; Lindahl, T. XRCC1 protein interacts with one of two distinct forms of DNA ligase III. Biochemistry 1997, 36, 5207–5211. [Google Scholar] [CrossRef]
- Dianov, G.; Price, A.; Lindahl, T. Generation of single-nucleotide repair patches following excision of uracil residues from DNA. Mol. Cell. Biol. 1992, 12, 1605–1612. [Google Scholar] [CrossRef]
- Zhou, J.; Chan, J.; Lambelé, M.; Yusufzai, T.; Stumpff, J.; Opresko, P.L.; Thali, M.; Wallace, S.S. NEIL3 Repairs Telomere Damage during S Phase to Secure Chromosome Segregation at Mitosis. Cell Rep. 2017, 20, 2044–2056. [Google Scholar] [CrossRef] [PubMed]
- Wiederhold, L.; Leppard, J.B.; Kedar, P.; Karimi-Busheri, F.; Rasouli-Nia, A.; Weinfeld, M.; Tomkinson, A.E.; Izumi, T.; Prasad, R.; Wilson, S.H.; et al. AP endonuclease-independent DNA base excision repair in human cells. Mol. Cell 2004, 15, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Doublie, S.; Wallace, S.S. Neil3, the final frontier for the DNA glycosylases that recognize oxidative damage. Mutat. Res. 2013, 743, 4–11. [Google Scholar] [CrossRef]
- Liu, M.; Bandaru, V.; Holmes, A.; Averill, A.M.; Cannan, W.; Wallace, S.S. Expression and purification of active mouse and human NEIL3 proteins. Protein Expr. Purif. 2012, 84, 130–139. [Google Scholar] [CrossRef]
- Krokeide, S.Z.; Laerdahl, J.K.; Salah, M.; Luna, L.; Cederkvist, F.H.; Fleming, A.M.; Burrows, C.J.; Dalhus, B.; Bjørås, M. Human NEIL3 is mainly a monofunctional DNA glycosylase removing spiroimindiohydantoin and guanidinohydantoin. DNA Repair (Amst.) 2013, 12, 1159–1164. [Google Scholar] [CrossRef]
