The Transcriptome of Verticillium dahliae Responds Differentially Depending on the Disease Susceptibility Level of the Olive (Olea europaea L.) Cultivar
Abstract
:1. Introduction
2. Materials and Methods
2.1. Verticillium dahliae–olive bioassays
2.2. Preparation of RNA Samples and Sequencing
2.3. RNA-seq Data Pre-Processing
2.4. Gene expression and Differentially Expressed Genes
2.5. Data and Materials Availability
3. Results
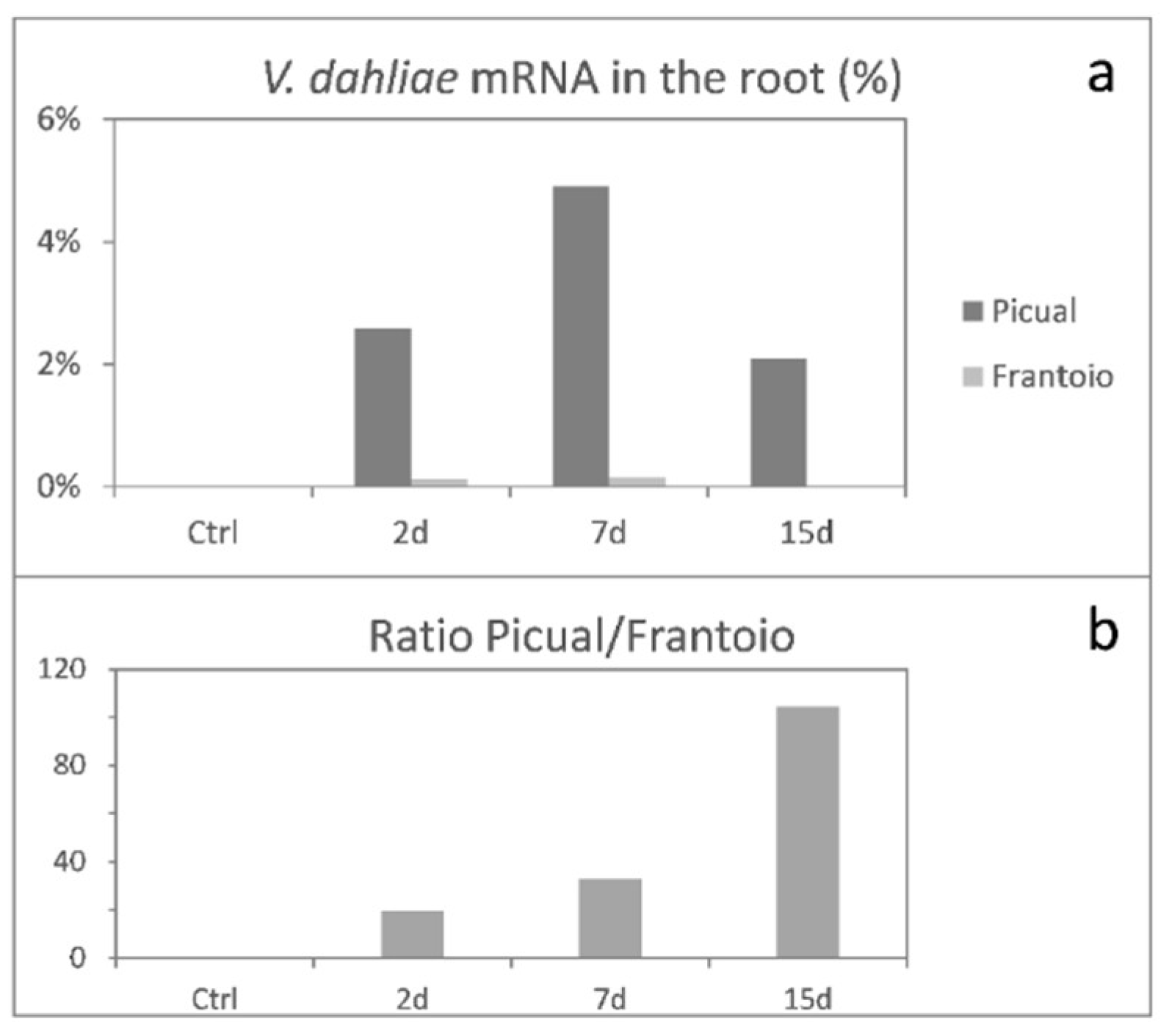
3.1. Global mRNA Amount of Verticillium Dahliae D pathotype is Strikingly Higher in ‘Picual’ (VWO-Susceptible) Than in ‘Frantoio’ (VWO-Tolerant) Roots
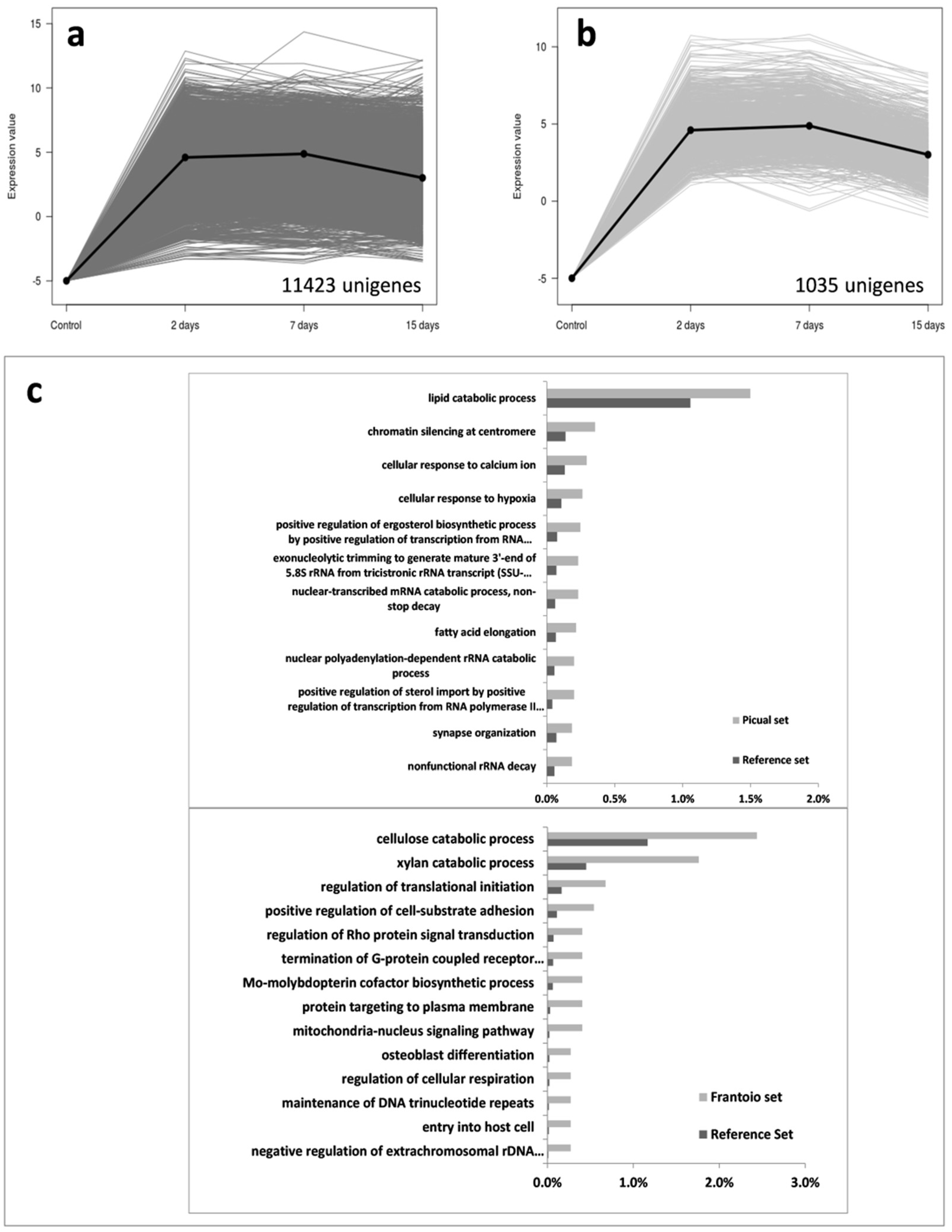
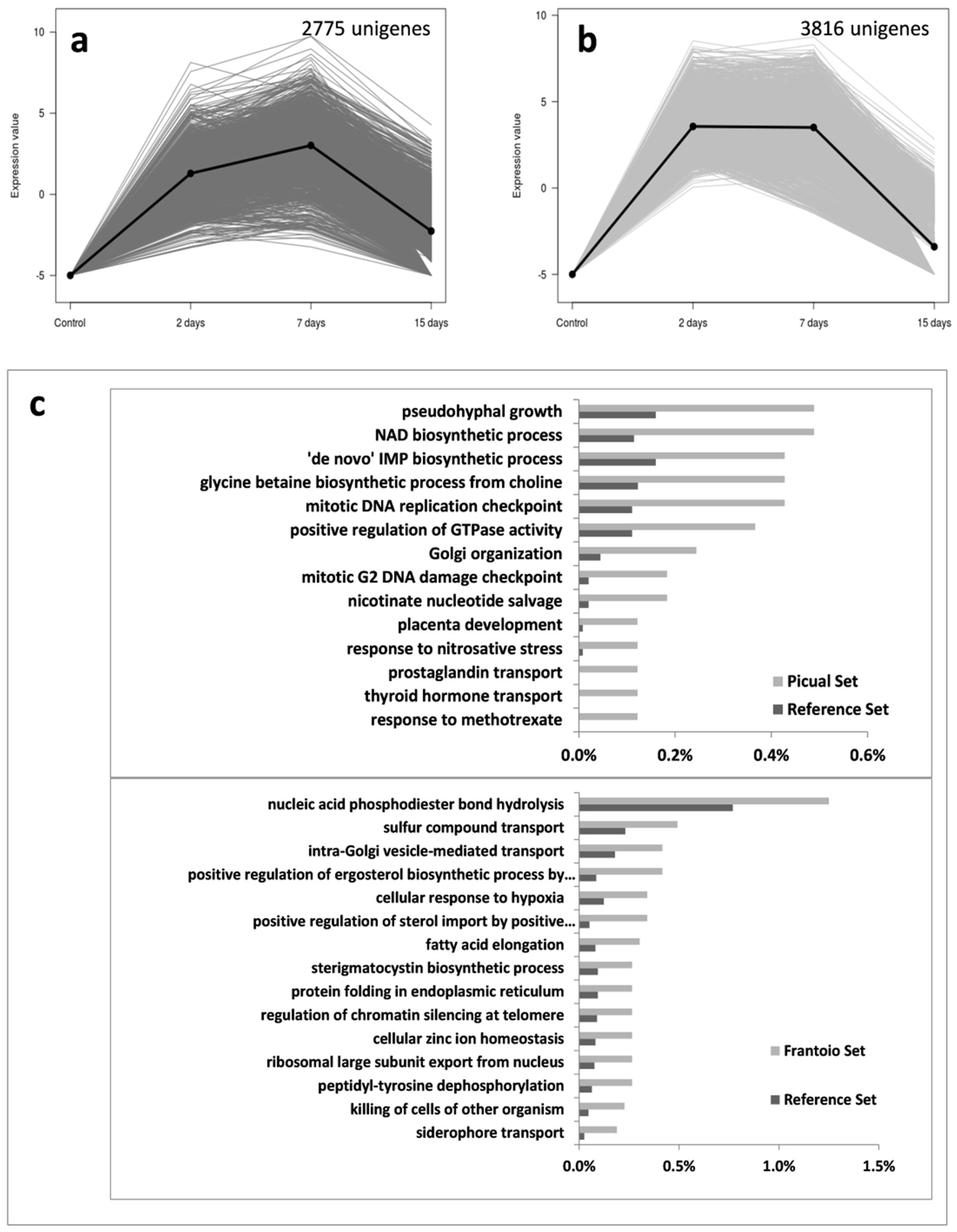
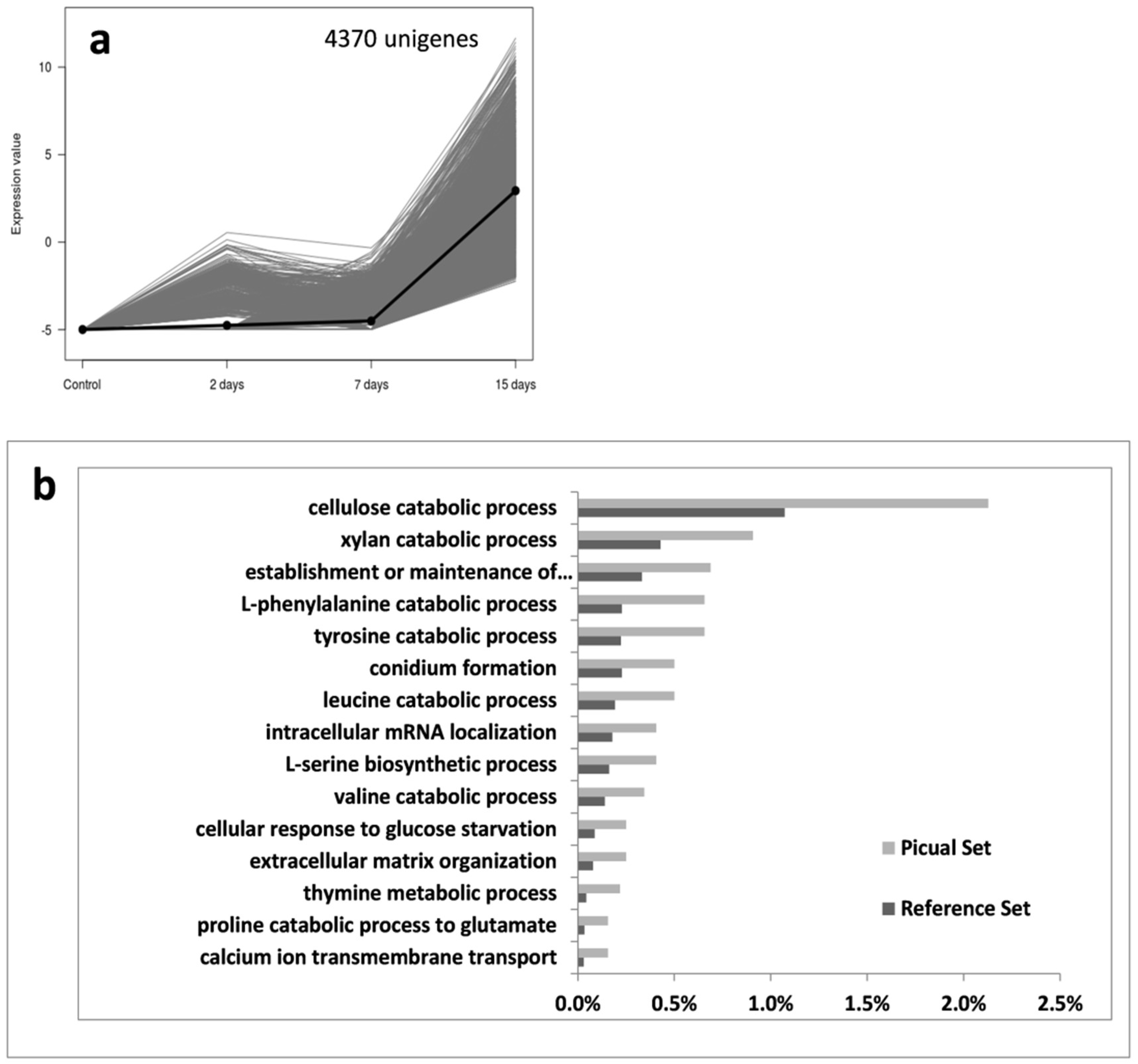
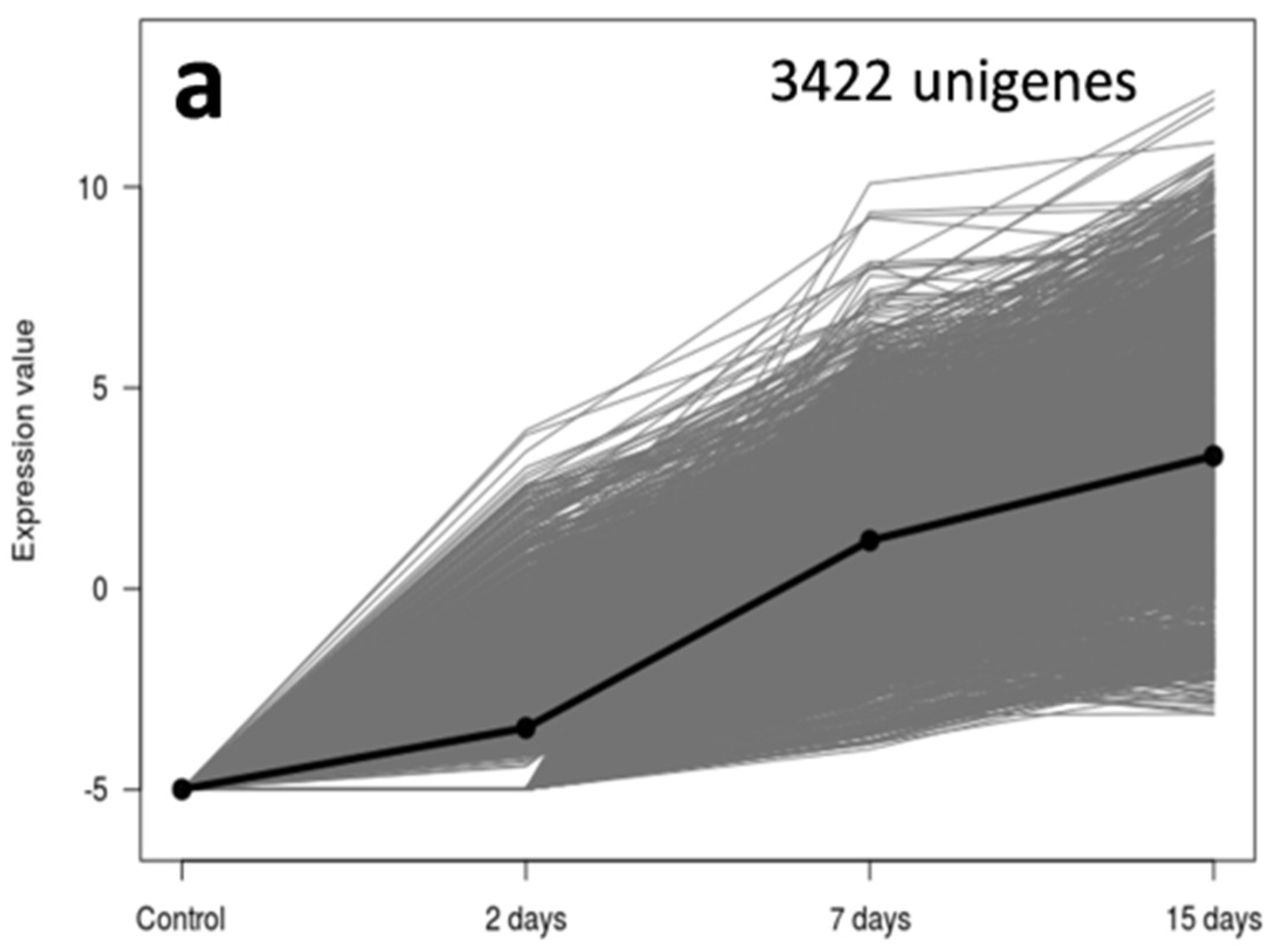
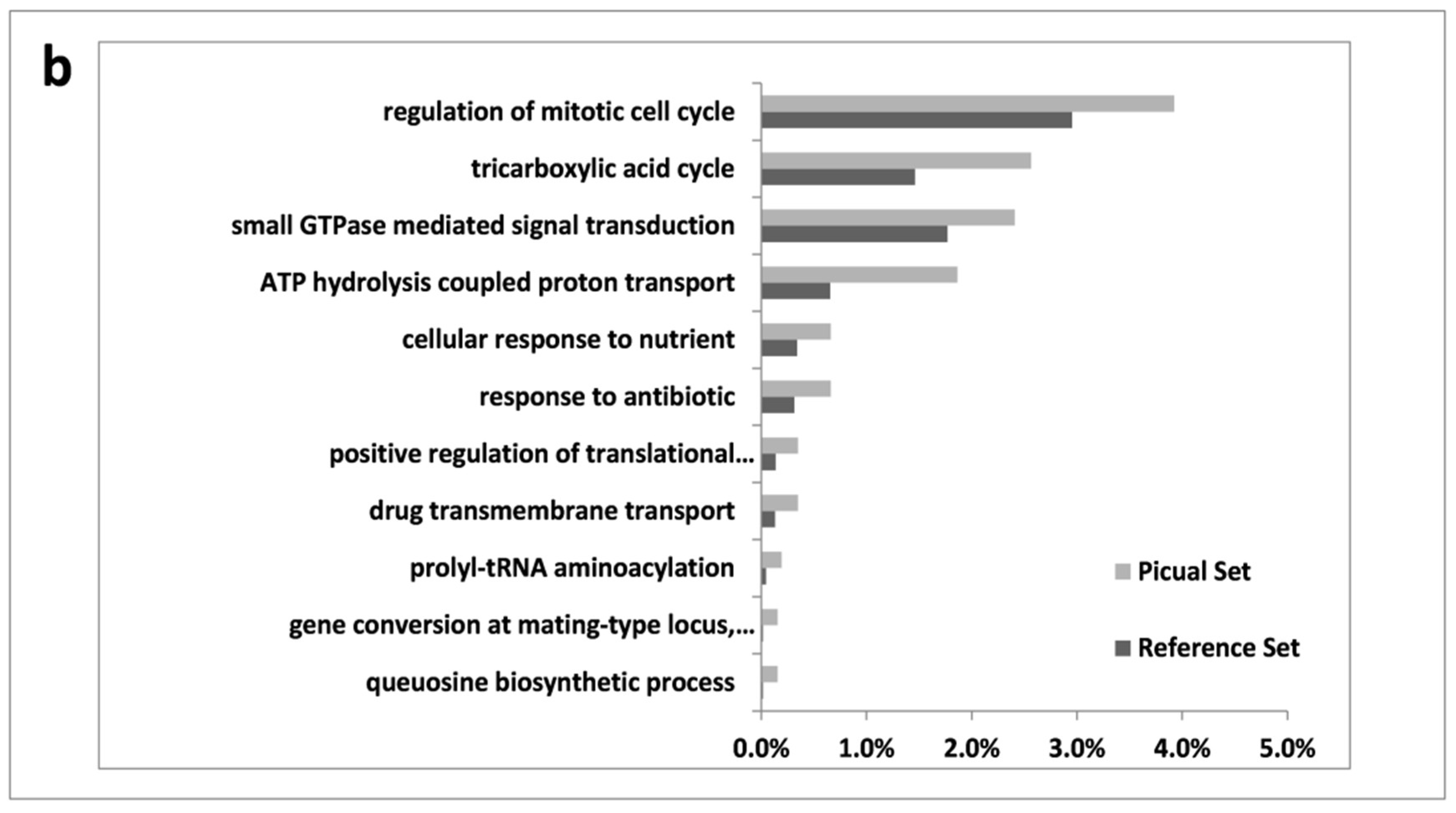
3.2. An Overwhelming Number of Verticillium Dahliae Unigenes Are Only Induced in Roots of the Susceptible Cultivar Picual
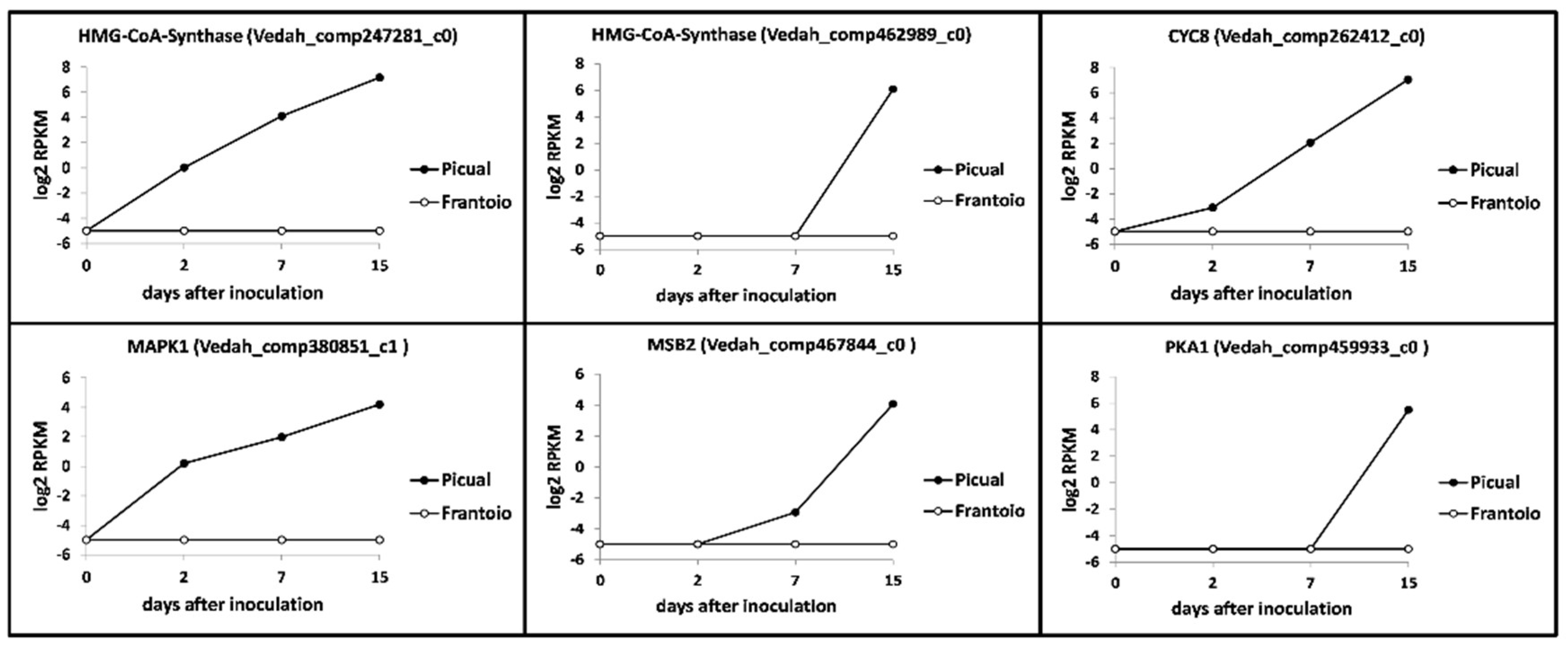
3.3. Targeting Verticillium Dahliae Niche Adaptation-Related Genes and Their Differential Expression Patterns in Cultivars Differing in VWO Susceptibility
3.4. Verticillium Dahliae Effector-Coding Unigenes Do Not Show Differential Expression Patterns in ‘Picual’ and ‘Frantoio’ Roots
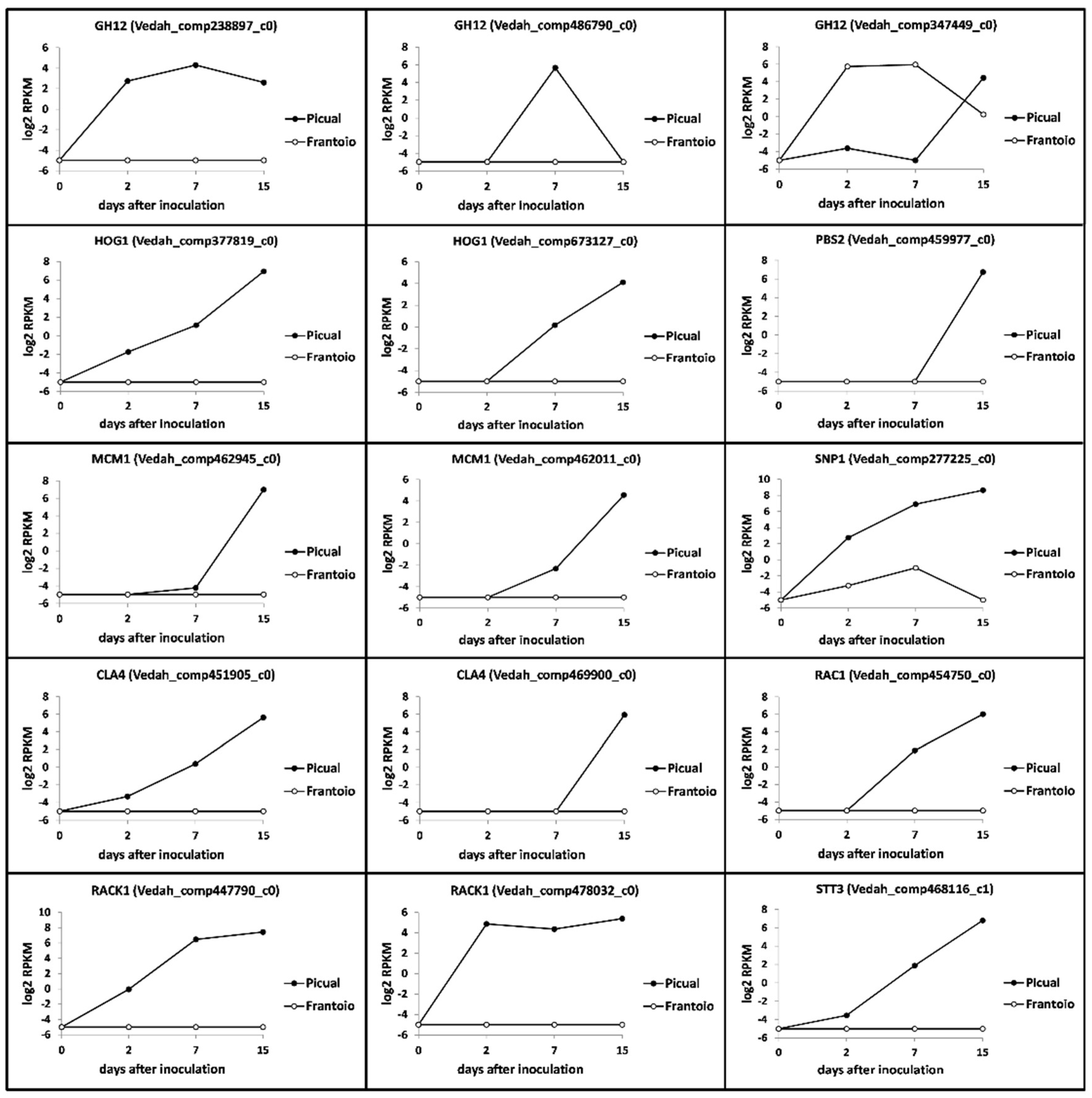
3.5. Verticillium Dahliae D pathotype Unigenes Involved in Pathogenicity, Virulence and Microsclerotia Development Showing Differential Expression Patterns
3.6. Multi-Task Verticillium Dahliae Unigenes Displaying Differential Expression Patterns
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fradin, E.F.; Thomma, B.P.H.J. Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol. Plant Pathol. 2006, 7, 71–86. [Google Scholar] [CrossRef]
- Klosterman, S.J.; Atallah, Z.K.; Vallad, G.E.; Subbarao, K.V. Diversity, pathogenicity and management of Verticillium species. Annu. Rev. Phytopathol. 2009, 47, 39–62. [Google Scholar] [CrossRef]
- Daayf, F. Verticillium wilts in crop plants: Pathogen invasion and host defense responses. Can. J. Plant Pathol. 2015, 37, 8–20. [Google Scholar] [CrossRef]
- Pegg, G.F.; Brady, B.L. Verticillium Wilts; CAB International: Wallingford, UK, 2002. [Google Scholar]
- Milgroom, M.G.; Jiménez-Gasco, M.M.; Olivares-García, C.; Jiménez-Díaz, R.M. Clonal expansion and migration of a highly virulent, defoliating lineage of Verticillium dahliae. Phytopathology 2016, 106, 1036–1046. [Google Scholar] [CrossRef]
