Genetic Effects of LPIN1 Polymorphisms on Milk Production Traits in Dairy Cattle
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Selection and Phenotypic Data Collection
2.2. Genomic DNA Extraction
2.3. SNP Identification and Estimation of Linkage Disequilibrium (LD)
2.4. Association Analyses on Milk Production Traits
2.5. Detection of LPIN1 Gene Expression by Quantitative Real-Time PCR (qRT-PCR)
2.6. Protein Structure Prediction
2.7. Ethics Approval
3. Results
3.1. SNPs Identification in LPIN1
3.2. Association Analysis Between SNPs and the Five Milk Production Traits
3.3. Associations Between Haplotype Blocks and the Five Milk Traits
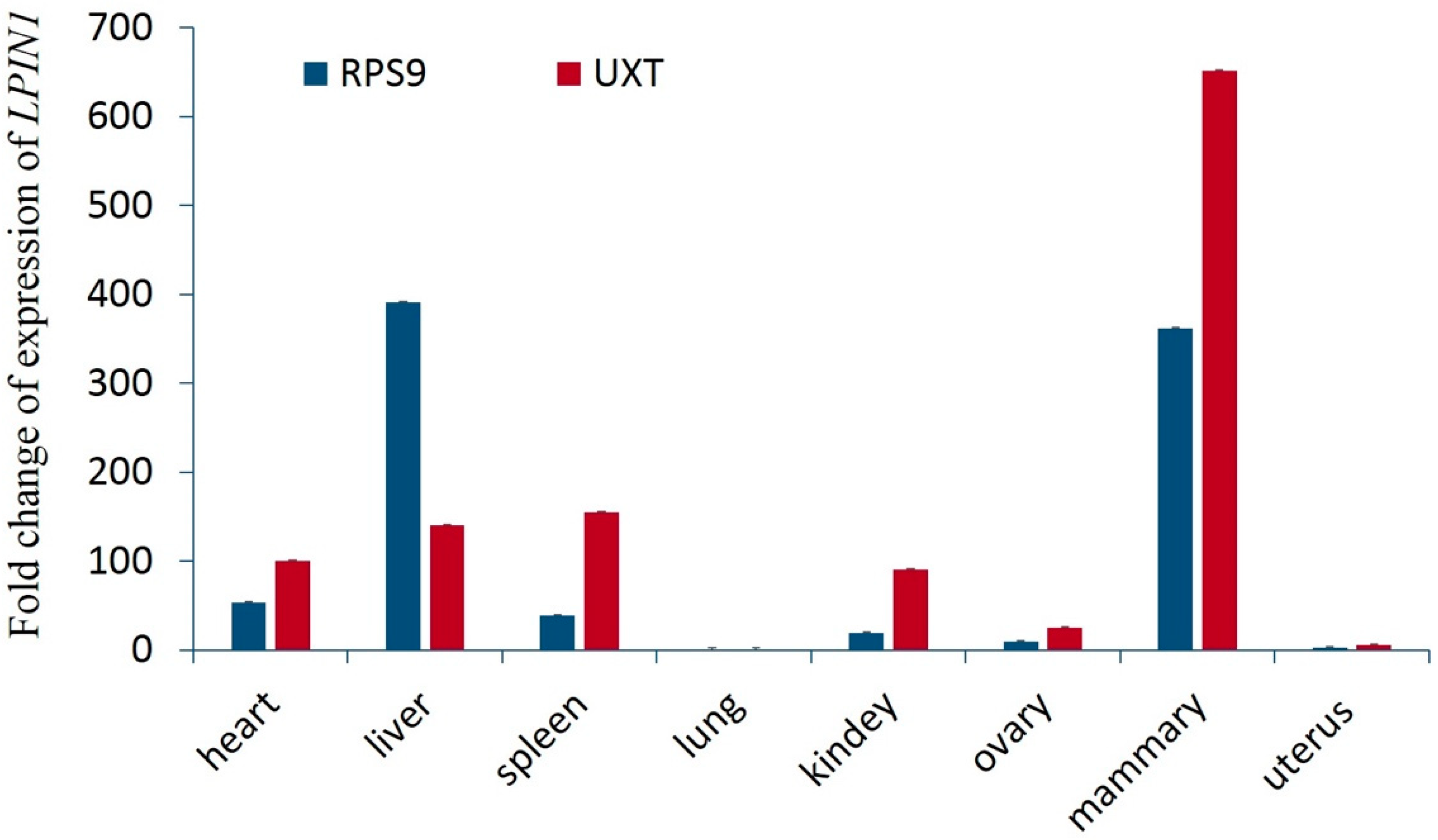
3.4. Expression of LPIN1 in Eight Tissues
3.5. Protein Structure Variation Caused by the Missense Mutation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Visentin, G.; Niero, G.; Berry, D.P.; Costa, A.; Cassandro, M.; De Marchi, M.; Penasa, M. Genetic (co)variances between milk mineral concentration and chemical composition in lactating Holstein-Friesian dairy cows. Animal 2019, 13, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Bastin, C.; Soyeurt, H.; Gengler, N. Genetic parameters of milk production traits and fatty acid contents in milk for Holstein cows in parity 1-3. J. Anim. Breed. Genet. 2013, 130, 118–127. [Google Scholar] [CrossRef]
- Georges, M.; Nielsen, D.; Mackinnon, M.; Mishra, A.; Okimoto, R.; Pasquino, A.T.; Sargeant, L.S.; Sorensen, A.; Steele, M.R.; Zhao, X.; et al. Mapping quantitative trait loci controlling milk production in dairy cattle by exploiting progeny testing. Genetics 1995, 139, 907–920. [Google Scholar]
- Wang, D.; Ning, C.; Liu, J.F.; Zhang, Q.; Jiang, L. Short communication: Replication of genome-wide association studies for milk production traits in Chinese Holstein by an efficient rotated linear mixed model. J. Dairy Sci. 2019, 102, 2378–2383. [Google Scholar] [CrossRef]
- Fang, Z.H.; Bovenhuis, H.; van Valenberg, H.J.F.; Martin, P.; Duchemin, S.I.; Huppertz, T.; Visker, M. Genome-wide association study for alphaS1- and alphaS2-casein phosphorylation in Dutch Holstein Friesian. J. Dairy Sci. 2019, 102, 1374–1385. [Google Scholar] [CrossRef] [PubMed]
