Tubulin-Based DNA Barcode: Principle and Applications to Complex Food Matrices
Abstract
:1. Introduction
- Along the food chain, species identification for raw material purity assessment and food authentication is fundamental to grant products identity and quality and to protect consumers from adulterations and fraud;
- Species identification by DNA analyses are becoming widely accepted in the food sector and now many analytical tools are available;
- Among different methods, DNA barcoding has become increasingly popular and metabarcoding has attracted attention for its applications to complex food matrices.
2. Barcoding by Sequencing
- Bacteria—16S rRNA is the indisputable gold standard for microbial phylogenesis.
- Animals—The mitochondrial gene for the subunit 1 of cytochrome oxydase (COI or Cox1) is the generally accepted marker for almost all animal species. A huge number of primers have been designed for the amplification of COI from various animal groups (1016 COI primers in the BOLsystem database, accessed on 23 February 2019). ‘Universal’ primers amplifying the COI barcode region have also been described, but in silico analysis shows that they are poorly conserved [12]. For historical reasons, the mitochondrial gene encoding cytochrome b (cytb) is still used in the food sector, particularly for game meat [13].
- Plants—It is now accepted that in plants, universal species discrimination may never be possible with a single locus-based approach. Neither plastid data alone, nor in combination with information obtained from the nuclear genome will be able to cover all plant species. The combination of the two plastid markers, ribulose 1,5-bisphosphate carboxylase gene (rbcL) and maturase K (matK), accepted as the core barcoding regions [14], do not grant a suitable coverage of plant species and must be often implemented with the use of other hypervariable sequences, mainly the plastid interspacer region trnH-psbA and/or the nuclear ITS [10,15]. Also, the plastid trnL intron have been largely applied to plant species identification [16,17]. Other plastid spacer sequences are often used for problematic taxa. Several studies have shown that about 75%–85% of plant species, up to more than 90% in some floras, can be identified at species level using a DNA barcoding approach that is based on these different combinations of markers [15].