- Takao, M.; Oohata, Y.; Kitadokoro, K.; Kobayashi, K.; Iwai, S.; Yasui, A.; Yonei, S.; Zhang, Q.M. Human Nei-like protein NEIL3 has AP lyase activity specific for single-stranded DNA and confers oxidative stress resistance in Escherichia coli mutant. Genes Cells 2009, 14, 261–270. [Google Scholar] [CrossRef]
- Martin, P.R.; Couve, S.; Zutterling, C.; Albelazi, M.S.; Groisman, R.; Matkarimov, B.T.; Parsons, J.L.; Elder, R.H.; Saparbaev, M.K. The Human DNA glycosylases NEIL1 and NEIL3 Excise Psoralen-Induced DNA-DNA Cross-Links in a Four-Stranded DNA Structure. Sci. Rep. 2017, 7, 17438. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Fleming, A.M.; Averill, A.M.; Burrows, C.J.; Wallace, S.S. The NEIL glycosylases remove oxidized guanine lesions from telomeric and promoter quadruplex DNA structures. Nucleic Acids Res. 2015, 43, 4039–4054. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Liu, M.; Fleming, A.M.; Burrows, C.J.; Wallace, S.S. Neil3 and NEIL1 DNA glycosylases remove oxidative damages from quadruplex DNA and exhibit preferences for lesions in the telomeric sequence context. J. Biol. Chem. 2013, 288, 27263–27272. [Google Scholar] [CrossRef]
- Neurauter, C.G.; Luna, L.; Bjoras, M. Release from quiescence stimulates the expression of human NEIL3 under the control of the Ras dependent ERK-MAP kinase pathway. DNA Repair (Amst.) 2012, 11, 401–409. [Google Scholar] [CrossRef]
- Hazra, T.K.; Mitra, S. Purification and characterization of NEIL1 and NEIL2, members of a distinct family of mammalian DNA glycosylases for repair of oxidized bases. Methods Enzymol. 2006, 408, 33–48. [Google Scholar] [PubMed]
