DNA Replication Timing Enters the Single-Cell Era
Abstract
:1. Introduction
2. Replication Timing and Transcriptional Activity: A Longstanding Relationship
3. Developmental Regulation of Replication Timing
4. Replication Foci and the ~1 Mb Chromatin Domain Model
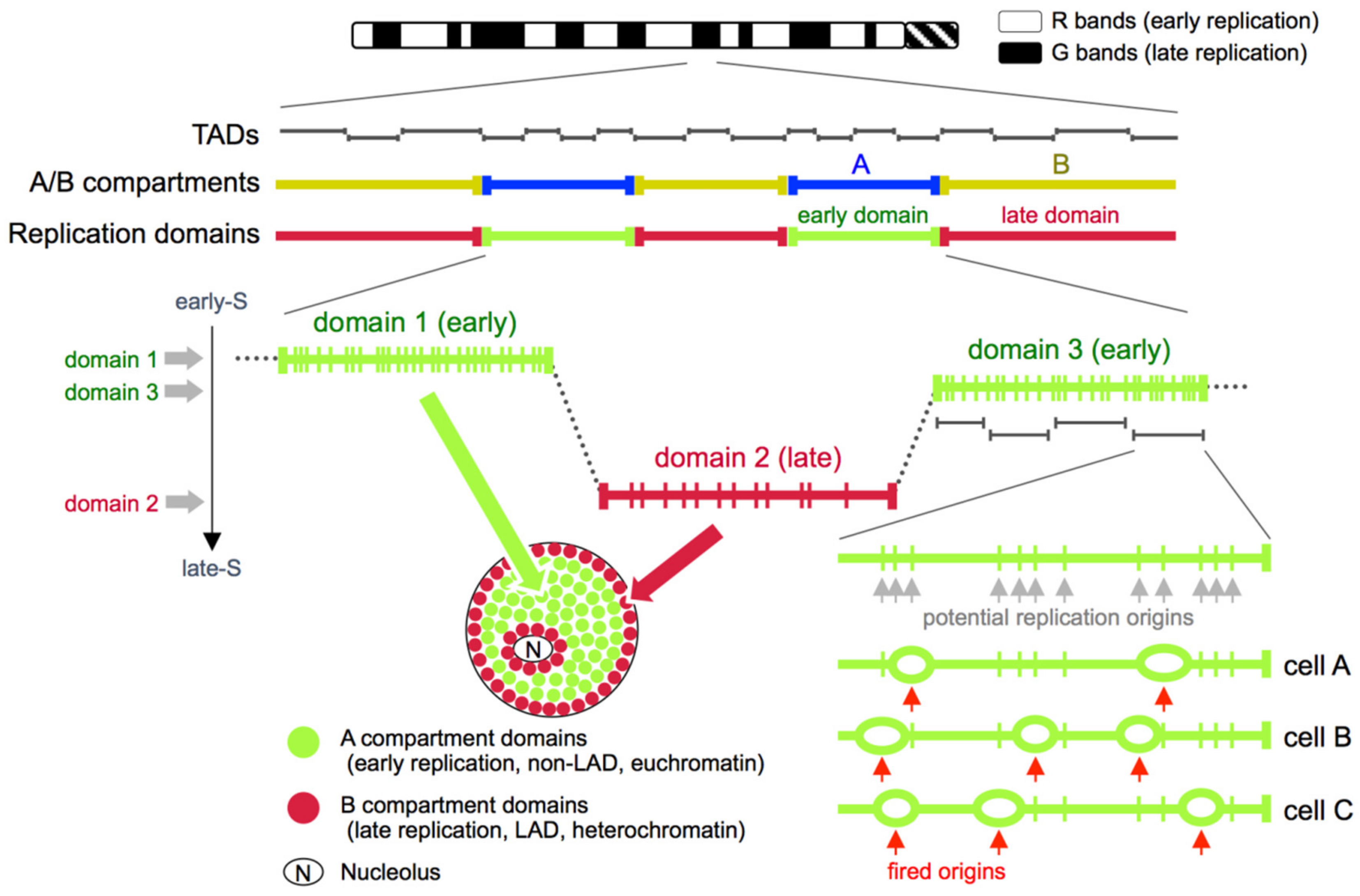
5. Replication Domains, Topologically Associating Domains (TADs), and A/B Compartments
6. Replication Timing and Cell Cycle