- Dervis, S.; Mercado-Blanco, J.; Erten, L.; Valverde-Corredor, A.; Pérez-Artés, E. Verticillium wilt of olive in Turkey: A survey on disease importance, pathogen diversity and susceptibility of relevant olive cultivars. Eur. J. Plant Pathol. 2010, 127, 287–301. [Google Scholar] [CrossRef]
- López-Escudero, F.J.; Mercado-Blanco, J.; Roca, J.M.; Valverde-Corredor, A.; Blanco-López, M.A. Verticillium wilt of olive in the Guadalquivir Valley (southern Spain): Relations with some agronomical factors and spread of Verticillium dahliae. Phytopathol. Mediterr. 2010, 49, 370–380. [Google Scholar]
- Triki, M.A.; Krid, S.; Hsairi, H.; Hammemi, I.; Ioos, R.; Gdoura, R.; Rhouma, A. Occurrence of Verticillium dahliae defoliating pathotypes on olive trees in Tunisia. Phytopathol. Mediterr. 2011, 50, 267–272. [Google Scholar]
- López-Escudero, F.J.; Mercado-Blanco, J. Verticillium wilt of olive: A case study to implement an integrated strategy to control a soil-borne pathogen. Plant Soil 2011, 344, 1–50. [Google Scholar] [CrossRef]
- Prieto, P.; Navarro-Raya, C.; Valverde-Corredor, A.; Amyotte, S.G.; Dobinson, K.F.; Mercado-Blanco, J. Colonization process of olive tissues by Verticillium dahliae and its in planta interaction with the biocontrol root endophyte Pseudomonas fluorescens PICF7. Microb. Biotechnol. 2009, 2, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Calderón, R.; Lucena, C.; Trapero-Casas, J.L.; Zarco-Tejada, P.J.; Navas-Cortés, J.A. Soil temperature determines the reaction of olive cultivars to Verticillium dahliae pathotypes. PLoS ONE 2014, 9, e110664. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Lama Cabanás, C.; Schilirò, E.; Valverde-Corredor, A.; Mercado-Blanco, J. Systemic responses in a tolerant olive (Olea europaea L.) cultivar upon root colonization by the vascular pathogen Verticillium dahliae. Front. Microbiol. 2015, 6, 928. [Google Scholar]
- Baídez, A.G.; Gómez, P.; Del Rio, J.A.; Ortuño, A. Dysfunctionality of the xylem in Olea europaea L. plants associated with the infection process by Verticillium dahliae Kleb. Role of phenolic compounds in plant defense mechanism. J. Agric. Food Chem. 2007, 55, 3373–3377. [Google Scholar] [CrossRef] [PubMed]
- Markakis, E.A.; Tjamos, S.E.; Antoniou, P.P.; Roussos, P.A.; Paplomatas, E.J.; Tjamos, E.C. Phenolic responses of resistant and susceptible olive cultivars induced by defoliating and non-defoliating Verticillium dahliae pathotypes. Plant Dis. 2010, 94, 1156–1162. [Google Scholar] [CrossRef] [PubMed]