- Bouwman, A.C.; Bovenhuis, H.; Visker, M.H.; van Arendonk, J.A. Genome-wide association of milk fatty acids in Dutch dairy cattle. Bmc Genet. 2011, 12, 43. [Google Scholar] [CrossRef]
- Matsumoto, H.; Sasaki, K.; Bessho, T.; Kobayashi, E.; Abe, T.; Sasazaki, S.; Oyama, K.; Mannen, H. The SNPs in the ACACA gene are effective on fatty acid composition in Holstein milk. Mol. Biol. Rep. 2012, 39, 8637–8644. [Google Scholar] [CrossRef]
- Bhattarai, D.; Chen, X.; Ur Rehman, Z.; Hao, X.; Ullah, F.; Dad, R.; Talpur, H.S.; Kadariya, I.; Cui, L.; Fan, M.; et al. Association of MAP4K4 gene single nucleotide polymorphism with mastitis and milk traits in Chinese Holstein cattle. J. Dairy. Res. 2017, 84, 76–79. [Google Scholar] [CrossRef]
- Han, B.; Liang, W.; Liu, L.; Li, Y.; Sun, D. Determination of genetic effects of ATF3 and CDKN1A genes on milk yield and compositions in Chinese Holstein population. Bmc Genet. 2017, 18, 47. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Liang, W.; Liu, L.; Li, Y.; Sun, D. Genetic association of the ACACB gene with milk yield and composition traits in dairy cattle. Anim. Genet. 2018. [Google Scholar] [CrossRef] [PubMed]
- Liang, R.; Han, B.; Li, Q.; Yuan, Y.; Li, J.; Sun, D. Using RNA sequencing to identify putative competing endogenous RNAs (ceRNAs) potentially regulating fat metabolism in bovine liver. Sci. Rep. 2017, 7, 6396. [Google Scholar] [CrossRef]
- Chen, Y.; Rui, B.B.; Tang, L.Y.; Hu, C.M. Lipin family proteins--key regulators in lipid metabolism. Ann. Nutr. Metab. 2015, 66, 10–18. [Google Scholar] [CrossRef]
- Peterson, T.R.; Sengupta, S.S.; Harris, T.E.; Carmack, A.E.; Kang, S.A.; Balderas, E.; Guertin, D.A.; Madden, K.L.; Carpenter, A.E.; Finck, B.N.; et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 2011, 146, 408–420. [Google Scholar] [CrossRef]
- Laplante, M.; Sabatini, D.M. Regulation of mTORC1 and its impact on gene expression at a glance. J. Cell Sci. 2013, 126, 1713–1719. [Google Scholar] [CrossRef]
- Kennedy, B.K.; Lamming, D.W. The Mechanistic Target of Rapamycin: The Grand ConducTOR of Metabolism and Aging. Cell Metab. 2016, 23, 990–1003. [Google Scholar] [CrossRef]
- Goldstein, J.L.; DeBose-Boyd, R.A.; Brown, M.S. Protein sensors for membrane sterols. Cell 2006, 124, 35–46. [Google Scholar] [CrossRef]