3. General Issues about Metabarcoding: Power, Applications and Limits
Food Metabarcoding
4. Barcoding by Fragment Analysis: When Less Is More
4.1. Species Identification by Insertions/Deletions (SpinDel) Profiling
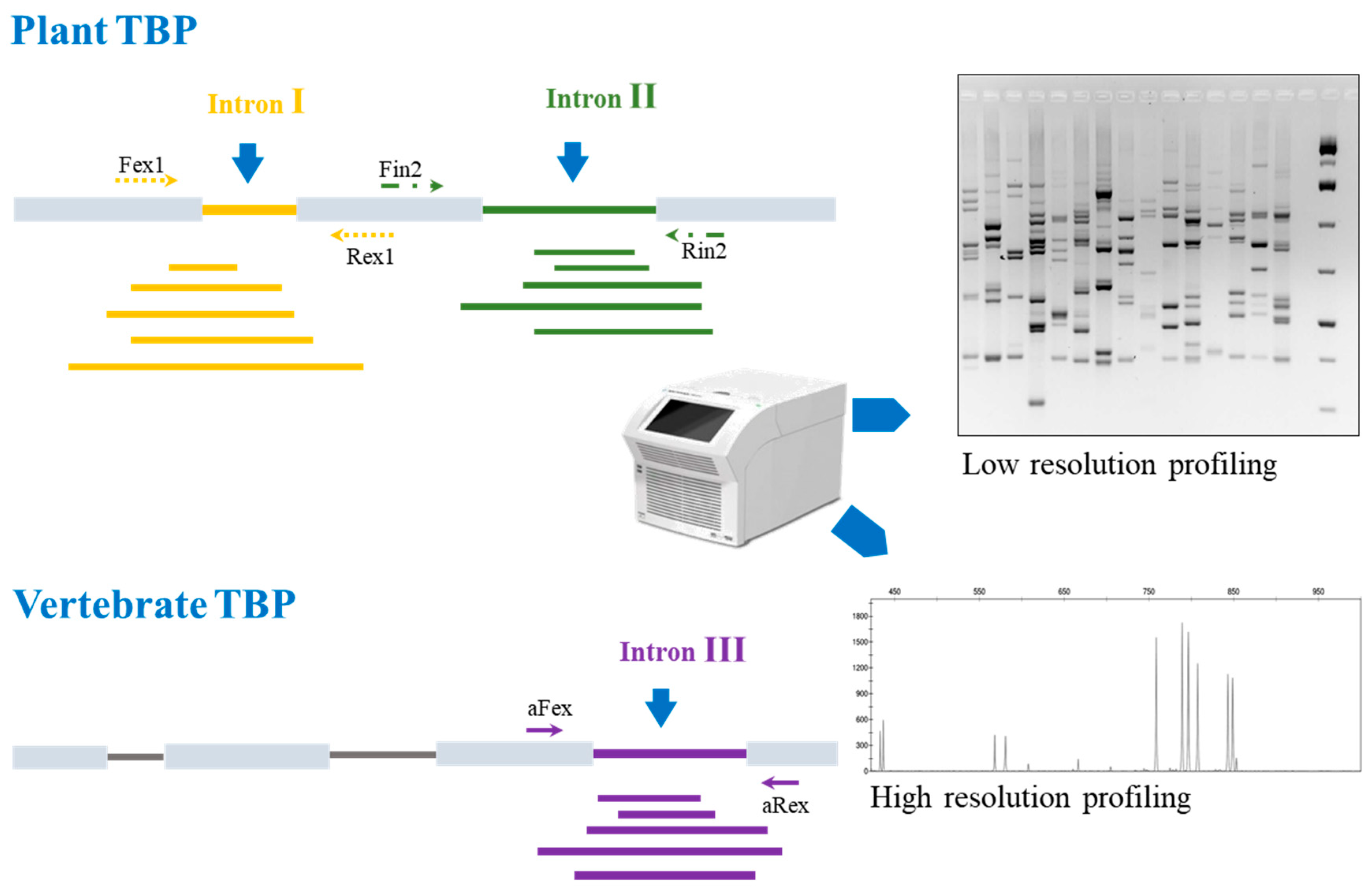
4.2. Tubulin-Based Polymorphism (TBP) Barcoding: The Principle
4.3. Reference Database and Bionformatic Needs
5. TBP Application’s Survey
5.1. Plant Species Identification and Raw Material Purity
5.2. Feed Composition by TBP Metabarcoding
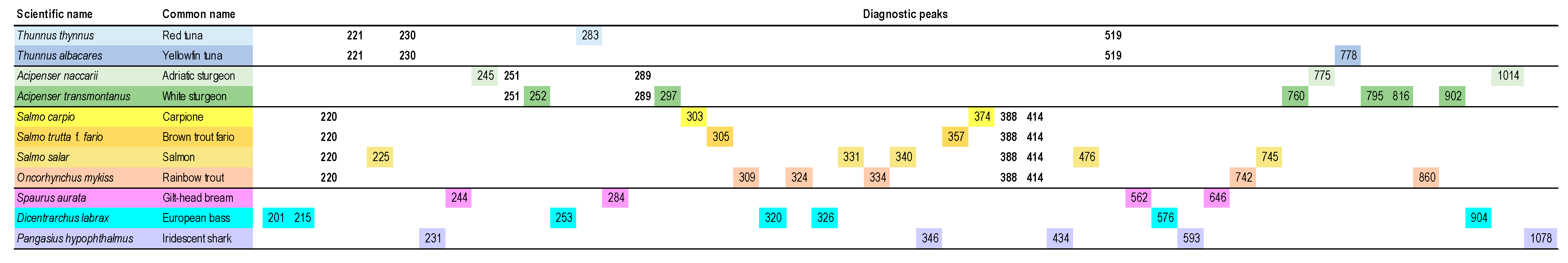
5.3. Meat and Fish Food Authentication
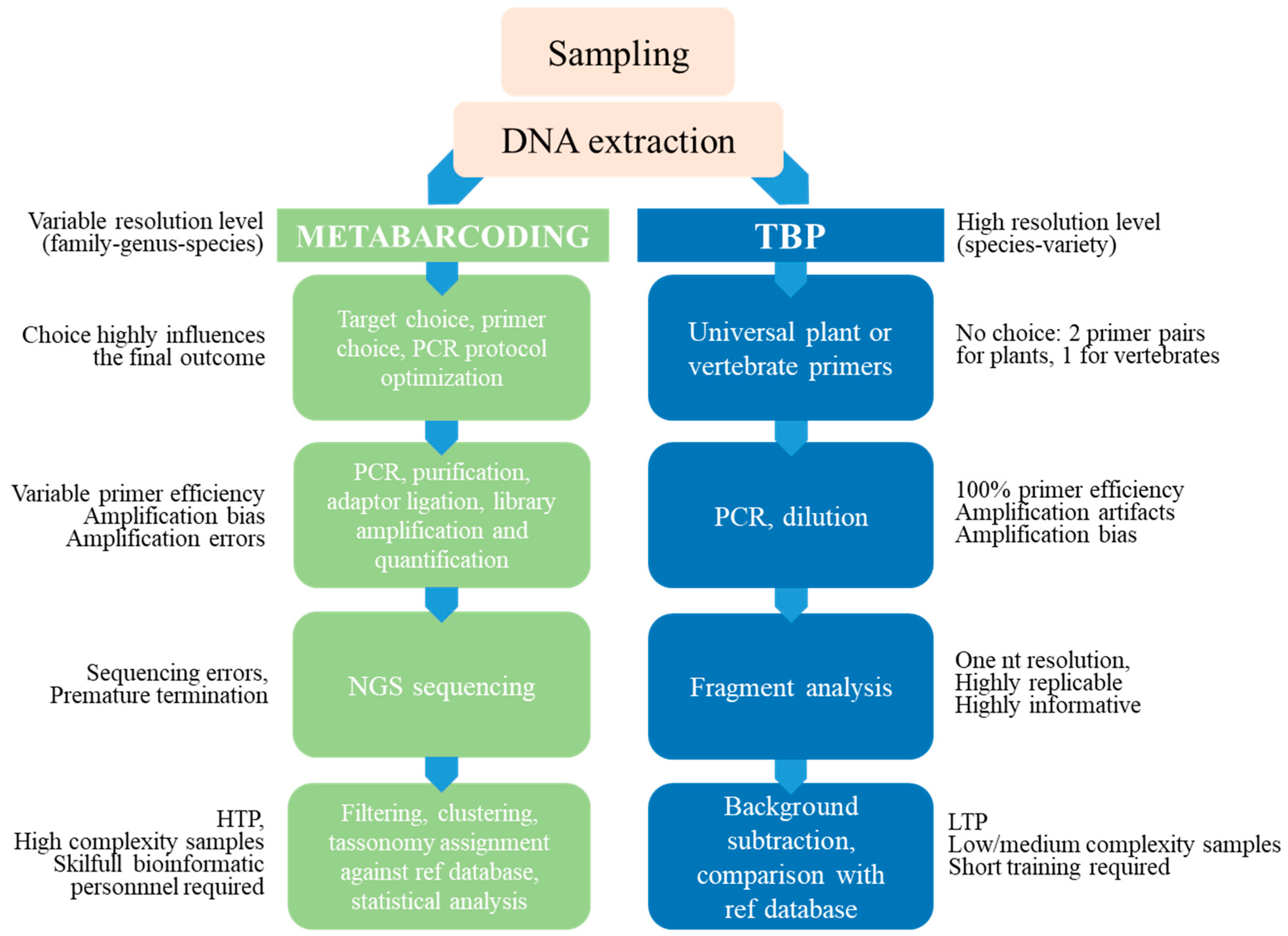
6. Authenticity Testing along the Food Chain: Advantages and Limits of Sequence-Based and Fragment-Based DNA Metabarcoding
7. Patents
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Woese, C.R.; Fox, G.E. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc. Natl. Acad. Sci. USA 1977, 74, 5088–5090. [Google Scholar] [CrossRef]
- Hebert, P.D.; Cywinska, A.; Ball, S.L.; de Waard, J.R. Biological identifications through DNA barcodes. Proc. Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef]
- International Barcode of Life. Available online: https://ibol.org (accessed on 25 February 2019).