- Bubici, G.; Cirulli, M. Control of Verticillium wilt of olive by resistant rootstocks. Plant Soil 2012, 352, 363–376. [Google Scholar] [CrossRef]
- Gharbi, Y.; Barkallah, M.; Bouazizi, E.; Cheffi, M.; Gdoura, R.; Triki, M.A. Differential fungal colonization and physiological defense responses of new olive cultivars infected by the necrotrophic fungus Verticillium dahliae. Acta Physiol. Plant 2016, 38, 242. [Google Scholar] [CrossRef]
- Gharbi, Y.; Barkallah, M.; Bouazizi, E.; Hibar, K.; Gdoura, R.; Triki, M.A. Lignification, phenols accumulation, induction of PR proteins and antioxidant-related enzymes are key factors in the resistance of Olea europaea to Verticillium wilt of olive. Acta Physiol. Plant 2017, 39, 43. [Google Scholar] [CrossRef]
- Duressa, D.; Anchieta, A.; Chen, D.M.; Klimes, A.; García-Pedrajas, M.D.; Dobinson, K.F.; Klosterman, S.J. RNA-seq analyses of gene expression in the microsclerotia of Verticillium dahliae. BMC Genomics 2013, 14, 607. [Google Scholar] [CrossRef]
- Hu, D.; Wang, C.; Tao, F.; Cui, Q.; Xu, X.; Shang, W.; Hu, X. Whole genome wide expression profiles on germination of Verticillium dahliae microsclerotia. PLoS ONE 2014, 9, e100046. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Jiang, H.; Zhu, X.; Wang, W.; He, X.; Shi, Y.; Yuan, Y.; Du, X.; Cai, Y. Analysis of sea-island cotton and upland cotton in response to Verticillium dahliae infection by RNA sequencing. BMC Genomics 2013, 14, 852. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X.F.; Ding, Z.G.; Ma, Q.; Zhang, G.R.; Zhang, S.L.; Li, Z.K.; Wu, L.Q.; Zhang, G.Y.; Ma, Z.Y. Transcriptome profiling of Gossypium barbadense inoculated with Verticillium dahliae provides a resource for cotton improvement. BMC Genomics 2013, 14, 637. [Google Scholar] [CrossRef]
- Faino, L.; de Jonge, R.; Thomma, B.P.H.J. The transcriptome of Verticillium dahliae-infected Nicotiana benthamiana determined by deep RNA sequencing. Plant Signal. Behav. 2012, 7, 1065–1069. [Google Scholar] [CrossRef]
- Tan, G.; Liu, K.; Kang, J.; Xu, K.; Zhang, Y.; Hu, L.; Zhang, J.; Li, C. Transcriptome analysis of the compatible interaction of tomato with Verticillium dahliae using RNA-sequencing. Front. Plant Sci. 2015, 6, 428. [Google Scholar] [CrossRef]