- Shao, W.; Espenshade, P.J. Expanding roles for SREBP in metabolism. Cell Metab. 2012, 16, 414–419. [Google Scholar] [CrossRef] [PubMed]
- Donkor, J.; Zhang, P.; Wong, S.; O’Loughlin, L.; Dewald, J.; Kok, B.P.; Brindley, D.N.; Reue, K. A conserved serine residue is required for the phosphatidate phosphatase activity but not the transcriptional coactivator functions of lipin-1 and lipin-2. J. Biol. Chem. 2009, 284, 29968–29978. [Google Scholar] [CrossRef]
- Hashimoto, T.; Cook, W.S.; Qi, C.; Yeldandi, A.V.; Reddy, J.K.; Rao, M.S. Defect in peroxisome proliferator-activated receptor alpha-inducible fatty acid oxidation determines the severity of hepatic steatosis in response to fasting. J. Biol. Chem. 2000, 275, 28918–28928. [Google Scholar] [CrossRef] [PubMed]
- Ashwell, M.S.; Heyen, D.W.; Sonstegard, T.S.; Van Tassell, C.P.; Da, Y.; VanRaden, P.M.; Ron, M.; Weller, J.I.; Lewin, H.A. Detection of quantitative trait loci affecting milk production, health, and reproductive traits in Holstein cattle. J. Dairy. Sci. 2004, 87, 468–475. [Google Scholar] [CrossRef]
- Cole, J.B.; Wiggans, G.R.; Ma, L.; Sonstegard, T.S.; Lawlor, T.J., Jr.; Crooker, B.A.; Van Tassell, C.P.; Yang, J.; Wang, S.; Matukumalli, L.K.; et al. Genome-wide association analysis of thirty one production, health, reproduction and body conformation traits in contemporary U.S. Holstein cows. BMC Genomics 2011, 12, 408. [Google Scholar] [CrossRef] [PubMed]
- Weikard, R.; Kuhn, C.; Goldammer, T.; Freyer, G.; Schwerin, M. The bovine PPARGC1A gene: molecular characterization and association of an SNP with variation of milk fat synthesis. Physiol. Genomics 2005, 21, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Pegolo, S.; Cecchinato, A.; Mele, M.; Conte, G.; Schiavon, S.; Bittante, G. Effects of candidate gene polymorphisms on the detailed fatty acids profile determined by gas chromatography in bovine milk. J. Dairy. Sci. 2016, 99, 4558–4573. [Google Scholar] [CrossRef] [PubMed]
- Nafikov, R.A.; Schoonmaker, J.P.; Korn, K.T.; Noack, K.; Garrick, D.J.; Koehler, K.J.; Minick-Bormann, J.; Reecy, J.M.; Spurlock, D.E.; Beitz, D.C. Polymorphisms in lipogenic genes and milk fatty acid composition in Holstein dairy cattle. Genomics 2014, 104, 572–581. [Google Scholar] [CrossRef] [PubMed]
