An Integrated Analysis of Cashmere Fineness lncRNAs in Cashmere Goats
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation and Ethics Statement
2.2. RNA Extraction, Library Preparation, Sequencing, and Quality Analysis
2.3. Identification of lncRNAs
2.4. Expression Profiling and Analysis of Differential Expression
2.5. LncRNA Target Gene Prediction and Enrichment Analysis
2.6. Validation of lncRNA Expression by Quantitative Real-Time PCR
2.7. Construction of the lncRNAs–Targets Network
3. Results
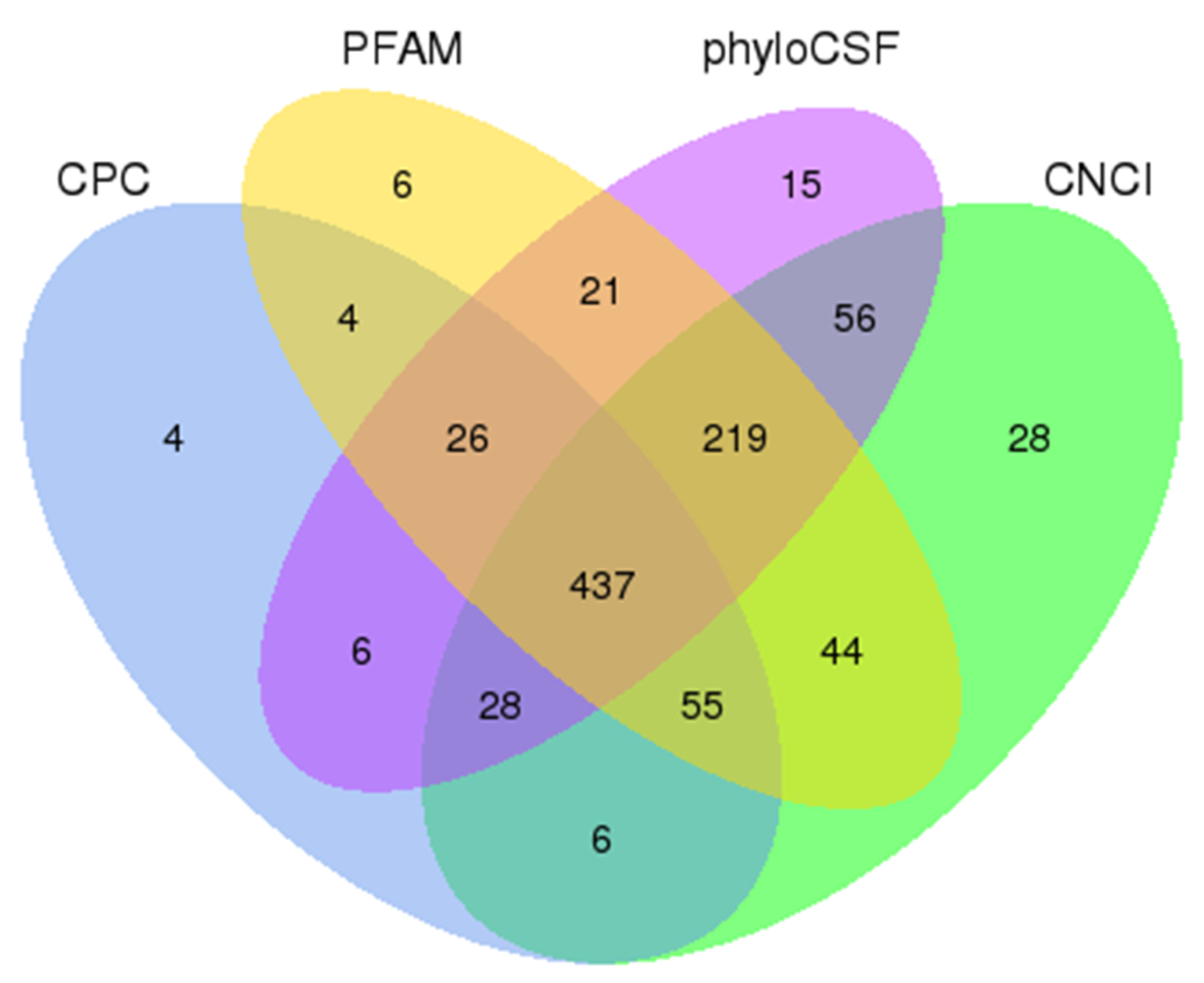
3.1. Identification of mRNA and lncRNAs in LCG and MCG
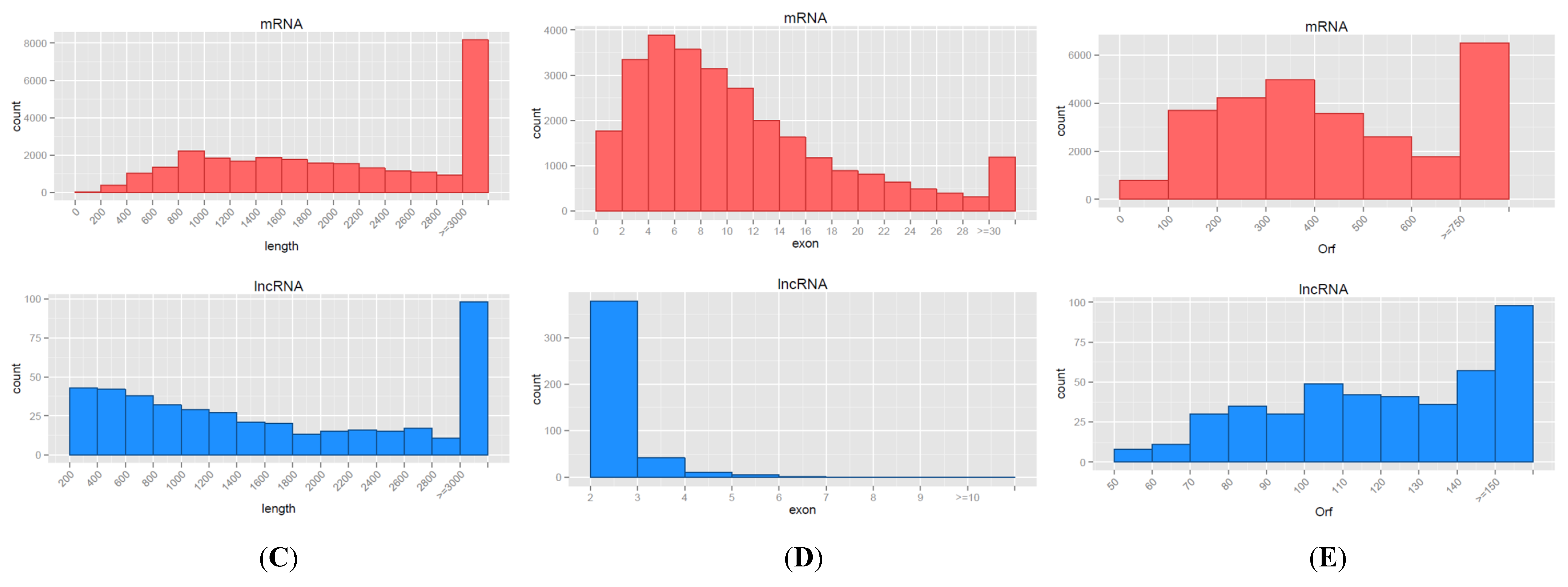
3.2. Characteristics of mRNAs and lncRNAs
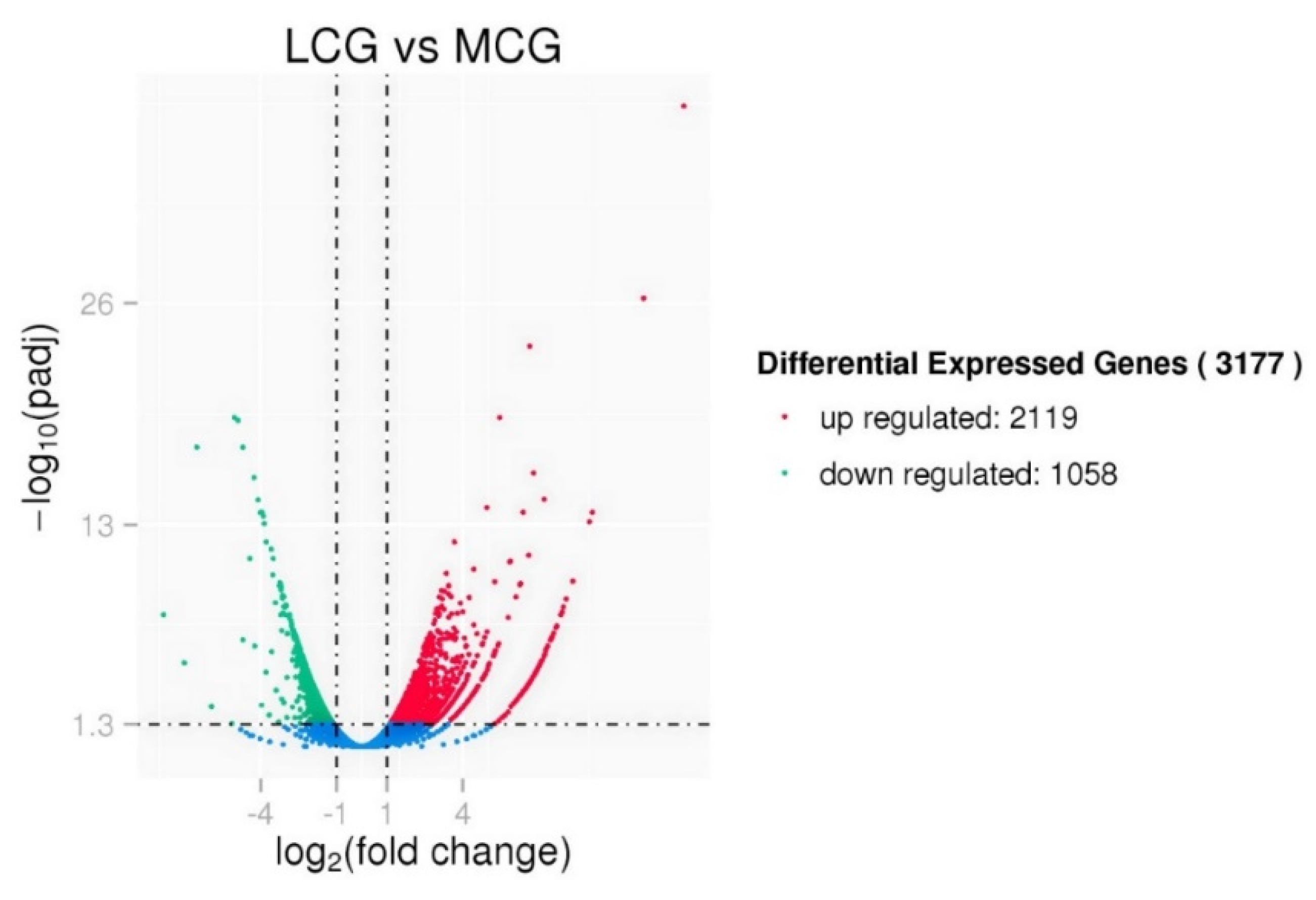
3.3. Differential Expression Analysis between lncRNAs and mRNAs
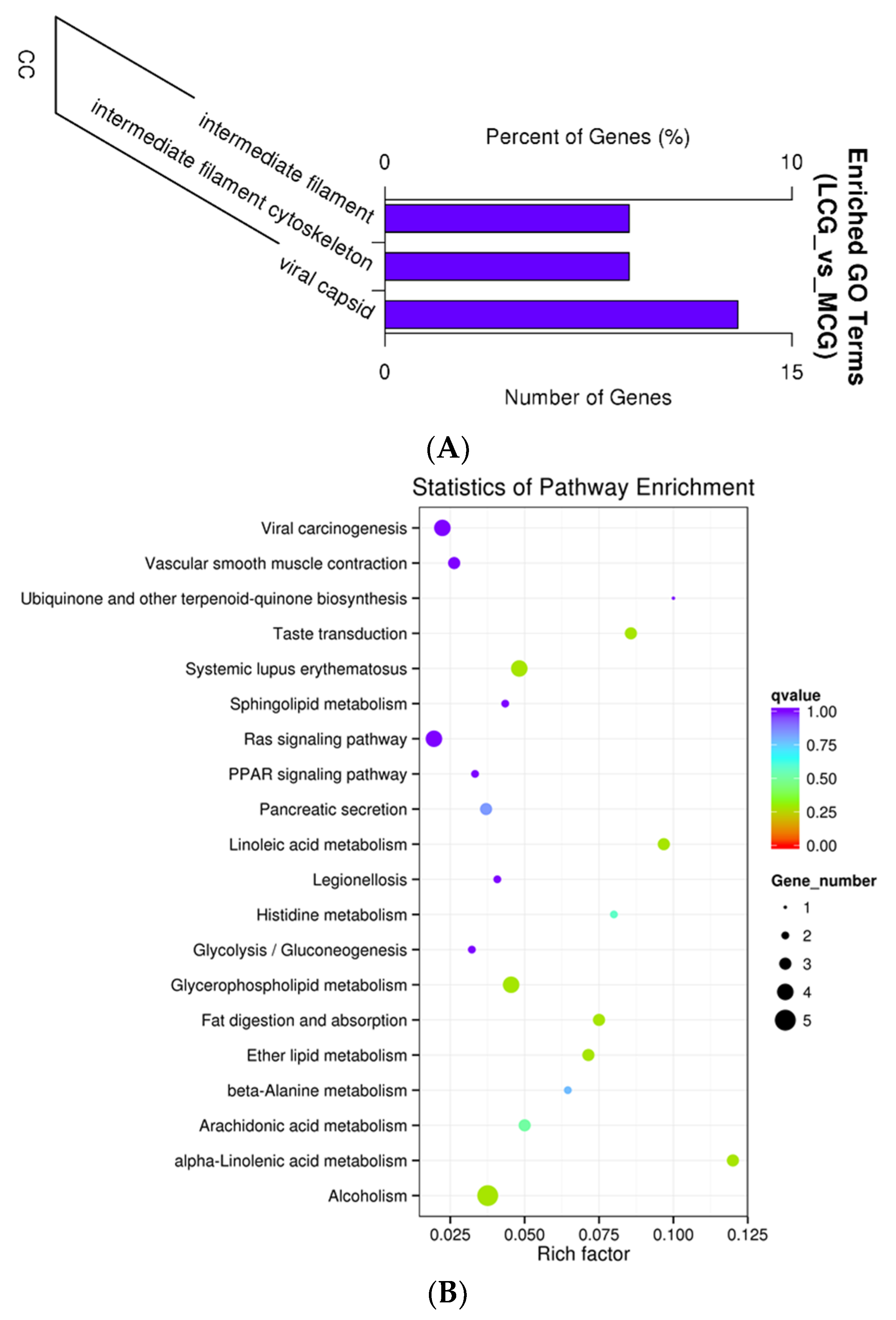
3.4. GO and KEGG Functional Enrichment Analyses
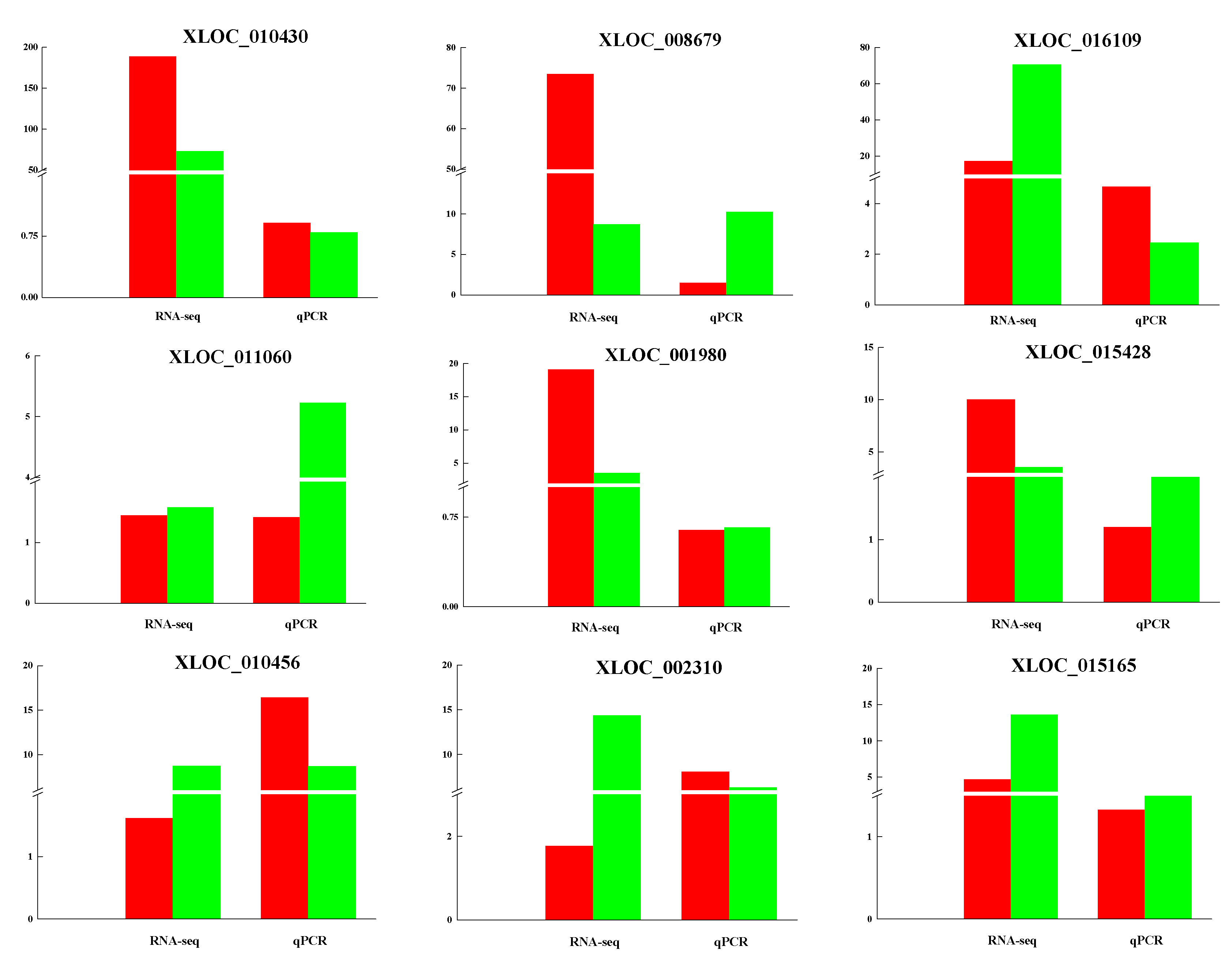
3.5. Validation of lncRNAs by qRT-PCR
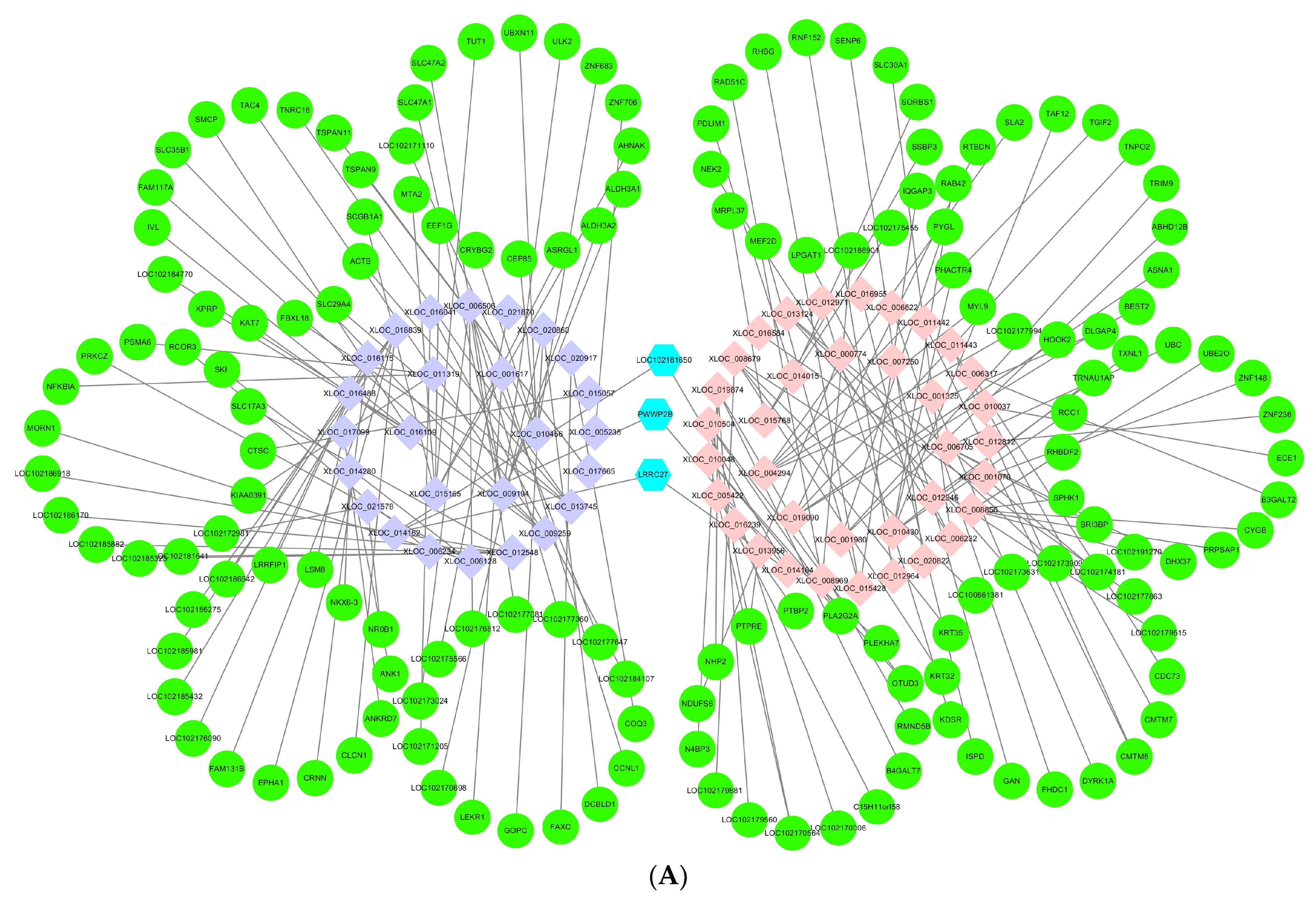
3.6. Networks of lncRNAs–mRNAs
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Zhang, B.; Chang, L.; Lan, Y.X.; Asif, N.; Guan, F.L.; Fu, D.K.; Li, B.; Yan, X.Y.; Zhang, H.B.; Zhang, X.Y.; et al. Genome-wide definition of selective sweeps reveals molecular evidence of trait-driven domestication among elite goat (Capra species) breeds for the production of dairy, cashmere, and meat. Gigascience 2018, 7. [Google Scholar] [CrossRef]
- Bai, W.L.; Dang, Y.L.; Wang, J.J.; Yin, R.H.; Wang, Z.Y.; Zhu, Y.B.; Cong, Y.Y.; Xue, H.L.; Deng, L.; Guo, D.; et al. Molecular characterization, expression and methylation status analysis of BMP4 gene in skin tissue of Liaoning cashmere goat during hair follicle cycle. Genetica 2016, 144, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Xia, T.; Chen, S.C.; Jiang, Z.; Shao, Y.F.; Jiang, X.M.; Li, P.F.; Xiao, B.X.; Guo, J.M. Long noncoding RNA FER1L4 suppresses cancer cell growth by acting as a competing endogenous RNA and regulating PTEN expression. Sci. Rep. 2015, 5, 13445. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Lu, X.; Yuan, L. LncRNA: A link between RNA and cancer. Biochim. Biophys. Acta 2014, 1839, 1097–1109. [Google Scholar] [CrossRef]
- Liang, L.; Ai, L.; Qian, J.; Fang, J.Y.; Xu, J. Long noncoding RNA expression profiles in gut tissues constitute molecular signatures that reflect the types of microbes. Sci. Rep. 2015, 5, 11763. [Google Scholar] [CrossRef]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Orom, U.A.; Derrien, T.; Beringer, M.; Gumireddy, K.; Gardini, A.; Bussotti, G.; Lai, F.; Zytnicki, M.; Notredame, C.; Huang, Q.; et al. Long noncoding RNAs with enhancer-like function in human cells. Cell 2010, 143, 46–58. [Google Scholar] [CrossRef]