7. Unraveling the Complex Relationship: Replication Timing, 3D Genome Architecture, and Transcription
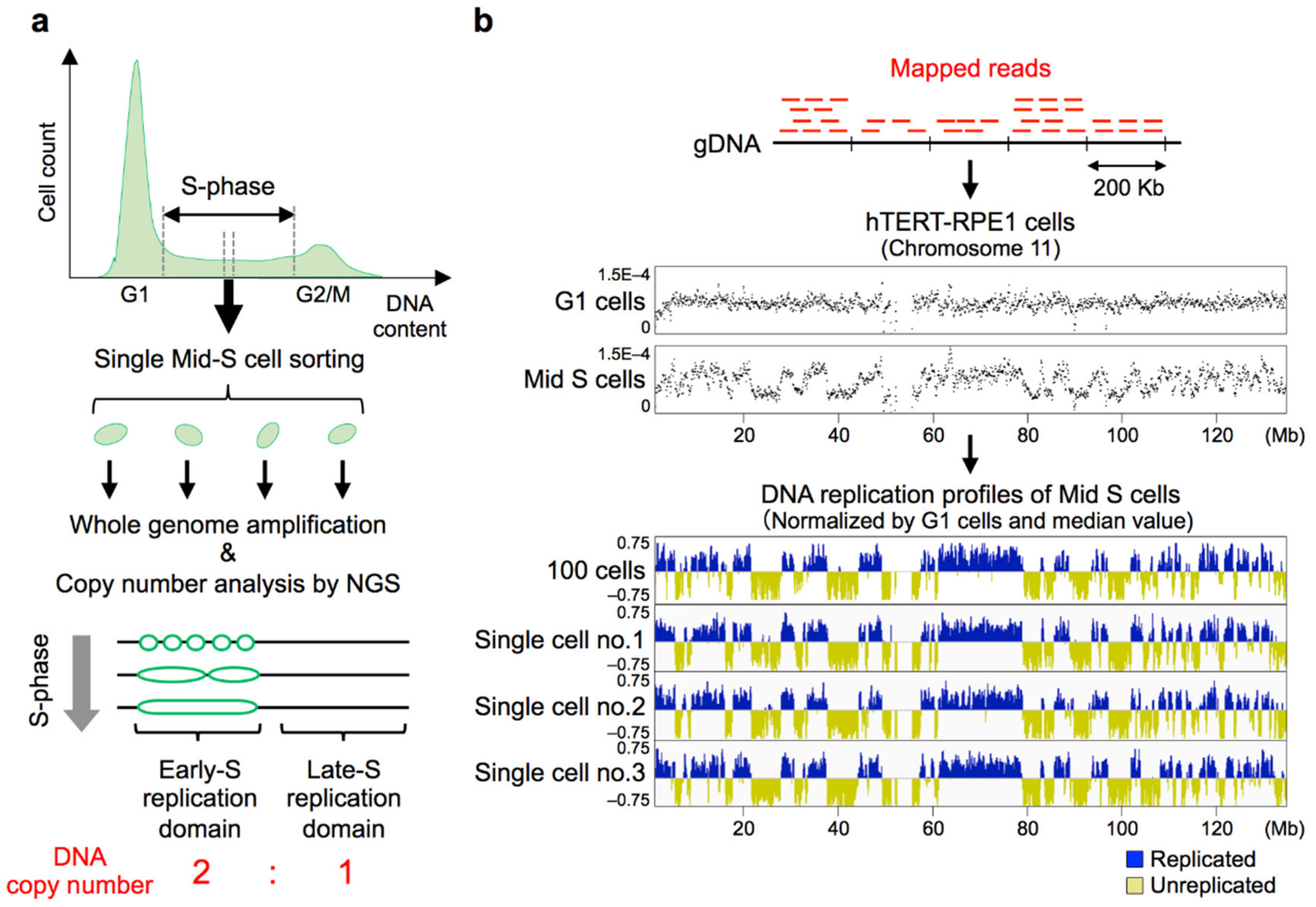
8. Establishment of Genome-Wide Single-Cell DNA Replication Profiling Methods
8.1. Replication Profiles Are Stable Among Cells and Are Cell-Type Specific Even at the Single-Cell Level
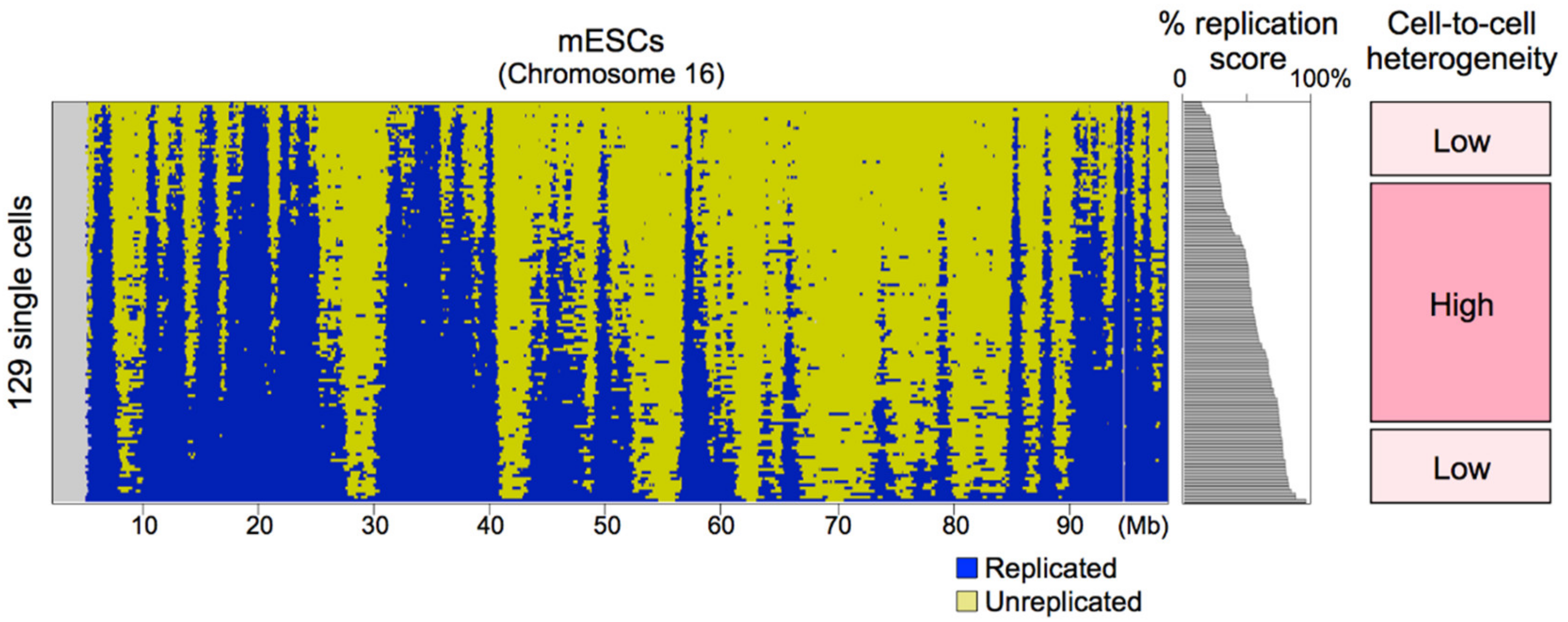
8.2. Sources of Cell-to-Cell Replication Timing Heterogeneity
8.3. Haplotype-Resolved Analysis, Allelic Expression Imbalance, and Replication Asynchrony
8.4. Single-Cell DNA Replication Profiles Correlate with A/B Compartment Organization
9. Future Perspectives of Single-Cell DNA Replication Profiling
9.1. Any Cell, Many Cells, Even Heterogeneous Cell Populations
9.2. scRepli-Seq in Combination with Copy Number Variation (CNV) Analysis
9.3. scRepli-seq in Combination with scRNA-seq
9.4. Imaging and scRepli-seq
9.5. scRepli-seq: Do We Need Higher Resolution?
10. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Watson, J.D.; Crick, F.H.C. Molecular structure of nucleic acids: A structure for deoxyribose nucleic acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.H. DNA synthesis in chromosomes: Implications of early experiments. BioEssays 1989, 10, 121–124. [Google Scholar] [CrossRef] [PubMed]
- Hiratani, I.; Gilbert, D.M. Autosomal lyonization of replication domains during early mammalian development. Adv. Exp. Med. Biol. 2010, 695, 41–58. [Google Scholar] [PubMed]
- Rhind, N.; Gilbert, D.M. DNA Replication Timing. Cold Spring Harb. Perspect. Med. 2013, 5, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Prioleau, M.-N.; MacAlpine, D.M. DNA replication origins—Where do we begin? Genes Dev. 2016, 30, 1683–1697. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Mulia, J.C.; Gilbert, D.M. Replication timing and transcriptional control: Beyond cause and effect—Part III. Curr. Opin. Cell Biol. 2016, 40, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Hiratani, I.; Ryba, T.; Itoh, M.; Yokochi, T.; Schwaiger, M.; Chang, C.W.; Lyou, Y.; Townes, T.M.; Schübeler, D.; Gilbert, D.M. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 2008, 6, 2220–2236. [Google Scholar] [CrossRef] [PubMed]
- Hiratani, I.; Ryba, T.; Itoh, M.; Rathjen, J.; Kulik, M.; Papp, B.; Fussner, E.; Bazett-Jones, D.P.; Plath, K.; Dalton, S.; et al. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res. 2010, 20, 155–169. [Google Scholar] [CrossRef]
- Ryba, T.; Hiratani, I.; Lu, J.; Itoh, M.; Kulik, M.; Zhang, J.; Schulz, T.C.; Robins, A.J.; Dalton, S.; Gilbert, D.M. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 2010, 20, 761–770. [Google Scholar] [CrossRef]
- Pope, B.D.; Ryba, T.; Dileep, V.; Yue, F.; Wu, W.; Denas, O.; Vera, D.L.; Wang, Y.; Hansen, R.S.; Canfield, T.K.; et al. Topologically associating domains are stable units of replication-timing regulation. Nature 2014, 515, 402–405. [Google Scholar] [CrossRef]
- Dileep, V.; Gilbert, D.M. Single-cell replication profiling to measure stochastic variation in mammalian replication timing. Nat. Commun. 2018, 9, 427. [Google Scholar] [CrossRef]
- Takahashi, S.; Miura, H.; Shibata, T.; Nagao, K.; Okumura, K.; Ogata, M.; Obuse, C.; Takebayashi, S.; Hiratani, I. Genome-wide stability of the DNA replication program in single mammalian cells. Nat. Genet. 2019, 51, 529–540. [Google Scholar] [CrossRef]
- Gilbert, D.M. Replication timing and transcriptional control: Beyond cause and effect. Curr. Opin. Cell Biol. 2002, 14, 377–383. [Google Scholar] [CrossRef]
- Schwaiger, M.; Schübeler, D. A question of timing: Emerging links between transcription and replication. Curr. Opin. Genet. Dev. 2006, 16, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Hiratani, I.; Takebayashi, S.; Lu, J.; Gilbert, D.M. Replication timing and transcriptional control: Beyond cause and effect-part II. Curr. Opin. Genet. Dev. 2009, 19, 142–149. [Google Scholar] [CrossRef]
- Taylor, J.H. Asynchronous Duplication of Chromosomes in Cultured Cells of Chinese Hamster. J. Cell Biol. 1960, 7, 455–463. [Google Scholar] [CrossRef]
- Comings, D. Mechanisms of chromosome banding and implications for chromosome structure. Annu. Rev. Genet. 1978, 12, 25–46. [Google Scholar] [CrossRef]