- Han, G.S.; Wu, W.I.; Carman, G.M. The Saccharomyces cerevisiae lipin homolog is a Mg2+ dependent phosphatidate phosphatase enzyme. J. Biol. Chem. 2006, 281, 9210–9218. [Google Scholar] [CrossRef]
- Bionaz, M.; Loor, J.J. ACSL1, AGPAT6, FABP3, LPIN1, and SLC27A6 are the most abundant isoforms in bovine mammary tissue and their expression is affected by stage of lactation. J. Nutr. 2008, 138, 1019–1024. [Google Scholar] [CrossRef]
- Reue, K.; Zhang, P. The lipin protein family: dual roles in lipid biosynthesis and gene expression. FEBS Lett. 2008, 582, 90–96. [Google Scholar] [CrossRef]
- Reue, K. The lipin family: mutations and metabolism. Curr. Opin. Lipidol. 2009, 20, 165–170. [Google Scholar] [CrossRef]
- Fawcett, K.A.; Grimsey, N.; Loos, R.J.; Wheeler, E.; Daly, A.; Soos, M.; Semple, R.; Syddall, H.; Cooper, C.; Siniossoglou, S.; et al. Evaluating the role of LPIN1 variation in insulin resistance, body weight, and human lipodystrophy in U.K. Populations. Diabetes 2008, 57, 2527–2533. [Google Scholar] [CrossRef]
- Jia, M.; Luo, L.; Liu, C. Statistical correlation between protein secondary structure and messenger RNA stem-loop structure. Biopolymers 2004, 73, 16–26. [Google Scholar] [CrossRef]
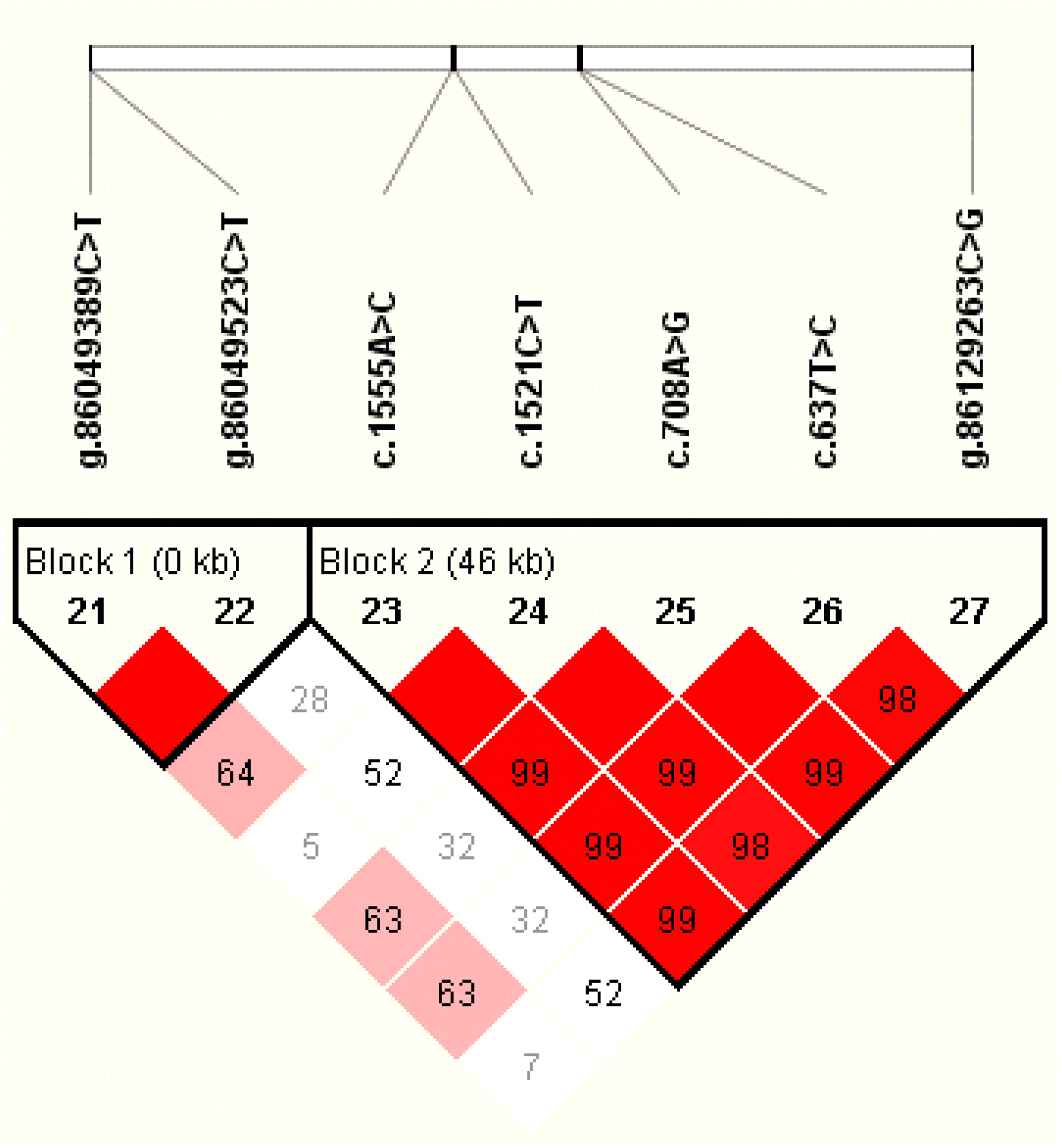
| SNP name | Alias | Location | Position (UMD3.1) | GenBank no. | Genotype | Genotypic Frequency | Allele | Allelic Frequency | Codon | Amino Acid | Alpha Helix (%) | Extended Strand (%) | Beta Turn (%) | Random Coil (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| g.86129263C > G | 5′ flanking region | chr11:86129263 | rs211527179 | CC | 0.52 | C | 0.72 | |||||||
| CG | 0.40 | G | 0.28 | |||||||||||
| GG | 0.08 | |||||||||||||
| c.637T > C | p.Met103Thr | exon 5 | chr11:86093896 | rs110871255 | TT | 0.35 | T | 0.58 | ATG | Methionine | 25.7 | 18.44 | 7.04 | 48.83 |
| TC | 0.46 | C | 0.42 | ACG | Threonine | 25.81 | 17.88 | 7.04 | 49.27 | |||||
| CC | 0.19 | |||||||||||||
| c.708A > G | p.Thr127Ala | exon 5 | chr11:86093825 | rs110161110 | AA | 0.18 | A | 0.42 | ACA | Threonine | 25.7 | 18.44 | 7.04 | 48.83 |
| AG | 0.47 | G | 0.58 | GCA | Alanine | 26.03 | 18.32 | 7.04 | 48.6 | |||||
| GG | 0.35 | |||||||||||||
| c.1521C > T | p.Pro398Ser | exon 8 | chr11:86082411 | rs207681322 | CC | 0.52 | C | 0.72 | CCC | Proline | 25.7 | 18.44 | 7.04 | 48.83 |