- Jiménez-Ruiz, J.; de la O Leyva-Pérez, M.; Schilirò, E.; Barroso, J.B.; Bombarely, A.; Mueller, L.; Mercado-Blanco, J.; Luque, F. Transcriptomic analysis of the Olea europaea L. roots during the Verticillium dahliae early infection process. Plant Genome 2017, 10. [Google Scholar] [CrossRef]
- Leyva-Pérez, M.O.; Jiménez-Ruiz, J.; Gómez-Lama Cabanás, C.; Valverde-Corredor, A.; Barroso, J.B.; Luque, F.; Mercado-Blanco, J. Tolerance of olive (Olea europaea) cv Frantoio to Verticillium dahliae relies on both basal and pathogen-induced differential transcriptomic responses. New Phytol 2018, 217, 671–686. [Google Scholar] [CrossRef]
- Ajengui, A.; Bertolini, E.; Ligorio, A.; Chebil, S.; Ippolito, A.; Sanzani, S.M. Comparative transcriptome analysis of two citrus germplasms with contrasting susceptibility to Phytophthora nicotianae provides new insights into tolerance mechanisms. Plant Cell Rep. 2017, 37, 483–499. [Google Scholar] [CrossRef]
- Hu, Y.; Zhong, X.; Liu, X.; Lou, B.; Zhou, C.; Wang, X. Comparative transcriptome analysis unveils the tolerance mechanisms of Citrus hystrix in response to ‘Candidatus Liberibacter asiaticus’ infection. PLoS ONE 2017, 12, e0189229. [Google Scholar] [CrossRef]
- Matić, S.; Bagnaresi, P.; Biselli, C.; Orru’, L.; Carneiro, G.A.; Siciliano, I.; Valé, G.; Gullino, M.L.; Spadaro, D. Comparative transcriptome profiling of resistant and susceptible rice genotypes in response to the seedborne pathogen Fusarium fujikuroi. BMC Genomics 2016, 17, 608. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Wang, Y.; Yang, J.; Yang, W. Comparative transcriptome analysis of resistant and susceptible tomato Lines in response to infection by Xanthomonas perforans Race T3. Front. Plant Sci. 2015, 6, 1173. [Google Scholar] [CrossRef]
- Giampetruzzi, A.; Morelli, M.; Saponari, M.; Loconsole, G.; Chiumenti, M.; Boscia, D.; Savino, V.N.; Martelli, G.P.; Saldarelli, P. Transcriptome profiling of two olive cultivars in response to infection by the CoDiRO strain of Xylella fastidiosa subsp. pauca. BMC Genomics 2016, 17, 475. [Google Scholar] [CrossRef]
- Progar, V.; Jakše, J.; Štajner, N.; Radišek, S.; Javornik, B.; Berne, S. Comparative transcriptional analysis of hop responses to infection with Verticillium nonalfalfae. Plant Cell Rep. 2017, 36, 1599–1613. [Google Scholar] [CrossRef] [PubMed]
- Stergiopoulos, I.; de Wit, P.J.G.M. Fungal effector proteins. Annu. Rev. Phytopathol. 2009, 47, 233–263. [Google Scholar] [CrossRef]