- Casiraghi, M.; Labra, M.; Ferri, E.; Galimberti, A.; De Mattia, F. DNA barcoding: A six-question tour to improve users’ awareness about the method. Brief. Bioinform. 2010, 11, 440–453. [Google Scholar] [CrossRef] [PubMed]
- Collins, R.A.; Cruickshank, R.H. The seven deadly sins of DNA barcoding. Mol. Ecol. Resour. 2013, 13, 969–975. [Google Scholar] [CrossRef]
- Taylor, H.R.; Harris, W.E. An emergent science on the brink of irrelevance: A review of the past 8 years of DNA barcoding. Mol. Ecol. Resour. 2012, 12, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Kress, W.J.; García-Robledo, C.; Uriarte, M.; Erickson, D.L. DNA barcodes for ecology, evolution, and conservation. Trends Ecol. Evol. 2015, 30, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Staats, M.; Arulandhu, A.J.; Gravendeel, B.; Holst-Jensen, A.; Scholtens, I.; Peelen, T.; Prins, T.W.; Kok, E. Advances in DNA metabarcoding for food and wildlife forensic species identification. Anal. Bioanal. Chem. 2016, 408, 4615–4630. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Consortium, F.B.; List, F.B.C.A. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Hollingsworth, P.; Graham, S.; Little, D. Choosing and Using a Plant DNA Barcode. PLoS ONE 2011, 6. [Google Scholar] [CrossRef]
- Wang, X.C.; Liu, C.; Huang, L.; Bengtsson-Palme, J.; Chen, H.; Zhang, J.H.; Cai, D.; Li, J.Q. ITS1: A DNA barcode better than ITS2 in eukaryotes? Mol. Ecol. Resour. 2015, 15, 573–586. [Google Scholar] [CrossRef]
- Deagle, B.; Jarman, S.; Coissac, E.; Pompanon, F.; Taberlet, P. DNA metabarcoding and the cytochrome c oxidase subunit I marker: Not a perfect match. Biol. Lett. 2014, 10. [Google Scholar] [CrossRef]
- D’Amato, M.E.; Alechine, E.; Cloete, K.W.; Davison, S.; Corach, D. Where is the game? Wild meat products authentication in South Africa: A case study. Investig. Genet. 2013, 4, 6. [Google Scholar] [CrossRef]
- Group, C.P.W. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar] [CrossRef]
- Kress, W. Plant DNA barcodes: Applications today and in the future. J. Syst. Evol. 2017, 55, 291–307. [Google Scholar] [CrossRef]
- James, D.; Schmidt, A. Use of an intron region of a chloroplast tRNA gene (trnL) as a target for PCR identification of specific food crops including sources of potential allergens. Food Res. Int. 2004, 37, 395–402. [Google Scholar] [CrossRef]
- Taberlet, P.; Coissac, E.; Pompanon, F.; Gielly, L.; Miquel, C.; Valentini, A.; Vermat, T.; Corthier, G.; Brochmann, C.; Willerslev, E. Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res. 2007, 35. [Google Scholar] [CrossRef]
- Coissac, E.; Hollingsworth, P.M.; Lavergne, S.; Taberlet, P. From barcodes to genomes: Extending the concept of DNA barcoding. Mol. Ecol. 2016, 25, 1423–1428. [Google Scholar] [CrossRef]
- Hollingsworth, P.M.; Li, D.Z.; van der Bank, M.; Twyford, A.D. Telling plant species apart with DNA: From barcodes to genomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016, 371. [Google Scholar] [CrossRef]
- Deiner, K.; Bik, H.M.; Mächler, E.; Seymour, M.; Lacoursière-Roussel, A.; Altermatt, F.; Creer, S.; Bista, I.; Lodge, D.M.; de Vere, N.; et al. Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Mol. Ecol. 2017, 26, 5872–5895. [Google Scholar] [CrossRef]
- Abdelfattah, A.; Malacrinò, A.; Wisniewski, M.; Cacciola, S.O.; Schena, L. Metabarcoding: A powerful tool to investigate microbial communities and shape future plant protection strategies. Biol. Control 2017, 120, 1–10. [Google Scholar]
- Taberlet, P.; Coissac, E.; Pompanon, F.; Brochmann, C.; Willerslev, E. Towards next-generation biodiversity assessment using DNA metabarcoding. Mol. Ecol. 2012, 21, 2045–2050. [Google Scholar] [CrossRef]
- Aylagas, E.; Borja, A.; Irigoien, X.; Rodriguez-Ezpeleta, N. Benchmarking DNA Metabarcoding for Biodiversity-Based Monitoring and Assessment. Front. Mar. Sci. 2016, 3. [Google Scholar] [CrossRef]