| CT | 0.40 | T | 0.28 | TCC | Serine | 25.81 | 18.44 | 7.04 | 48.72 | |||||
| TT | 0.08 | |||||||||||||
| c.1555A > C | p.His409Pro | exon 8 | chr11:86082377 | rs137642654 | AA | 0.18 | A | 0.41 | CAT | Histidine | 25.7 | 18.44 | 7.04 | 48.83 |
| AC | 0.46 | C | 0.59 | CCT | Proline | 25.7 | 18.44 | 7.04 | 48.83 | |||||
| CC | 0.35 | |||||||||||||
| g.86049523C > T | 3′ flanking region | chr11:86049523 | rs135886289 | CC | 0.80 | C | 0.89 | |||||||
| TC | 0.19 | T | 0.11 | |||||||||||
| TT | 0.01 | |||||||||||||
| g.86049389C > T | 3′ flanking region | chr11:86049389 | rs109039955 | CC | 0.57 | C | 0.76 | |||||||
| CT | 0.38 | T | 0.24 | |||||||||||
| TT | 0.05 |
| SNP | Lactation | Genotype (No.) | Milk Yield (kg) | Fat Yield (kg) | Fat Percentage (%) | Protein Yield (kg) | Protein Percentage (%) |
|---|---|---|---|---|---|---|---|
| g.86129263C > G | 1 | CC (549) | 10,277 ± 64.99 | 339.78 ± 2.89 a | 3.33 ± 0.03 | 302.37 ± 2.1 a | 2.95 ± 0.01 |
| GC (418) | 10,373 ± 66.2 | 343.97 ± 2.93 b | 3.33 ± 0.03 | 305.94 ± 2.13 b | 2.96 ± 0.01 | ||
| GG (84) | 10,427 ± 102 | 348.19 ± 4.28 b | 3.36 ± 0.04 | 308.25 ± 3.12 b | 2.96 ± 0.01 | ||
| p | 0.0897 | 0.0264 * | 0.7395 | 0.0171 * | 0.6191 | ||
| 2 | CC (393) | 10,830 ± 66.31 | 394.13 ± 2.94 | 3.65 ± 0.03 a | 322.86 ± 2.14 | 2.98 ± 0.01 | |
| GC (279) | 10,909 ± 70.55 | 389.92 ± 3.1 | 3.57 ± 0.03 b | 323.77 ± 2.26 | 2.96 ± 0.01 | ||
| GG (56) | 11,027 ± 121.7 | 396.24 ± 5.05 | 3.58 ± 0.05 ab | 328.01 ± 3.68 | 2.97 ± 0.02 | ||
| p | 0.1742 | 0.1778 | 0.0114 * | 0.321 | 0.3621 | ||
| c.637T > C | 1 | CC (355) | 10,306 ± 67.75 | 341.19 ± 2.99 | 3.33 ± 0.03 | 304.44 ± 2.18 | 2.96 ± 0.01 |
| CT (471) | 10,298 ± 65.56 | 342.24 ± 2.91 | 3.34 ± 0.03 | 303.55 ± 2.12 | 2.95 ± 0.01 | ||
| TT (195) | 10,260 ± 79.47 | 340.9 ± 3.43 | 3.34 ± 0.03 | 301.61 ± 2.5 | 2.95 ± 0.01 | ||
| p | 0.8001 | 0.8326 | 0.8354 | 0.3977 | 0.3462 | ||
| 2 | CC (245) | 10,822 ± 74.61 ab | 387.99 ± 3.26 | 3.59 ± 0.03 a | 320.57 ± 2.38 ab | 2.96 ± 0.01 | |
| CT (334) | 10,915 ± 69.65 a | 393.46 ± 3.08 | 3.61 ± 0.03 ab | 324.8 ± 2.24 a | 2.98 ± 0.01 | ||
| TT (134) | 10,695 ± 88.29 b | 391.37 ± 3.79 | 3.68 ± 0.04 b | 318.59 ± 2.76 b | 2.98 ± 0.01 | ||