- Hegde, M.L.; Hegde, P.M.; Bellot, L.J.; Mandal, S.M.; Hazra, T.K.; Li, G.M.; Boldogh, I.; Tomkinson, A.E.; Mitra, S. Prereplicative repair of oxidized bases in the human genome is mediated by NEIL1 DNA glycosylase together with replication proteins. Proc. Natl. Acad. Sci. USA 2013, 110, E3090–E3099. [Google Scholar] [CrossRef]
- Rangaswamy, S.; Pandey, A.; Mitra, S.; Hedge, M.L. Pre-Replicative Repair of Oxidized Bases Maintains Fidelity in Mammalian Genomes: The Cowcatcher Role of NEIL1 DNA Glycosylase. Genes (Basel) 2017, 8, 175. [Google Scholar] [CrossRef]
- Olsen, M.B.; Hildrestrand, G.A.; Scheffler, K.; Vinge, L.E.; Alfsnes, K.; Palibrk, V.; Wang, J.; Neurauter, C.G.; Luna, L.; Johansen, J.; et al. NEIL3-Dependent Regulation of Cardiac Fibroblast Proliferation Prevents Myocardial Rupture. Cell Rep. 2017, 18, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Semlow, D.R.; Zhang, J.; Budzowska, M.; Drohat, A.C.; Walter, J.C. Replication-Dependent Unhooking of DNA Interstrand Cross-Links by the NEIL3 Glycosylase. Cell 2016, 167, 498–511. [Google Scholar] [CrossRef] [PubMed]
- Couve-Privat, S.; Mace, G.; Rosselli, F.; Saparbaev, M.K. Psoralen-induced DNA adducts are substrates for the base excision repair pathway in human cells. Nucleic Acids Res. 2007, 35, 5672–5682. [Google Scholar] [CrossRef]
- Liu, M.; Bandaru, V.; Bond, J.P.; Jaruga, P.; Zhao, X.; Christov, P.P.; Burrows, C.J.; Rizzo, C.J.; Dizdaroglu, M.; Wallace, S.S. The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. Proc. Natl. Acad. Sci. USA 2010, 107, 4925–4930. [Google Scholar] [CrossRef] [PubMed]
- Edmonds, M.J.; Carter, R.J.; Nickson, C.M.; Williams, S.C.; Parsons, J.L. Ubiquitylation-dependent regulation of NEIL1 by Mule and TRIM26 is required for the cellular DNA damage response. Nucleic Acids Res. 2017, 45, 726–738. [Google Scholar] [CrossRef] [PubMed]