- Fahner, N.A.; Shokralla, S.; Baird, D.J.; Hajibabaei, M. Large-Scale Monitoring of Plants through Environmental DNA Metabarcoding of Soil: Recovery, Resolution, and Annotation of Four DNA Markers. PLoS ONE 2016, 11, e0157505. [Google Scholar] [CrossRef]
- Pompanon, F.; Deagle, B.; Symondson, W.; Brown, D.; Jarman, S.; Taberlet, P. Who is eating what: Diet assessment using next generation sequencing. Mol. Ecol. 2012, 21, 1931–1950. [Google Scholar] [CrossRef]
- Moorhouse-Gann, R.J.; Dunn, J.C.; de Vere, N.; Goder, M.; Cole, N.; Hipperson, H.; Symondson, W.O.C. New universal ITS2 primers for high-resolution herbivory analyses using DNA metabarcoding in both tropical and temperate zones. Sci. Rep. 2018, 8, 8542. [Google Scholar] [CrossRef]
- Elbrecht, V.; Leese, F. Can DNA-Based Ecosystem Assessments Quantify Species Abundance? Testing Primer Bias and Biomass—Sequence Relationships with an Innovative Metabarcoding Protocol. PLoS ONE 2015, 10, e0130324. [Google Scholar] [CrossRef]
- Tillmar, A.; Dell’Amico, B.; Welander, J.; Holmlund, G. A Universal Method for Species Identification of Mammals Utilizing Next Generation Sequencing for the Analysis of DNA Mixtures. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Soininen, E.; Valentini, A.; Coissac, E.; Miquel, C.; Gielly, L.; Brochmann, C.; Brysting, A.; Sonstebo, J.; Ims, R.; Yoccoz, N.; et al. Analysing diet of small herbivores: The efficiency of DNA barcoding coupled with high-throughput pyrosequencing for deciphering the composition of complex plant mixtures. Front. Zool. 2009, 6. [Google Scholar] [CrossRef]
- Valentini, A.; Miquel, C.; Nawaz, M.A.; Bellemain, E.; Coissac, E.; Pompanon, F.; Gielly, L.; Cruaud, C.; Nascetti, G.; Wincker, P.; et al. New perspectives in diet analysis based on DNA barcoding and parallel pyrosequencing: The trnL approach. Mol. Ecol. Resour. 2009, 9, 51–60. [Google Scholar] [CrossRef]
- Cheng, X.; Su, X.; Chen, X.; Zhao, H.; Bo, C.; Xu, J.; Bai, H.; Ning, K. Biological ingredient analysis of traditional Chinese medicine preparation based on high-throughput sequencing: The story for Liuwei Dihuang Wan. Sci. Rep. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Coissac, E.; Riaz, T.; Puillandre, N. Bioinformatic challenges for DNA metabarcoding of plants and animals. Mol. Ecol. 2012, 21, 1834–1847. [Google Scholar] [CrossRef]
- Galimberti, A.; Labra, M.; Sandionigi, A.; Bruno, A.; Mezzasalma, V.; DeMattia, F. DNA Barcoding for Minor Crops and Food Traceability. Adv. Agric. 2014, 2014, 831875. [Google Scholar] [CrossRef]
- Ferri, E.; Galimberti, A.; Casiraghi, M.; Airoldi, C.; Ciaramelli, C.; Palmioli, A.; Mezzasalma, V.; Bruni, I.; Labra, M. Towards a Universal Approach Based on Omics Technologies for the Quality Control of Food. Biomed. Res. Int. 2015. [Google Scholar] [CrossRef]
- Barcaccia, G.; Lucchin, M.; Cassandro, M. DNA Barcoding as a Molecular Tool to Track Down Mislabeling and Food Piracy. Diversity 2016, 8, 2. [Google Scholar] [CrossRef]
- Littlefair, J.; Clare, E. Barcoding the food chain: From Sanger to high-throughput sequencing. Genome 2016, 59, 946–958. [Google Scholar] [CrossRef] [PubMed]
- Coghlan, M.; Haile, J.; Houston, J.; Murray, D.; White, N.; Moolhuijzen, P.; Bellgard, M.; Bunce, M. Deep Sequencing of Plant and Animal DNA Contained within Traditional Chinese Medicines Reveals Legality Issues and Health Safety Concerns. PLoS Genet. 2012, 8, 436–446. [Google Scholar] [CrossRef]
- Raclariu, A.C.; Paltinean, R.; Vlase, L.; Labarre, A.; Manzanilla, V.; Ichim, M.C.; Crisan, G.; Brysting, A.K.; de Boer, H. Comparative authentication of Hypericum perforatum herbal products using DNA metabarcoding, TLC and HPLC-MS. Sci. Rep. 2017, 7, 1291. [Google Scholar] [CrossRef] [PubMed]