| p | 0.0239 * | 0.1384 | 0.0457 * | 0.0135 * | 0.3878 | ||
| c.708A > G | 1 | AA (189) | 10,306 ± 79.73 | 341.1 ± 3.43 | 3.33 ± 0.03 | 302.25 ± 2.5 | 2.94 ± 0.01 |
| GA (483) | 10,283 ± 65.18 | 341.71 ± 2.9 | 3.34 ± 0.03 | 302.53 ± 2.11 | 2.95 ± 0.01 | ||
| GG (363) | 10,288 ± 67.68 | 339.86 ± 2.99 | 3.33 ± 0.03 | 302.91 ± 2.18 | 2.96 ± 0.01 | ||
| p | 0.9415 | 0.6967 | 0.7041 | 0.9482 | 0.4343 | ||
| 2 | AA (131) | 10,585 ± 87.91 a | 385.53 ± 3.75 | 3.67 ± 0.04 | 315.15 ± 2.73 a | 2.98 ± 0.01 | |
| GA (345) | 10,820 ± 67.54 b | 388.86 ± 2.99 | 3.61 ± 0.03 | 321.28 ± 2.17 b | 2.98 ± 0.01 | ||
| GG (249) | 10,700 ± 74.6 ab | 382.4 ± 3.27 | 3.59 ± 0.03 | 315.81 ± 2.38 a | 2.96 ± 0.01 | ||
| p | 0.0108 * | 0.06 | 0.0578 | 0.0047 ** | 0.1921 | ||
| c.1521C > T | 1 | CC (548) | 10,353 ± 64.39 | 342.88 ± 2.86 a | 3.33 ± 0.03 | 305.29 ± 2.08 a | 2.95 ± 0.01 |
| CT (424) | 10,416 ± 66.09 | 346.9 ± 2.93 ab | 3.34 ± 0.03 | 308.3 ± 2.13 b | 2.96 ± 0.01 | ||
| TT (82) | 10,529 ± 102.6 | 351.26 ± 4.3 b | 3.35 ± 0.04 | 311.14 ± 3.13 b | 2.96 ± 0.01 | ||
| P | 0.1277 | 0.0298 * | 0.8969 | 0.0326 * | 0.5631 | ||
| 2 | CC (391) | 10,794 ± 66.73 Aa | 389.16 ± 2.92 | 3.66 ± 0.03 a | 320.29 ± 2.13 | 2.98 ± 0.01 | |
| CT (286) | 10,752 ± 71.54 Aa | 383.27 ± 3.13 | 3.59 ± 0.03 b | 319.05 ± 2.28 | 2.97 ± 0.01 | ||
| TT (56) | 11,321 ± 125.69 B | 387.58 ± 5.11 | 3.56 ± 0.05 ab | 324.06 ± 3.73 | 2.95 ± 0.02 | ||
| p | <0.0001 ** | 0.0734 | 0.0087 ** | 0.343 | 0.4776 | ||
| c.1555A > C | 1 | AA (190) | 10,318 ± 79.45 | 342.83 ± 3.42 | 3.34 ± 0.03 | 303.86 ± 2.49 | 2.95 ± 0.01 |
| CA (484) | 10,276 ± 64.88 | 341.58 ± 2.88 | 3.34 ± 0.03 | 303.41 ± 2.1 | 2.96 ± 0.01 | ||
| CC (367) | 10,289 ± 68.17 | 341.3 ± 3.01 | 3.33 ± 0.03 | 304.43 ± 2.19 | 2.96 ± 0.01 | ||
| p | 0.8129 | 0.8592 | 0.9428 | 0.8119 | 0.3199 | ||
| 2 | AA (129) | 10,608 ± 88.23 Aa | 390.24 ± 3.77 | 3.7 ± 0.04 a | 316.59 ± 2.74 a | 2.99 ± 0.01 | |
| CA (349) | 10,875 ± 68.16 Bb | 393.78 ± 3.01 | 3.63 ± 0.03 ab | 322.98 ± 2.19 b | 2.98 ± 0.01 | ||
| CC (249) | 10,772 ± 73.77 ab | 387.22 ± 3.22 | 3.6 ± 0.03 b | 318.36 ± 2.35 a | 2.96 ± 0.01 | ||
| p | 0.005 ** | 0.0515 | 0.023 * | 0.0087 ** | 0.1266 | ||
| g.86049523C > T | 1 | CC (849) | 10,240 ± 60.9 Aa | 338 ± 2.73 Aa | 3.32 ± 0.03 | 302.19 ± 1.99 Aa | 2.96 ± 0.01 |