- Xiong, D.; Wang, Y.; Tian, L.; Tian, C. MADS-Box transcription factor VdMcm1 regulates conidiation, microsclerotia formation, pathogenicity, and secondary metabolism of Verticillium dahliae. Front. Microbiol. 2016, 7, 1192. [Google Scholar] [CrossRef]
- Maldonado-González, M.M.; Bakker, P.A.H.M.; Prieto, P.; Mercado-Blanco, J. Arabidopsis thaliana as a tool to identify traits involved in Verticillium dahliae biocontrol by the olive root endophyte Pseudomonas fluorescens PICF7. Front. Microbiol. 2015, 6, 266. [Google Scholar] [CrossRef]
- Jiménez-Díaz, R.M.; Olivares-García, C.; Trapero-Casas, J.L.; Jiménez-Gasco, M.M.; Navas-Cortés, J.A.; Landa, B.B.; Milgroom, M.G. Variation of pathotypes and races and their correlations with clonal lineages in Verticillium dahliae. Plant Pathol. 2016, 66, 651–666. [Google Scholar] [CrossRef]
- Mercado-Blanco, J.; Rodríguez-Jurado, D.; Pérez-Artés, E.; Jiménez-Díaz, R.M. Detection of the defoliating pathotype of Verticillium dahliae in infected olive plants by nested PCR. Eur. J. Plant Pathol. 2002, 108, 1–13. [Google Scholar] [CrossRef]
- Prieto, P.; Mercado-Blanco, J. Endophytic colonization of olive roots by the biocontrol strain Pseudomonas fluorescens PICF7. FEMS Microbiol. Ecol. 2008, 64, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Aronesty, E. Ea-utils: “Command-Line Tools for Processing Biological Sequencing Data”. 2011. Available online: https://github.com/ExpressionAnalysis/ea-utils (accessed on 10 May 2013).
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Mercado-Blanco, J.; Collado-Romero, M.; Parrilla-Araujo, S.; Rodríguez-Jurado, D.; Jiménez-Díaz, R.M. Quantitative monitoring of colonization of olive genotypes by Verticillium dahliae pathotypes with real-time polymerase chain reaction. Physiol. Mol. Plant Pathol. 2003, 63, 91–105. [Google Scholar] [CrossRef]
- Markakis, E.A.; Tjamos, S.E.; Antoniou, P.P.; Paplomatas, E.J.; Tjamos, E.C. Symptom development, pathogen solation and Real-Time qPCR quantification as factors for evaluating the resistance of olive cultivars to Verticillium pathotypes. Eur. J. Plant Pathol. 2009, 124, 603–611. [Google Scholar] [CrossRef]
- Trapero, C.; Alcántara, E.; Jiménez, J.; Amaro-Ventura, M.C.; Romero, J.; Koopmann, B.; Karlovsky, P.; von Tiedemann, A.; Pérez-Rodríguez, M.; López-Escudero, F.J. Starch hydrolysis and vessel occlusion related to wilt symptoms in olive stems of susceptible cultivars infected by Verticillium dahliae. Front. Plant Sci. 2018, 9, 72. [Google Scholar] [CrossRef] [PubMed]