- Rinn, J.L.; Chang, H. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef]
- Spurlock, C.F.; Tossberg, J.T.; Guo, Y.; Collier, S.P.; Crooke, P.S.; Aune, T.M. Expression and functions of long noncoding RNAs during human T helper cell differentiation. Nat. Commun. 2015, 6, 6932. [Google Scholar] [CrossRef]
- Ren, H.X.; Wang, G.F.; Chen, L.; Jiang, J.; Liu, L.J.; Li, N.F.; Zhao, J.H.; Sun, X.Y.; Zhou, P. Genome-wide analysis of long non-coding RNAs at early stage of skin pigmentation in goats (Capra hircus). BMC Genom. 2016, 17, 67. [Google Scholar] [CrossRef]
- Wang, S.H.; Ge, W.; Luo, Z.X.; Guo, Y.; Jiao, B.L.; Qu, L.; Zhang, Z.Y.; Wang, X. Integrated analysis of coding genes and non-coding RNAs during hair follicle cycle of cashmere goat (Capra hircus). BMC Genom. 2017, 18, 767. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Yang, M.; Li, Y.F.; Rouzi, M.; Zhao, Q.J.; Pu, Y.B.; He, X.H.; Mwacharo, J.M.; Yang, N.; Ma, Y.H.; et al. Genome-wide discovery of lincRNAs with spatiotemporal expression patterns in the skin of goat during the cashmere growth cycle. BMC Genom. 2018, 19, 495. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Guo, X.; Wang, H.; Hao, F.; Liu, D. Differential gene expression analysis between anagen and telogen of Capra hircus skin based on the de novo assembled transcriptome sequence. Gene 2013, 520, 30–38. [Google Scholar] [CrossRef]
- Zheng, Y.Y.; Wang, Z.Y.; Zhu, Y.B.; Wang, W.; Bai, M.; Jiao, Q.; Wang, Y.R.; Zhao, S.J.; Yin, X.B.; Guo, D.; et al. LncRNA-000133 from secondary hair follicle of Cashmere goat: Identification, regulatory network and its effects on inductive property of dermal papilla cells. Anim. Biotechnol. 2019, 1532–2378. [Google Scholar] [CrossRef]
- Bai, W.L.; Dang, Y.L.; Yin, R.H.; Jiang, W.Q.; Wang, Z.Y.; Zhu, Y.B.; Wang, S.Q.; Zhao, Y.Y.; Deng, L.; Luo, G.B.; et al. Differential Expression of microRNAs and their Regulatory Networks in Skin Tissue of Liaoning Cashmere Goat during Hair Follicle Cycles. Anim. Biotechnol. 2016, 27, 104–112. [Google Scholar] [CrossRef]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000 Res. 2018, 7, 1338. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Dong, Y.; Xie, M.; Jiang, Y.; Xiao, N.Q.; Du, X.Y.; Zhang, W.G.; Tosser-Klopp, G.; Wang, J.H.; Yang, S.; Liang, J.; et al. Sequencing and automated whole-genome optical mapping of the genome of a domestic goat (Capra hircus). Nat. Biotechnol. 2012, 31, 135–141. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Sun, L.; Luo, H.T.; Bu, D.C.; Zhao, G.G.; Yu, K.T.; Zhang, C.H.; Liu, Y.M.; Chen, R.M.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166e166. [Google Scholar] [CrossRef]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2017, 36, W345–W349. [Google Scholar] [CrossRef] [PubMed]