| TC (200) | 10,560 ± 77.78 Bb | 352.15 ± 3.36 Bb | 3.35 ± 0.03 | 311.83 ± 2.45 Bb | 2.96 ± 0.01 | ||
| TT (12) | 10,457 ± 242.54 ab | 330.84 ± 9.83 ab | 3.21 ± 0.1 | 303.49 ± 7.17 ab | 2.92 ± 0.04 | ||
| p | <0.0001 ** | <.0001 ** | 0.2535 | <0.0001 ** | 0.4739 | ||
| 2 | CC (595) | 10,802 ± 63.12 a | 388.88 ± 2.84 a | 3.61 ± 0.03 | 322.08 ± 2.05 a | 2.97 ± 0.01 a | |
| TC (137) | 10,647 ± 86.89 ab | 385.9 ± 3.71 ab | 3.65 ± 0.04 | 320.52 ± 2.73 a | 3 ± 0.01 Bb | ||
| TT (5) | 9933.02 ± 349.64 b | 351.83 ± 14.12 b | 3.54 ± 0.14 | 294.92 ± 10.4 b | 2.98 ± 0.05 ab | ||
| p | 0.0091 ** | 0.0236 * | 0.4408 | 0.0281 * | 0.0144 * | ||
| g.86049389C > T | 1 | CC (590) | 10,320 ± 62.88 Aa | 343.46 ± 2.8 Aa | 3.34 ± 0.03 | 305.67 ± 2.04 Aa | 2.96 ± 0.01 a |
| CT (394) | 10,562 ± 66.66 Bb | 350.18 ± 2.95 Bb | 3.32 ± 0.03 | 313.12 ± 2.15 Bb | 2.96 ± 0.01 a | ||
| TT (56) | 10,518 ± 118.53 ab | 341.71 ± 4.9 ab | 3.27 ± 0.05 | 306.2 ± 3.57 ab | 2.92 ± 0.02 b | ||
| p | <0.0001 ** | 0.0023 ** | 0.2859 | <0.0001 ** | 0.0237 * | ||
| 2 | CC (416) | 10,783 ± 65.86 | 391.43 ± 2.93 a | 3.64 ± 0.03 | 319.02 ± 2.14 | 2.97 ± 0.01 | |
| CT (277) | 10,725 ± 71.14 | 384.63 ± 3.13 b | 3.61 ± 0.03 | 318.2 ± 2.28 | 2.98 ± 0.01 | ||
| TT (35) | 10,916 ± 149.18 | 399.1 ± 6.14 a | 3.68 ± 0.06 | 326.33 ± 4.48 | 3 ± 0.02 | ||
| p | 0.3522 | 0.0053 ** | 0.2585 | 0.1662 | 0.1656 |
| Block | Lactation | Haplotype Combination (No.) | Milk Yield (kg) | Fat Yield (kg) | Fat Percentage (%) | Protein Yield (kg) | Protein Percentage (%) |
|---|---|---|---|---|---|---|---|
| LPIN1-1 | 1 | H1H1 (614) | 10,244 ± 63.83 Aa | 341.58 ± 2.85 Aa | 3.35 ± 0.03 | 302.9 ± 2.08 Aa | 2.96 ± 0.01 |
| H1H2 (174) | 10,592 ± 80.33 Bb | 355.63 ± 3.45 B | 3.36 ± 0.03 | 314.22 ± 2.52 B | 2.96 ± 0.01 | ||
| H1H3 (223) | 10,374 ± 76.44 a | 340.98 ± 3.32 Aa | 3.31 ± 0.03 | 306.43 ± 2.42 Aa | 2.96 ± 0.01 | ||
| p | <0.0001 ** | <0.0001 ** | 0.1356 | <0.0001 ** | 0.7979 | ||
| 2 | H1H1 (426) | 10,820 ± 66.83 | 390.47 ± 2.98 a | 3.62 ± 0.03 | 320.89 ± 2.17 | 2.97 ± 0.01 a | |
| H1H2 (121) | 10,695 ± 92.24 | 384.18 ± 3.95 ab | 3.61 ± 0.04 | 320.77 ± 2.87 | 3 ± 0.01 b | ||
| H1H3 (158) | 10,797 ± 84.63 | 382.69 ± 3.64 b | 3.56 ± 0.03 | 318.51 ± 2.65 | 2.96 ± 0.01 a | ||