- Raghuraman, M.K.; Winzeler, E.A.; Collingwood, D.; Hunt, S.; Wodicka, L.; Conway, A.; Lockhart, D.J.; Davis, R.W.; Brewer, B.J.; Fangman, W.L. Replication dynamics of the yeast genome. Science 2001, 294, 115–121. [Google Scholar] [CrossRef]
- Schübeler, D.; Scalzo, D.; Kooperberg, C.; van Steensel, B.; Delrow, J.; Groudine, M. Genome-wide DNA replication profile for Drosophila melanogaster: A link between transcription and replication timing. Nat. Genet. 2002, 32, 438–442. [Google Scholar] [CrossRef]
- MacAlpine, D.M.; Rodríguez, H.K.; Bell, S.P. Coordination of replication and transcription along a Drosophila chromosome. Genes Dev. 2004, 18, 3094–3105. [Google Scholar] [CrossRef]
- Woodfine, K.; Fiegler, H.; Beare, D.M.; Collins, J.E.; McCann, O.T.; Young, B.D.; Debernardi, S.; Mott, R.; Dunham, I.; Carter, N.P. Replication timing of the human genome. Hum. Mol. Genet. 2004, 13, 191–202. [Google Scholar] [CrossRef] [PubMed]
- White, E.J.; Emanuelsson, O.; Scalzo, D.; Royce, T.; Kosak, S.; Oakeley, E.J.; Weissman, S.; Gerstein, M.; Groudine, M.; Snyder, M.; et al. DNA replication-timing analysis of human chromosome 22 at high resolution and different developmental states. Proc. Natl. Acad. Sci. USA 2004, 101, 17771–17776. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.; Bekiranov, S.; Karnani, N.; Kapranov, P.; Ghosh, S.; MacAlpine, D.; Lee, C.; Hwang, D.S.; Gingeras, T.R.; Dutta, A. Temporal profile of replication of human chromosomes. Proc. Natl. Acad. Sci. USA 2005, 102, 6419–6424. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, D.M. Replication timing and metazoan evolution. Nat. Genet. 2002, 32, 336–337. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, A.D. Shaping time: Chromatin structure and the DNA replication programme. Trends Genet. 2005, 21, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Bechhoefer, J.; Rhind, N. Replication timing and its emergence from stochastic processes. Trends Genet. 2012, 28, 374–381. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Poveda, A.; Pasero, P. Time to be versatile: Regulation of the replication timing program in budding yeast. J. Mol. Biol. 2013, 425, 4696–4705. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, O.M. Location, location, location: It’s all in the timing for replication origins. Genes Dev. 2013, 27, 117–128. [Google Scholar] [CrossRef]
- De Moura, A.P.S.; Retkute, R.; Hawkins, M.; Nieduszynski, C.A. Mathematical modelling of whole chromosome replication. Nucleic Acids Res. 2010, 38, 5623–5633. [Google Scholar] [CrossRef]
- Lebofsky, R.; Heilig, R.; Sonnleitner, M.; Weissenbach, J.; Bensimon, A. DNA replication origin interference increases the spacing between initiation events in human cells. Mol. Biol. Cell 2006, 17, 5337–5345. [Google Scholar] [CrossRef]
- Norio, P.; Kosiyatrakul, S.; Yang, Q.; Guan, Z.; Brown, N.M.; Thomas, S.; Riblet, R.; Schildkraut, C.L. Progressive activation of DNA replication initiation in large domains of the immunoglobulin heavy chain locus during B cell development. Mol. Cell 2005, 20, 575–587. [Google Scholar] [CrossRef] [PubMed]
- Demczuk, A.; Gauthier, M.G.; Veras, I.; Kosiyatrakul, S.; Schildkraut, C.L.; Busslinger, M.; Bechhoefer, J.; Norio, P. Regulation of DNA replication within the immunoglobulin heavy-chain locus during B cell commitment. PLoS Biol. 2012, 10, e1001360. [Google Scholar] [CrossRef] [PubMed]