- Klattenhoff, A.W.; Thakur, M.; Chu, C.S.; Ray, D.; Habib, S.L.; Kidane, D. Loss of NEIL3 DNA glycosylase markedly increases replication associated double strand breaks and enhances sensitivity to ATR inhibitor in glioblastoma cells. Oncotarget 2017, 8, 112942–112958. [Google Scholar] [CrossRef]
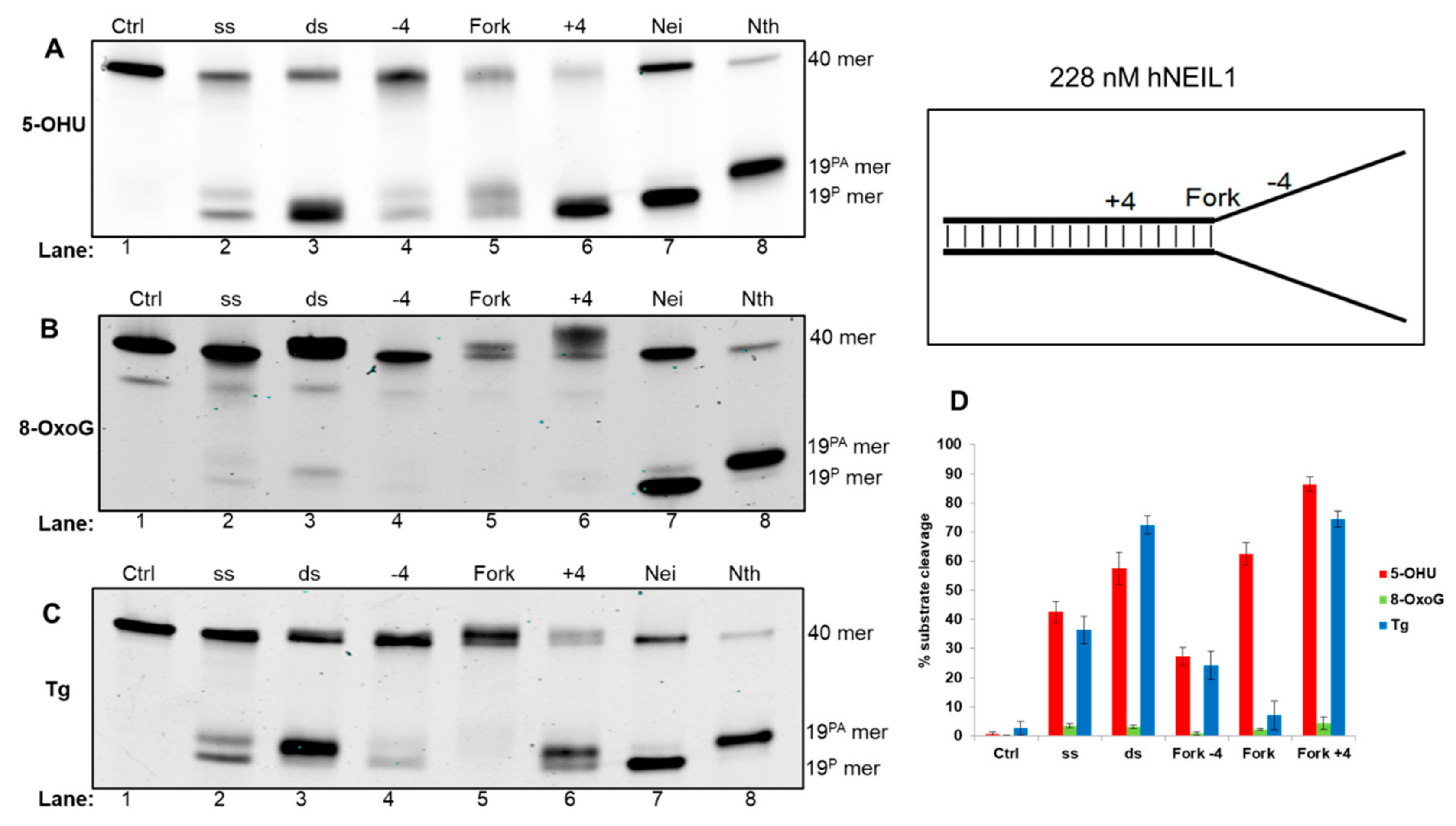
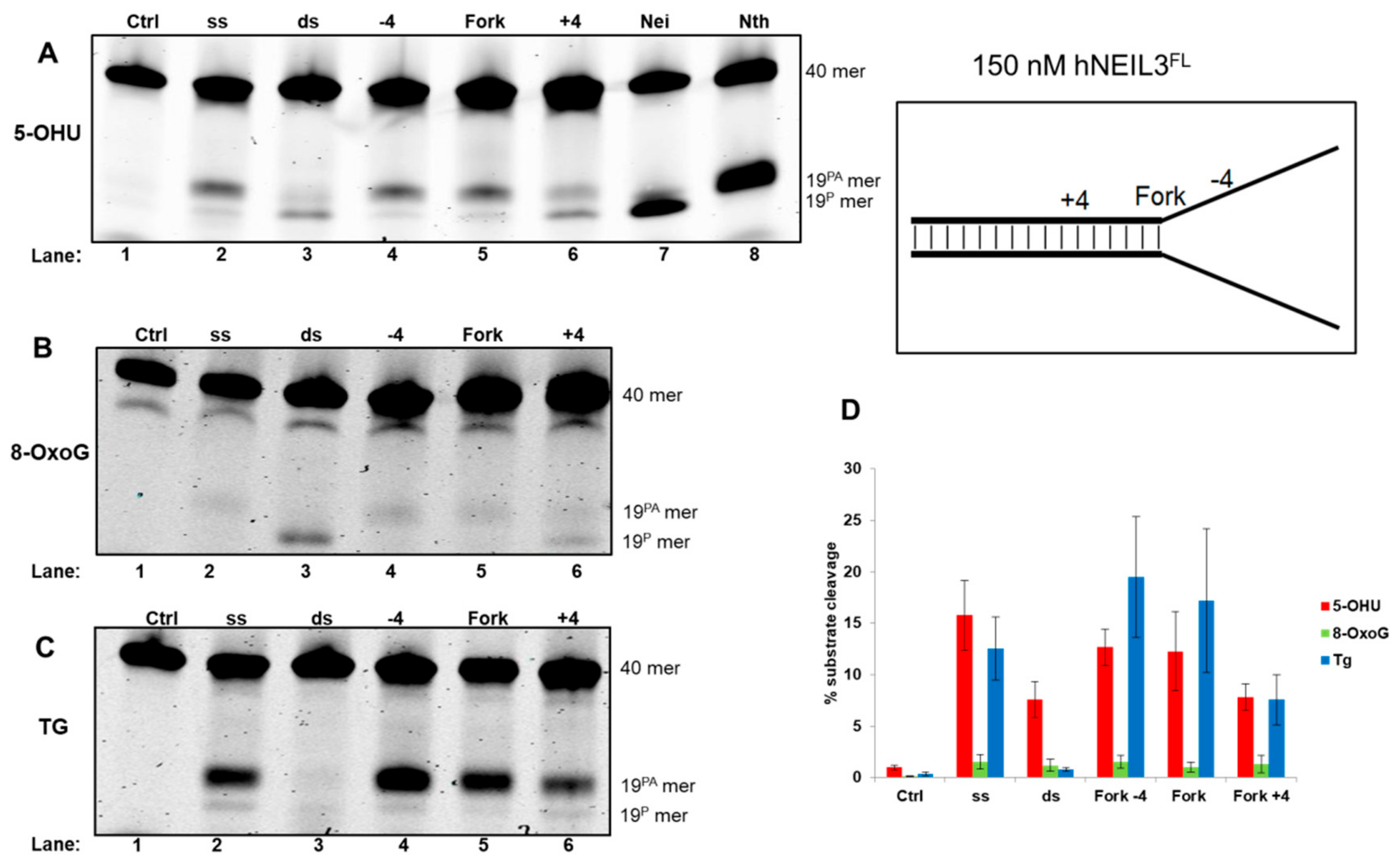
| Fork−4 | Fork | Fork+4 |
|---|---|---|
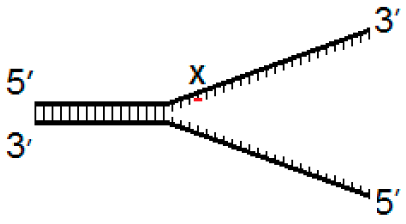 | 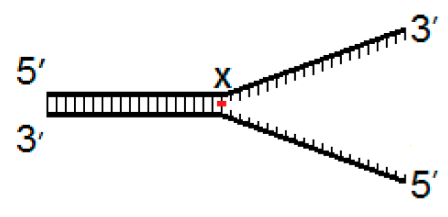 |  |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Albelazi, M.S.; Martin, P.R.; Mohammed, S.; Mutti, L.; Parsons, J.L.; Elder, R.H. The Biochemical Role of the Human NEIL1 and NEIL3 DNA Glycosylases on Model DNA Replication Forks. Genes 2019, 10, 315. https://doi.org/10.3390/genes10040315
Albelazi MS, Martin PR, Mohammed S, Mutti L, Parsons JL, Elder RH. The Biochemical Role of the Human NEIL1 and NEIL3 DNA Glycosylases on Model DNA Replication Forks. Genes. 2019; 10(4):315. https://doi.org/10.3390/genes10040315
Chicago/Turabian StyleAlbelazi, Mustafa S., Peter R. Martin, Soran Mohammed, Luciano Mutti, Jason L. Parsons, and Rhoderick H. Elder. 2019. "The Biochemical Role of the Human NEIL1 and NEIL3 DNA Glycosylases on Model DNA Replication Forks" Genes 10, no. 4: 315. https://doi.org/10.3390/genes10040315
APA StyleAlbelazi, M. S., Martin, P. R., Mohammed, S., Mutti, L., Parsons, J. L., & Elder, R. H. (2019). The Biochemical Role of the Human NEIL1 and NEIL3 DNA Glycosylases on Model DNA Replication Forks. Genes, 10(4), 315. https://doi.org/10.3390/genes10040315