| p | 0.3532 | 0.0185 * | 0.1696 | 0.5462 | 0.01 * | ||
| LPIN1-2 | 1 | H1H1 (201) | 10,195 ± 81.19 Aa | 339.42 ± 3.38 | 3.32 ± 0.03 | 303.29 ± 2.49 ac | 2.94 ± 0.01 |
| H1H2 (227) | 10,498 ± 78.47 Bb | 346.15 ± 3.3 | 3.35 ± 0.03 | 307.55 ± 2.39 bc | 2.95 ± 0.01 | ||
| H1H3 (247) | 10,299 ± 76.23 ab | 340.2 ± 3.19 | 3.33 ± 0.03 | 302.72 ± 2.35 a | 2.95 ± 0.01 | ||
| H2H2 (80) | 10,398 ± 110.37 ab | 348.02 ± 4.38 | 3.36 ± 0.04 | 307.48 ± 3.2 abc | 2.96 ± 0.01 | ||
| H2H3 (183) | 10,332 ± 80.05 ab | 342.19 ± 3.37 | 3.32 ± 0.03 | 308.35 ± 2.47 b | 2.96 ± 0.01 | ||
| H3H3 (93) | 10,178 ± 104.38 a | 341.58 ± 4.09 | 3.36 ± 0.04 | 302.3 ± 3.04 ac | 2.95 ± 0.01 | ||
| p | 0.002 ** | 0.1276 | 0.8047 | 0.0383 * | 0.8859 | ||
| 2 | H1H1 (134) | 10,632 ± 88.27 a | 387.72 ± 3.79 | 3.66 ± 0.04 | 315.83 ± 2.76 Aa | 2.98 ± 0.01 | |
| H1H2 (154) | 10,907 ± 84.15 ab | 390.43 ± 3.63 | 3.58 ± 0.03 | 323.03 ± 2.64 Cc | 2.97 ± 0.01 | ||
| H1H3 (184) | 10,944 ± 80.55 b | 394.99 ± 3.49 | 3.62 ± 0.03 | 323.2 ± 2.54 BCbc | 2.96 ± 0.01 | ||
| H2H2 (54) | 10,909 ± 123.9 ab | 390.82 ± 5.15 | 3.58 ± 0.05 | 322.46 ± 3.76 abc | 2.97 ± 0.02 | ||
| H2H3 (121) | 10,748 ± 89.14 ab | 383.7 ± 3.81 | 3.57 ± 0.04 | 315.33 ± 2.78 Aa | 2.94 ± 0.01 | ||
| H3H3 (68) | 10,700 ± 109.38 ab | 387.14 ± 4.57 | 3.61 ± 0.04 | 317.31 ± 3.33 abc | 2.97 ± 0.02 | ||
| p | 0.0044 ** | 0.0708 | 0.2162 | 0.0052 ** | 0.3756 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, B.; Yuan, Y.; Liang, R.; Li, Y.; Liu, L.; Sun, D. Genetic Effects of LPIN1 Polymorphisms on Milk Production Traits in Dairy Cattle. Genes 2019, 10, 265. https://doi.org/10.3390/genes10040265
Han B, Yuan Y, Liang R, Li Y, Liu L, Sun D. Genetic Effects of LPIN1 Polymorphisms on Milk Production Traits in Dairy Cattle. Genes. 2019; 10(4):265. https://doi.org/10.3390/genes10040265
Chicago/Turabian StyleHan, Bo, Yuwei Yuan, Ruobing Liang, Yanhua Li, Lin Liu, and Dongxiao Sun. 2019. "Genetic Effects of LPIN1 Polymorphisms on Milk Production Traits in Dairy Cattle" Genes 10, no. 4: 265. https://doi.org/10.3390/genes10040265
APA StyleHan, B., Yuan, Y., Liang, R., Li, Y., Liu, L., & Sun, D. (2019). Genetic Effects of LPIN1 Polymorphisms on Milk Production Traits in Dairy Cattle. Genes, 10(4), 265. https://doi.org/10.3390/genes10040265