- Raclariu, A.C.; Mocan, A.; Popa, M.O.; Vlase, L.; Ichim, M.C.; Crisan, G.; Brysting, A.K.; de Boer, H. Product Authentication Using DNA Metabarcoding and HPLC-MS Reveals Widespread Adulteration with Veronica chamaedrys. Front. Pharmacol. 2017, 8, 378. [Google Scholar] [CrossRef]
- Carvalho, D.; Palhares, R.; Drummond, M.; Gadanho, M. Food metagenomics: Next generation sequencing identifies species mixtures and mislabeling within highly processed cod products. Food Control 2017, 80, 183–186. [Google Scholar] [CrossRef]
- Kappel, K.; Haase, I.; Kappel, C.; Sotelo, C.; Schroder, U. Species identification in mixed tuna samples with next-generation sequencing targeting two short cytochrome b gene fragments. Food Chem. 2017, 234, 212–219. [Google Scholar] [CrossRef]
- Giusti, A.; Tinacci, L.; Sotelo, C.; Marchetti, M.; Guidi, A.; Zheng, W.; Armani, A. Seafood Identification in Multispecies Products: Assessment of 16SrRNA, cytb, and COI Universal Primers’ Efficiency as a Preliminary Analytical Step for Setting up Metabarcoding Next-Generation Sequencing Techniques. J. Agric. Food Chem. 2017, 65, 2902–2912. [Google Scholar] [CrossRef]
- Bertolini, F.; Ghionda, M.; D’Alessandro, E.; Geraci, C.; Chiofalo, V.; Fontanesi, L. A Next Generation Semiconductor Based Sequencing Approach for the Identification of Meat Species in DNA Mixtures. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Dobrovolny, S.; Blaschitz, M.; Weinmaier, T.; Pechatschek, J.; Cichna-Markl, M.; Indra, A.; Hufnagl, P.; Hochegger, R. Development of a DNA metabarcoding method for the identification of fifteen mammalian and six poultry species in food. Food Chem. 2019, 272, 354–361. [Google Scholar] [CrossRef]
- Ripp, F.; Krombholz, C.; Liu, Y.; Weber, M.; Schafer, A.; Schmidt, B.; Koppel, R.; Hankeln, T. All-Food-Seq (AFS): A quantifiable screen for species in biological samples by deep DNA sequencing. BMC Genom. 2014, 15. [Google Scholar] [CrossRef] [PubMed]
- Mueller, U.G.; Wolfenbarger, L.L. AFLP genotyping and fingerprinting. Trends Ecol. Evol. 1999, 14, 389–394. [Google Scholar] [CrossRef]
- Lo, Y.; Shaw, P. DNA-based techniques for authentication of processed food and food supplements. Food Chem. 2018, 240, 767–774. [Google Scholar] [CrossRef]
- Zhao, X.; Yang, L.; Zheng, Y.; Xu, Z.; Wu, W. Subspecies-specific intron length polymorphism markers reveal clear genetic differentiation in common wild rice (Oryza rufipogon L.) in relation to the domestication of cultivated rice (O. sativa L.). J. Genet. Genom. 2009, 36, 435–442. [Google Scholar] [CrossRef]
- Poczai, P.; Cernák, I.; Gorji, A.M.; Nagy, S.; Taller, J.; Polgár, Z. Development of intron targeting (IT) markers for potato and cross-species amplification in Solanum nigrum (Solanaceae). Am. J. Bot. 2010, 97, e142–e145. [Google Scholar] [CrossRef] [PubMed]
- Mafra, I.; Ferreira, I.; Oliveira, M. Food authentication by PCR-based methods. Eur. Food Res. Technol. 2008, 227, 649–665. [Google Scholar] [CrossRef]
- Druml, B.; Cichna-Markl, M. High resolution melting (HRM) analysis of DNA-its role and potential in food analysis. Food Chem. 2014, 158, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Bardini, M.; Lee, D.; Donini, P.; Mariani, A.; Giani, S.; Toschi, M.; Lowe, C.; Breviario, D. Tubulin-based polymorphism (TBP): A new tool, based on functionally relevant sequences, to assess genetic diversity in plant species. Genome 2004, 47, 281–291. [Google Scholar] [CrossRef]
- Pereira, F.; Carneiro, J.; Matthiesen, R.; van Asch, B.; Pinto, N.; Gusmão, L.; Amorim, A. Identification of species by multiplex analysis of variable-length sequences. Nucleic Acids Res. 2010, 38, e203. [Google Scholar] [CrossRef] [PubMed]
- SPInDel, Species Identification by Insertions/Delections. Available online: http://www.portugene.com/SPInDel/SPInDel_web.html (accessed on 25 February 2019).