- Musungu, B.M.; Bhatnagar, D.; Brown, R.L.; Payne, G.A.; O’Brian, G.; Fakhoury, A.M.; Geisler, M. A network approach of gene co-expression in the Zea mays/Aspergillus flavus pathosystem to map host/pathogen interaction pathways. Front. Genet. 2016, 7, 206. [Google Scholar] [CrossRef] [PubMed]
- Lanver, D.; Muller, A.N.; Happel, P.; Schweizer, G.; Haas, F.B.; Franitza, M.; Pellegrin, C.; Reissmann, S.; Altmuller, J.; Rensing, S.A.; et al. The biotrophic development of Ustilago maydis studied by RNA-Seq analysis. Plant Cell 2018, 30, 300–323. [Google Scholar] [CrossRef] [PubMed]
- McClure, R.S.; Overall, C.C.; Hill, E.A.; Song, H.S.; Charania, M.; Bernstein, H.C.; McDermott, J.E.; Beliaev, A.S. Species-specific transcriptomic network inference of interspecies interactions. ISME J. 2018, 12, 2011–2023. [Google Scholar] [CrossRef] [PubMed]
- Nobori, T.; Velasquez, A.C.; Wu, J.; Kvitko, B.H.; Kremer, J.M.; Wang, Y.; He, S.Y.; Tsuda, K. Transcriptome landscape of a bacterial pathogen under plant immunity. Proc. Natl. Acad. Sci. USA 2018, 115, E3064. [Google Scholar] [CrossRef]
- Yu, J.; Paterson, N.; Blamey, J.; Millan, M. Cellulose, xylan and lignin interactions during pyrolysis of lignocellulosic biomass. Fuel 2017, 191, 140–149. [Google Scholar] [CrossRef]
- Sexton, A.C.; Paulsen, M.; Woestemeyer, J.; Howlett, B.J. Cloning, characterization and chromosomal location of three genes encoding host-cell-wall degrading enzymes in Leptosphaeria maculans, a fungal pathogen of Brassica spp. Gene 2000, 248, 89–97. [Google Scholar] [CrossRef]
- Eshel, D.; Miyara, I.; Ailing, T.; Dinoor, A.; Prusky, D. pH regulates endoglucanase expression and virulence of Alternaria alternata in persimmon fruit. Mol. Plant Microbe Interact. 2002, 15, 774–779. [Google Scholar] [CrossRef]
- Novo, M.; Pomar, F.; Gayoso, C.; Merino, F. Cellulase activity in isolates of Verticillium dahliae differing in aggressiveness. Plant Dis. 2006, 90, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Van Vu, B.; Itoh, K.; Nguyen, Q.B.; Tosa, Y.; Nakayashiki, H. Cellulases belonging to glycoside hydrolase families 6 and 7 contribute to the virulence of Magnaporthe oryzae. Mol. Plant Microbe Interact. 2012, 25, 1135–1141. [Google Scholar] [CrossRef]