- Punta, M.; Coggill, P.C.; Eberhardt, R.Y.; Mistry, J.; Tate, J.; Boursnell, C.; Pang, N.; Forslund, K.; Ceric, G.; Clements, J.; et al. The Pfam protein families database. Nucleic Acids Res. 2012, 40, D290–D301. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.F.; Jungreis, I.; Kellis, M. PhyloCSF: A comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 2011, 27, i275–i282. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Ideker, T. Cytoscape 2.8. New features for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.X.; Ye, J.Y.C.; Zhang, K.; Li, X.M.; Luo, L.; Ding, J.P.; Li, Y.S.; Cao, H.G.; Ling, Y.H.; Zhang, X.R.; et al. Screening and evaluating of long noncoding RNAs in the puberty of goats. BMC Genom. 2017, 18, 164. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, B.T.; Landry, J.R. RNA-Seq quantitative measurement of expression through massively parallel RNA-sequencing. Methods 2009, 48, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Ozsolak, F.; Milos, P.M. RNA sequencing: Advances, challenges and opportunities. Nat. Rev. Genet. 2011, 12, 87–98. [Google Scholar] [CrossRef]
- Cabili, M.N.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011, 25, 1915–1927. [Google Scholar] [CrossRef]
- Pauli, A.; Valen, E.; Lin, M.F.; Garber, M.; Vastenhouw, N.L.; Levin, J.Z.; Fan, L.; Sandelin, A.; Rinn, J.L.; Regev, A.; et al. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012, 22, 577–591. [Google Scholar] [CrossRef]
- Duchstein, P.; Clark, T.; Zahn, D. Atomistic modeling of a KRT35/KRT85 keratin dimer: Folding in aqueous solution and unfolding under tensile load. Phys. Chem. Chem. Phys. 2015, 17, 21880–21884. [Google Scholar] [CrossRef]
- Giesen, M.; Gruedl, S.; Holtkoetter, O.; Fuhrmann, G.; Koerner, A.; Petersohn, D. Ageing processes influence keratin and KAP expression in human hair follicles. Exp. Dermatol. 2011, 20, 759–761. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wang, X.; Yan, H.; Zeng, J.; Ma, S.; Niu, Y.; Zhou, G.; Jiang, Y.; Chen, Y.; Zhang, M. Comparative Transcriptome Analysis of Fetal Skin Reveals Key Genes Related to Hair Follicle Morphogenesis in Cashmere Goats. PLoS ONE 2016, 11, e0151118. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.D.; Wildermoth, J.E.; Wallace, O.A.M.; Gordon, S.W.; Maqbool, N.J.; Maclean, P.H.; Nixon, A.J.; Pearson, A.J. Annotation of sheep keratin intermediate filament genes and their patterns of expression. Exp. Dermatol. 2011, 20, 582–588. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.J.; Guo, T.T.; Chao, Y.; Liu, J.B.; Guo, J.; Feng, R.L.; Niu, C.; Sun, X.P.; Yang, B.H. Integrated Analysis of the Roles of Long Noncoding RNA and Coding RNA Expression in Sheep (Ovis aries) Skin during Initiation of Secondary Hair Follicle. PLoS ONE 2016, 11, e0156890. [Google Scholar] [CrossRef] [PubMed]
- Ge, W.; Wang, S.H.; Sun, B.; Zhang, Y.L.; Shen, W.; Khatib, H.; Wang, X. Melatonin promotes Cashmere goat (Capra hircus) secondary hair follicle growth: A view from integrated analysis of long non-coding and coding RNAs. Cell Cycle 2018, 17, 1255–1267. [Google Scholar] [CrossRef]
- Zhu, Y.B.; Wang, Z.Y.; Yin, R.H.; Jiao, Q.; Zhao, S.J.; Cong, Y.Y.; Xue, H.L.; Guo, D.; Wang, S.Q.; Zhu, Y.X.; et al. A lncRNA-H19 transcript from secondary hair follicle of Liaoning cashmere goat: Identification, regulatory network and expression regulated potentially by its promoter methylation. Gene 2018, 641, 78–85. [Google Scholar] [CrossRef]
- Zhou, G.X.; Kang, D.J.; Ma, S.; Wang, X.T.; Gao, Y.; Yang, Y.X.; Wang, X.L.; Chen, Y.L. Integrative analysis reveals ncRNA-mediated molecular regulatory network driving secondary hair follicle regression in cashmere goats. BMC Genom. 2018, 19, 222. [Google Scholar] [CrossRef]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Jin, M.; Cao, M.; Cao, Q.; Piao, J.; Zhao, F.Q.; Piao, J. Long noncoding RNA and gene expression analysis of melatonin-exposed Liaoning cashmere goat fibroblasts indicating cashmere growth. Naturwissenschaften Sci. Nat. 2018, 105, 9–10. [Google Scholar] [CrossRef]
- Magin, T.M.; Vijayaraj, P.; Leube, R.E. Structural and regulatory functions of keratins. Exp. Cell Res. 2007, 313, 2021–2032. [Google Scholar] [CrossRef]
- Strnad, P.; Usachov, V.; Debes, C.; Gräter, F.; Parry, D.A.D.; Omary, M.B. Unique amino acid signatures that are evolutionarily conserved distinguish simple-type, epidermal and hair keratins. J. Cell Sci. 2011, 124, 4221–4232. [Google Scholar] [CrossRef] [PubMed]
- Kirfel, J.; Magin, T.M.; Reichelt, J. Keratins: A structural scaffold with emerging functions. Cell. Mol. Life Sci. 2003, 60, 56–71. [Google Scholar] [CrossRef] [PubMed]
- Wallace, L.; Roberts-Thompson, L.; Reichelt, J. Deletion of K1/K10 does not impair epidermal stratification but affects desmosomal structure and nuclear integrity. J. Cell Sci. 2012, 125, 1750–1758. [Google Scholar] [CrossRef]