- Carneiro, J.; Pereira, F.; Amorim, A. SPInDel: A multifunctional workbench for species identification using insertion/deletion variants. Mol. Ecol. Resour. 2012, 12, 1190–1195. [Google Scholar] [CrossRef]
- Santos, C.; Pereira, F. Identification of plant species using variable length chloroplast DNA sequences. Forensic Sci. Int. Genet. 2018, 36, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Findeisen, P.; Mühlhausen, S.; Dempewolf, S.; Hertzog, J.; Zietlow, A.; Carlomagno, T.; Kollmar, M. Six subgroups and extensive recent duplications characterize the evolution of the eukaryotic tubulin protein family. Genome Biol. Evol. 2014, 6, 2274–2288. [Google Scholar] [CrossRef] [PubMed]
- Silletti, S.; Morello, L.; Gavazzi, F.; Gianì, S.; Braglia, L.; Breviario, D. Untargeted DNA-based methods for the authentication of wheat species and related cereals in food products. Food Chem. 2019, 271, 410–418. [Google Scholar] [CrossRef]
- Wyler, S.; Naciri, Y. Evolutionary histories determine DNA barcoding success in vascular plants: Seven case studies using intraspecific broad sampling of closely related species. BMC Evolut. Biol. 2016, 16. [Google Scholar] [CrossRef]
- Gavazzi, F.; Braglia, L.; Mastromauro, F.; Gianì, S.; Morello, L.; Breviario, D. The Tubulin-Based-Polymorphism Method Provides a Simple and Effective Alternative to the Genomic Profiling of Grape. PLoS ONE 2016, 11, e0163335. [Google Scholar] [CrossRef]
- Braglia, L.; Manca, A.; Gianì, S.; Hatzopoulos, P.; Breviario, D. A Simplified Approach for Olive (Olea europaea L.) Genotyping andCultivars Traceability. Am. J. Plant Sci. 2017, 8, 3475–3489. [Google Scholar] [CrossRef]
- Breviario, D.; Baird, W.; Sangoi, S.; Hilu, K.; Blumetti, P.; Giani, S. High polymorphism and resolution in targeted fingerprinting with combined ss-tubulin introns. Mol. Breed. 2007, 20, 249–259. [Google Scholar] [CrossRef]
- Wu, G.A.; Terol, J.; Ibanez, V.; López-García, A.; Pérez-Román, E.; Borredá, C.; Domingo, C.; Tadeo, F.R.; Carbonell-Caballero, J.; Alonso, R.; et al. Genomics of the origin and evolution of Citrus. Nature 2018, 554, 311–316. [Google Scholar] [CrossRef]
- Mahadani, P.; Ghosh, S.K. Utility of indels for species-level identification of a biologically complex plant group: A study with intergenic spacer in Citrus. Mol. Biol. Rep. 2014, 41, 7217–7222. [Google Scholar] [CrossRef] [PubMed]
- Braglia, L.; Gavazzi, F.; Morello, L.; Breviario, D. TBP genomic profiling of Citrus x myrtyfolia “Chinotto di Savona”. 2019; unpublished. [Google Scholar]
- Gianì, S.; Morello, L.; Gavazzi, F.; Breviario, D. A Novel and Convenient Method for Animal Species Identification in Food Products. 2019; in preparation. [Google Scholar]
- Leggatt, R.; Iwama, G. Occurrence of polyploidy in the fishes. Rev. Fish Biol. Fish. 2003, 13, 237–246. [Google Scholar] [CrossRef]
- Grape Genome Database. Available online: http://genomes.cribi.unipd.it/grape (accessed on 25 February 2019).
- Braglia, L.; Manca, A.; Mastromauro, F.; Breviario, D. cTBP: A Successful Intron Length Polymorphis (ILP)-Based Genotyping Method Targeted to Well Defined Experimental Needs. Diversity 2010, 2, 572–585. [Google Scholar] [CrossRef]
- Braglia, L.; Gavazzi, F.; Giovannini, A.; Nicoletti, F.; De Benedetti, L.; Breviario, D. TBP-assisted species and hybrid identification in the genus Passiflora. Mol. Breed. 2014, 33, 209–219. [Google Scholar] [CrossRef]
- Galasso, I.; Manca, A.; Braglia, L.; Ponzoni, E.; Breviario, D. Genomic Fingerprinting of Camelina Species Using cTBP as Molecular Marker. Am. J. Plant Sci. 2015, 6, 1184–1200. [Google Scholar] [CrossRef]
- Brock, J.; Mandakova, T.; Lysak, M.; Al-Shehbaz, I. Camelina neglecta (Brassicaceae, Camelineae), a new diploid species from Europe. Phytokeys 2019, 51–57. [Google Scholar] [CrossRef]
- Casazza, A.; Morcia, C.; Ponzoni, E.; Gavazzi, F.; Benedettelli, S.; Breviario, D. A reliable assay for the detection of soft wheat adulteration in Italian pasta is based on the use of new DNA molecular markers capable of discriminating between Triticum aestivum and Triticum durum. J. Cereal Sci. 2012, 56, 733–740. [Google Scholar] [CrossRef]