- Gui, Y.-J.; Chen, J.-Y.; Zhang, D.-D.; Li, N.-Y.; Li, T.-G.; Zhang, W.Q.; Wang, X.Y.; Short, D.P.G.; Li, L.; Guo, W.; et al. Verticillium dahliae manipulates plant immunity by glycoside hydrolase 12 proteins in conjunction with carbohydrate-binding module 1. Environ. Microbiol. 2017, 19, 1914–1932. [Google Scholar] [CrossRef]
- Banfield, W.M. Distribution by the sap stream of spores of three fungi that induce vascular wilt diseases of elm. J. Agric. Res. 1941, 62, 637–681. [Google Scholar]
- Su, X.; Rehman, L.; Guo, H.; Li, X.; Cheng, H. The oligosaccharyl transferase subunit STT3 mediates fungal development and is required for virulence in Verticillium dahliae. Curr. Genet. 2018, 64, 235–246. [Google Scholar] [CrossRef]
- Klimes, A.; Dobinson, K.F.; Thomma, B.P.H.J.; Kosterman, S.J. Genomics spurs rapid advances in our understanding of the biology of vascular wilt pathogens in the genus Verticillium. Annu. Rev. Phytopathol. 2015, 53, 181–198. [Google Scholar] [CrossRef]
- Tzima, A.K.; Paplomatas, E.J.; Rauyaree, P.; Ospina-Giraldo, M.D.; Kang, S. VdSNF1, the sucrose nonfermenting protein kinase gene of Verticillium dahliae, is required for virulence and expression of genes involved in cell-wall degradation. Mol. Plant Microbe Interact. 2011, 24, 129–142. [Google Scholar] [CrossRef]
- Singh, S.; Braus-Stromeyer, S.A.; Timpner, C.; Tran, V.T.; Lohaus, G.; Reusche, M.; Knüfer, J.; Teichmann, T.; von Tiedemann, A.; Braus, G.H. Silencing of Vlaro2 for chorismate synthase revealed that the phytopathogen Verticillium longisporum induces the cross-pathway control in the xylem. Appl. Microbiol. Biotechnol. 2010, 85, 1961–1976. [Google Scholar] [CrossRef]
- Flajsman, M.; Mandelc, S.; Radisek, S.; Stajner, N.; Jakse, J.; Kosmelj, K.; Javornik, B. Identification of novel virulence-associated proteins secreted to xylem by Verticillium nonalfalfae during colonization of hop plants. Mol. Plant Microbe Interact. 2016, 29, 362–373. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, Y.; Liang, Y.; Dong, Y.; Yang, X.; Yuan, J.; Qiu, D. The Verticillium dahliae SnodProt1-like protein VdCP1 contributes to virulence and triggers the plant immune system. Front. Plant Sci. 2017, 8, 1880. [Google Scholar] [CrossRef]
- Tian, L.; Xu, J.; Zhou, L.; Guo, W. VdMsb regulates virulence and microsclerotia production in the fungal plant pathogen Verticillium dahliae. Gene 2014, 550, 238–244. [Google Scholar] [CrossRef]