- Pan, X.; Hobbs, R.P.; Coulombe, P.A. The expanding significance of keratin intermediate filaments in normal and diseased epithelia. Curr. Opin. Cell Biol. 2013, 25, 47–56. [Google Scholar] [CrossRef]
- Rogers, M.A.; Langbein, L.; Praetzel-Wunder, S.; Winter, H.; Schweizer, J. Human hair keratin-associated proteins (KAPs). Int. Rev. Cytol. 2006, 251, 209–263. [Google Scholar] [PubMed]
- Li, Y.; Zhou, G.X.; Zhang, R.; Guo, J.Z.; Li, C.; Martind, G.; Chen, Y.L.; Wang, X.L. Comparative proteomic analyses using iTRAQ-labeling provides insights into fiber diversity in sheep and goats. J. Proteom. 2017, 172, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Song, S.; Dong, K.Z.; Chen, X.F.; Liu, X.X.; Rouzi, M.; Zhao, Q.J.; He, X.L.; Pu, Y.B.; Guan, W.J.; et al. Skin transcriptome reveals the intrinsic molecular mechanisms underlying hair follicle cycling in Cashmere goats under natural and shortened photoperiod conditions. Sci. Rep. 2017, 7, 13502. [Google Scholar] [CrossRef]
- Green, K.; Simpson, C.L. Desmosomes: New perspectives on a classic. J. Investig. Dermatol. 2007, 127, 2499. [Google Scholar] [CrossRef]
- Owens, P.; Bazzi, H.; Engelking, E.; Han, G.X.; Christiano, A.M.; Wang, X.J. Smad4-dependent desmoglein-4 expression contributes to hair follicle integrity. Dev. Biol. 2008, 322, 156–166. [Google Scholar] [CrossRef]
- Bai, W.L.; Wang, J.J.; Yin, R.H.; Dang, Y.L.; Wang, Z.Y.; Zhu, Y.B.; Cong, Y.Y.; Deng, L.; Guo, D.; Wang, S.Q.; et al. Molecular characterization of HOXC8 gene and methylation status analysis of its exon 1 associated with the length of cashmere fiber in Liaoning cashmere goat. Genetica 2017, 145, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Lindner, G.; Menrad, A.; Gherardi, E.; Merlino, G.; Welker, P.; Handjiski, B.; Roloff, B.; Paus, R. Involvement of hepatocyte growth factor/scatter factor and met receptor signaling in hair follicle morphogenesis and cycling. FASEB J. 2000, 4, 319–332. [Google Scholar] [CrossRef]
- McElwee, K.J.; Huth, A.; Kissling, S.; Hoffmann, R. Macrophage-stimulating protein promotes hair growth ex vivo and induces anagen from telogen stage hair follicles in vivo. J. Investig. Dermatol. 2004, 123, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.M.; Zhang, S.B.; Lei, X.H.; Deng, Z.L.; Guo, W.X.; Qiu, Z.F.; Liu, S.; Wang, X.Y.; Zhang, H.; Duan, E.K. Estrogen leads to reversible hair cycle retardation through inducing premature catagen and maintaining telogen. PLoS ONE 2012, 7, e40124. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, K.; Fontanil, T.; Cal, S.; Mendoza-Revilla, J.; Fuentes-Guajardo, M.; Chacon-Duque, J.C.; Al-Saadi, F.; Johansson, J.A.; Quinto-Sanchez, M.; Acuna-Alonzo, V.; et al. A genome-wide association scan in admixed Latin Americans identifies loci influencing facial and scalp hair features. Nat. Commun. 2016, 7, 10815. [Google Scholar] [CrossRef]