- Braglia, L.; Gianì, S.; Breviario, D.; Gavazzi, F.; Mastromauro, F.; Morello, L. Development and validation of the modular Feed-code method for qualitative and quantitative determination of feed botanical composition. Anal. Bioanal. Chem. 2016, 408, 8299–8316. [Google Scholar] [CrossRef] [PubMed]
- Breviario, D. Is There Any Alternative to Canonical DNA Barcoding of Multicellular Eukaryotic Species? A Case for the Tubulin Gene Family. Int. J. Mol. Sci. 2017, 18, 827. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Vazquez, E.; Perez, J.; Martinez, J.; Pardinas, A.; Lopez, B.; Karaiskou, N.; Casa, M.; Machado-Schiaffino, G.; Triantafyllidis, A. High Level of Mislabeling in Spanish and Greek Hake Markets Suggests the Fraudulent Introduction of African Species. J. Agric. Food Chem. 2011, 59, 475–480. [Google Scholar] [CrossRef]
- Johnson, R. Food Fraud and “Economically Motivated Adulteration” of Food and Food Ingredients; Congressional Research Service: Washington, DC, USA, 2014. [Google Scholar]
- Pollack, S.; Kawalek, M.; Williams-Hill, D.; Hellberg, R. Evaluation of DNA barcoding methodologies for the identification of fish species in cooked products. Food Control 2018, 84, 297–304. [Google Scholar] [CrossRef]
- FAO (Food and Agriculture Organization of the United Nations). Overview of Food Fraud in the Fisheries Sector, by Alan Reilly; Fisheries and Aquaculture Circular No. 1165; FAO: Rome, Italy, 2018. [Google Scholar]
- European Commission. Commission Recommendation of 12.3.2015 on a Coordinated Control Plan with a View to Establishing the Prevalence of Fraudulent Practices in the Marketing of Certain Foods. 2015. Available online: http://ec.europa.eu/transparency/regdoc/rep/3/2015/EN/3-2015-1558-EN-F1-1.PDF (accessed on 8 March 2019).
- Gavazzi, F.; Pigna, G.; Braglia, L.; Gianì, S.; Breviario, D.; Morello, L. Evolutionary characterization and transcript profiling of β-tubulin genes in flax (Linum usitatissimum L.) during plant development. BMC Plant Biol. 2017, 17, 237. [Google Scholar] [CrossRef]
- McCafferty, J.; Reid, R.; Spencer, M.; Hamp, T.; Fodor, A. Peak Studio: A tool for the visualization and analysis of fragment analysis files. Environ. Microbiol. Rep. 2012, 4, 556–561. [Google Scholar] [CrossRef]
- Leonforte, A.; Sudheesh, S.; Cogan, N.O.; Salisbury, P.A.; Nicolas, M.E.; Materne, M.; Forster, J.W.; Kaur, S. SNP marker discovery, linkage map construction and identification of QTLs for enhanced salinity tolerance in field pea (Pisum sativum L.). BMC Plant Biol. 2013, 13, 161. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Hou, J.; Yin, T.; Chen, Y. An analytical toolkit for polyploid willow discrimination. Sci. Rep. 2016, 6, 37702. [Google Scholar] [CrossRef] [PubMed]
- Ramlee, M.K.; Yan, T.; Cheung, A.M.; Chuah, C.T.; Li, S. High-throughput genotyping of CRISPR/Cas9-mediated mutants using fluorescent PCR-capillary gel electrophoresis. Sci. Rep. 2015, 5, 15587. [Google Scholar] [CrossRef]
- Yang, Z.; Steentoft, C.; Hauge, C.; Hansen, L.; Thomsen, A.L.; Niola, F.; Vester-Christensen, M.B.; Frödin, M.; Clausen, H.; Wandall, H.H.; et al. Fast and sensitive detection of indels induced by precise gene targeting. Nucleic Acids Res. 2015, 43, e59. [Google Scholar] [CrossRef] [PubMed]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morello, L.; Braglia, L.; Gavazzi, F.; Gianì, S.; Breviario, D. Tubulin-Based DNA Barcode: Principle and Applications to Complex Food Matrices. Genes 2019, 10, 229. https://doi.org/10.3390/genes10030229
Morello L, Braglia L, Gavazzi F, Gianì S, Breviario D. Tubulin-Based DNA Barcode: Principle and Applications to Complex Food Matrices. Genes. 2019; 10(3):229. https://doi.org/10.3390/genes10030229
Chicago/Turabian StyleMorello, Laura, Luca Braglia, Floriana Gavazzi, Silvia Gianì, and Diego Breviario. 2019. "Tubulin-Based DNA Barcode: Principle and Applications to Complex Food Matrices" Genes 10, no. 3: 229. https://doi.org/10.3390/genes10030229