- Tzima, A.; Paplomatas, E.J.; Rauyaree, P.; Kang, S. Roles of the catalytic subunit of cAMP-dependent protein kinase a in virulence and development of the soilborne plant pathogen Verticillium dahliae. Fungal Genet. Biol. 2010, 47, 406–415. [Google Scholar] [CrossRef]
- Li, Z.-F.; Liu, Y.-J.; Feng, Z.-L.; Feng, H.-J.; Klosterman, S.J.; Zhou, F.-F.; Zhao, L.H.; Shi, Y.Q.; Zhu, H.Q. VdCYC8, encoding CYC8 glucose repression mediator protein, is required for microsclerotia formation and full virulence in Verticillium dahliae. PLoS ONE 2015, 10, e0144020. [Google Scholar] [CrossRef]
- Tian, L.; Wang, Y.; Yu, J.; Xiong, D.; Zhao, H.; Tian, C. The mitogen-activated protein kinase kinase VdPbs2 of Verticillium dahliae regulates microsclerotia formation, stress response, and plant infection. Front. Microbiol. 2016, 7, 1532. [Google Scholar] [CrossRef]
- Wang, Y.; Tian, L.; Xiong, D.; Klosterman, S.J.; Xiao, S.; Tian, C. The mitogen-activated protein kinase gene, VdHog1, regulates osmotic stress response, microsclerotia formation and virulence in Verticillium dahliae. Fungal Genet. Biol. 2016, 88, 13–23. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiménez-Ruiz, J.; Leyva-Pérez, M.d.l.O.; Gómez-Lama Cabanás, C.; Barroso, J.B.; Luque, F.; Mercado-Blanco, J. The Transcriptome of Verticillium dahliae Responds Differentially Depending on the Disease Susceptibility Level of the Olive (Olea europaea L.) Cultivar. Genes 2019, 10, 251. https://doi.org/10.3390/genes10040251
Jiménez-Ruiz J, Leyva-Pérez MdlO, Gómez-Lama Cabanás C, Barroso JB, Luque F, Mercado-Blanco J. The Transcriptome of Verticillium dahliae Responds Differentially Depending on the Disease Susceptibility Level of the Olive (Olea europaea L.) Cultivar. Genes. 2019; 10(4):251. https://doi.org/10.3390/genes10040251
Chicago/Turabian StyleJiménez-Ruiz, Jaime, María de la O Leyva-Pérez, Carmen Gómez-Lama Cabanás, Juan B. Barroso, Francisco Luque, and Jesús Mercado-Blanco. 2019. "The Transcriptome of Verticillium dahliae Responds Differentially Depending on the Disease Susceptibility Level of the Olive (Olea europaea L.) Cultivar" Genes 10, no. 4: 251. https://doi.org/10.3390/genes10040251
APA StyleJiménez-Ruiz, J., Leyva-Pérez, M. d. l. O., Gómez-Lama Cabanás, C., Barroso, J. B., Luque, F., & Mercado-Blanco, J. (2019). The Transcriptome of Verticillium dahliae Responds Differentially Depending on the Disease Susceptibility Level of the Olive (Olea europaea L.) Cultivar. Genes, 10(4), 251. https://doi.org/10.3390/genes10040251