- Wu, S.J.; Tan, J.Z.; Yang, Y.J.; Peng, Q.Q.; Zhang, M.F.; Li, J.X.; Lu, D.S.; Liu, Y.; Lou, H.Y.; Feng, Q.D.; et al. Genome-wide scans reveal variants at EDAR predominantly affecting hair straightness in Han Chinese and Uyghur populations. Hum. Genet. 2016, 135, 1279–1286. [Google Scholar] [CrossRef]
- Eriksson, N.; Macpherson, J.M.; Tung, J.Y.; Hon, L.S.; Naughton, B.; Saxonov, S.; Avey, L.; Wojcicki, A.; Pe’er, I.; Mountain, J. Web-based, participant-driven studies yield novel genetic associations for common traits. PLoS Genet. 2010, 6, e1000993. [Google Scholar] [CrossRef]
- Medland, S.E.; Nyholt, D.R.; Painter, J.N.; McEvoy, B.P.; McRae, A.F.; Zhu, G.; Gordon, S.D.; Ferreira, M.A.R.; Wright, M.J.; Henders, A.K.; et al. Common variants in the trichohyalin gene are associated with straight hair in Europeans. Am. J. Hum. Genet. 2009, 85, 750–755. [Google Scholar] [CrossRef]
- Fujimoto, A.; Kimura, R.; Ohashi, J.; Omi, K.; Yuliwulandari, R.; Batubara, L.; Mustofa, M.S.; Samakkarn, U.; SettheethamIshida, W.; Ishida, T.; et al. A scan for genetic determinants of human hair morphology: EDAR is associated with Asianhairthickness. Hum. Mol. Genet. 2008, 17, 835–843. [Google Scholar] [CrossRef] [PubMed]
- Qiao, X.; Su, R.; Wang, Y.; Wang, R.J.; Yang, T.; Li, X.K.; Chen, W.; He, S.Y.; Jiang, Y.; Xu, Q.W.; et al. Genome-wide Target Enrichment-aided Chip Design: A 66 K SNP Chip for Cashmere Goat. Sci. Rep. 2017, 7, 8621. [Google Scholar] [CrossRef]
- Zheng, Y.; Xu, Q.; Peng, Y.; Gong, Z.; Chen, H.; Lai, W.; Maibach, H.I. Expression Profiles of Long Noncoding RNA in UVA-Induced Human Skin Fibroblasts. Skin Pharmacol. Physiol. 2017, 30, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.R.; Schmidt-Ullrich, R.; Paus, R. The hair follicle as a dynamic miniorgan. Curr. Biol. 2009, 19, R132–R142. [Google Scholar] [CrossRef] [PubMed]
- Geyfman, M.; Andersen, B. Clock genes, hair growth and aging. Aging 2010, 2, 122–128. [Google Scholar] [CrossRef]
- Su, K.J.; Kim, S.T.; Lee, D.I. Decoction and Fermentation of Selected Medicinal Herbs Promote Hair Regrowth by Inducing Hair Follicle Growth in Conjunction with Wnts Signaling. Evid.-Based Complement. Altern. Med. 2016, 2016, 1–10. [Google Scholar]
- Botchkarev, V.A.; Fessing, M.Y. Edar signaling in the control of hair follicle development. J. Investig. Dermatol. Symp. Proc. 2005, 10, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Schmidt-Ullrich, R.; Paus, R. Molecular principles of hair follicle induction and morphogenesis. Bioessays 2005, 27, 247–261. [Google Scholar] [CrossRef]
- Mou, C.; Jackson, B.; Schneider, P.; Overbeek, P.A.; Headon, D.J. Generation of the primary hair follicle pattern. Proc. Natl. Acad. Sci. USA 2006, 103, 9075–9080. [Google Scholar] [CrossRef]
- Zhang, Y.; Tomann, P.; Andl, T.; Gallant, N.M.; Huelsken, J. Reciprocal requirements for EDA/EDAR/NF-κB and Wnt/b-catenin signaling pathways in hair follicle induction. Dev. Cell 2009, 17, 49–61. [Google Scholar] [CrossRef]


| Samples | LCG | MCG |
|---|---|---|
| Raw reads | 136,441,946 | 154,106,016 |
| Clean reads | 128,989,956 | 146,038,848 |
| Mapped reads | 111,249,567 | 123,211,813 |
| Mapping rate | 86.25% | 84.37% |
| Uniquely mapped reads | 107,287,774 | 117,506,303 |
| Unique mapping rate | 83.18% | 80.46% |
| Accession No. | FPKM_LCG | FPKM_MCG | p-Value | Target Genes |
|---|---|---|---|---|
| XLOC_010430 | 188.553 | 73.0454 | 0.001316309 | TRNAU1AP; RCC1; TAF12; RAB42; PHACTR4 |
| XLOC_001980 | 19.1252 | 3.50528 | 4.68 × 10−8 | PYGL; ABHD12B; TRIM9 |
| XLOC_016109 | 17.2188 | 70.6548 | 2.98 × 10−6 | LOC102184770; KPRP; SMCP; IVL; |
| XLOC_008679 | 84.16022883 | 10.1007685 | 1.92 × 10−11 | KRT32; KRT35; KRT36; KRT38; LOC102179515; LOC102179881; LOC100861381 |
| XLOC_008969 | 49.1703 | 2.20119 | 1.22 × 10−14 | LOC102186901; RAD51C |
| XLOC_016115 | 6.23184 | 55.5671 | 1.81 × 10−11 | LOC102186542; CRNN; LOC102176090 |
| XLOC_020822 | 12.6 | 4.6006 | 0.004947 | SUMO1 |
| XLOC_015428 | 10.0065 | 3.5299 | 0.00064 | IQGAP3; RHBG; MEF2D |
| XLOC_015165 | 4.64825 | 13.617 | 0.000989863 | SCGB1A1; TUT1; ASRGL1; AHNAK; MTA2; EEF1G |
| XLOC_011319 | 1.15849 | 2.77598 | 0.005641 | PSMA6; NFKBIA; KIAA0391 |
| Gene Name | FPKM_LCG | FPKM_MCG | log2FoldChange | p-Value |
|---|---|---|---|---|
| LOC102183488 | 12,694.8 | 1922.63 | −2.62355 | 1.90 × 10−9 |
| LOC102177231 | 7911.92 | 1767.08 | −2.14703 | 5.15 × 10−7 |
| TCHH | 6322.89 | 1638.67 | −1.93589 | 5.11 × 10−6 |
| LOC102185436 | 4516.72 | 767.495 | −2.54179 | 4.75 × 10−9 |
| LOC100861381 | 4044.28 | 858.502 | −2.22033 | 2.25 × 10−7 |
| KRTAP11-1 | 3930.26 | 697.121 | −2.47797 | 1.06 × 10−8 |
| KRTAP3-1 | 3190.44 | 431.082 | −2.86825 | 7.23 × 10−11 |
| LOC100861181 | 3186.97 | 162.853 | −4.27184 | 8.52 × 10−20 |
| LOC102184223 | 2111.34 | 361.071 | −2.45445 | 1.40 × 10−8 |
| KRT35 | 994.396 | 168.398 | −2.546514249 | 4.58 × 10−9 |
| HOXC13 | 135.246 | 26.9767 | −2.310349019 | 9.09 × 10−8 |
| KRT82 | 450.878 | 68.8984 | −2.270889688 | 1.30 × 10−7 |
| LOC100861183 | 1269.99 | 80.5535 | −3.957981688 | 1.59 × 10−17 |
| TCHHL1 | 196.326 | 45.1137 | −2.10699655 | 8.45 × 10−7 |
| DSG4 | 252.01 | 59.1967 | −2.075452034 | 1.18 × 10−6 |
| BMP2 | 17.1057 | 4.94124 | −1.775970627 | 4.34 × 10−5 |
| LOC102188576 | 58.099 | 732.179 | 3.672081 | 1.12 × 10−15 |
| ACMSD | 21.1133 | 213.701 | 3.353863 | 1.07 × 10−13 |
| CCDC80 | 34.9971 | 307.292 | 3.148606 | 1.61 × 10−12 |
| RCN1 | 28.2099 | 236.021 | 3.079448 | 4.55 × 10−12 |
| COL3A1 | 802.38 | 4,412.04 | 2.473234 | 1.11 × 10−8 |
| LOC102189356 | 53.903 | 231.887 | 2.120764 | 7.69 × 10−7 |
| LOC102187909 | 193.598 | 829.073 | 2.112533 | 7.61 × 10−7 |
| COL1A2 | 834.656 | 2821.44 | 1.771336234 | 2.74 × 10−5 |
| GO Term | lncRNA | Up-Target Gene | Down-Target Gene |
|---|---|---|---|
| intermediate filament | XLOC_008679 | LOC102179515; KRT32; KRT36; KRT35; LOC102176457; LOC102176457; KRT38; LOC100861381; LOC102179881 | |
| intermediate filament cytoskeleton | XLOC_008679 | LOC102179515; KRT32; KRT36; KRT35; LOC102176457; LOC102176457; KRT38; LOC100861381; LOC102179881 | |
| viral capsid | XLOC_008679 | KRT35; KRT36; LOC100861381; LOC102179515; KRT32; KRT38 | |
| XLOC_019874 | RMND5B | ||
| XLOC_020860 | GOPC | ||
| XLOC_010504 | LOC102170006; PLA2G2A | ||
| XLOC_001617 | LEKR1 | ||
| XLOC_015428 | IQGAP3 | ||
| XLOC_006506 | LOC102177360 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, Y.Y.; Sheng, S.D.; Hui, T.Y.; Yue, C.; Sun, J.M.; Guo, D.; Guo, S.L.; Li, B.J.; Xue, H.L.; Wang, Z.Y.; et al. An Integrated Analysis of Cashmere Fineness lncRNAs in Cashmere Goats. Genes 2019, 10, 266. https://doi.org/10.3390/genes10040266
Zheng YY, Sheng SD, Hui TY, Yue C, Sun JM, Guo D, Guo SL, Li BJ, Xue HL, Wang ZY, et al. An Integrated Analysis of Cashmere Fineness lncRNAs in Cashmere Goats. Genes. 2019; 10(4):266. https://doi.org/10.3390/genes10040266
Chicago/Turabian StyleZheng, Yuan Y., Sheng D. Sheng, Tai Y. Hui, Chang Yue, Jia M. Sun, Dan Guo, Su L. Guo, Bo J. Li, Hui L. Xue, Ze Y. Wang, and et al. 2019. "An Integrated Analysis of Cashmere Fineness lncRNAs in Cashmere Goats" Genes 10, no. 4: 266. https://doi.org/10.3390/genes10040266
APA StyleZheng, Y. Y., Sheng, S. D., Hui, T. Y., Yue, C., Sun, J. M., Guo, D., Guo, S. L., Li, B. J., Xue, H. L., Wang, Z. Y., & Bai, W. L. (2019). An Integrated Analysis of Cashmere Fineness lncRNAs in Cashmere Goats. Genes, 10(4), 266. https://doi.org/10.3390/genes10040266