- Labit, H.; Perewoska, I.; Germe, T.; Hyrien, O.; Marheineke, K. DNA replication timing is deterministic at the level of chromosomal domains but stochastic at the level of replicons in Xenopus egg extracts. Nucleic Acids Res. 2017, 36, 5623–5634. [Google Scholar] [CrossRef]
- Das, S.P.; Borrman, T.; Liu, V.W.T.; Yang, S.C.H.; Bechhoefer, J.; Rhind, N. Replication timing is regulated by the number of MCMs loaded at origins. Genome Res. 2015, 25, 1886–1892. [Google Scholar] [CrossRef] [PubMed]
- Hiratani, I.; Leskovar, A.; Gilbert, D.M. Differentiation-induced replication-timing changes are restricted to AT-rich/long interspersed nuclear element (LINE)-rich isochores. Proc. Natl. Acad. Sci. USA 2004, 101, 16861–16866. [Google Scholar] [CrossRef] [PubMed]
- Perry, P.; Sauer, S.; Billon, N.; Richardson, W.D.; Spivakov, M.; Warnes, G.; Livesey, F.J.; Merkenschlager, M.; Fisher, A.G.; Azuara, V. A dynamic switch in the replication timing of key regulator genes in embryonic stem cells upon neural induction. Cell Cycle 2004, 3, 1645–1650. [Google Scholar] [CrossRef]
- Hansen, R.S.; Thomas, S.; Sandstrom, R.; Canfield, T.K.; Thurman, R.E.; Weaver, M.; Dorschner, M.O.; Gartler, S.M.; Stamatoyannopoulos, J.A. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc. Natl. Acad. Sci. USA 2010, 107, 139–144. [Google Scholar] [CrossRef]
- Desprat, R.; Thierry-Mieg, D.; Lailler, N.; Lajugie, J.; Schildkraut, C.; Thierry-Mieg, J.; Bouhassira, E.E. Predictable dynamic program of timing of DNA replication in human cells. Genome Res. 2009, 19, 2288–2299. [Google Scholar] [CrossRef]
- Marchal, C.; Sasaki, T.; Vera, D.; Wilson, K.; Sima, J.; Rivera-Mulia, J.C.; Trevilla-García, C.; Nogues, C.; Nafie, E.; Gilbert, D.M. Genome-wide analysis of replication timing by next-generation sequencing with E/L Repli-seq. Nat. Protoc. 2018, 13, 819–839. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Mulia, J.C.; Buckley, Q.; Sasaki, T.; Zimmerman, J.; Didier, R.A.; Nazor, K.; Loring, J.F.; Lian, Z.; Weissman, S.; Robins, A.J.; et al. Dynamic changes in replication timing and gene expression during lineage specification of human pluripotent stem cells. Genome Res. 2015, 25, 1091–1103. [Google Scholar] [CrossRef]
- Huberman, J.A.; Riggs, A.D. On the mechanism of DNA replication in mammalian chromosomes. J. Mol. Biol. 1968, 32, 327–341. [Google Scholar] [CrossRef]
- Berezney, R.; Dubey, D.D.; Huberman, J.A. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma 2000, 108, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Morita, T.; Sato, C. Structural organizations of replicon domains during DNA synthetic phase in the mammalian nucleus. Exp. Cell Res. 1986, 165, 291–297. [Google Scholar] [CrossRef]
- Nakayasu, H.; Berezney, R. Mapping replication sites in the eucaryotic nucleus. J. Cell Biol. 1989, 108, 1–11. [Google Scholar] [CrossRef]
- O’Keefe, R.T.; Henderson, S.C.; Spector, D.L. Dynamic organization of DNA replication in mammalian cell nuclei: Spatially and temporally defined replication of chromosome-specific α-satellite DNA sequences. J. Cell Biol. 1992, 116, 1095–1110. [Google Scholar] [CrossRef] [PubMed]
- Jackson, D.A.; Pombo, A. Replicon clusters are stable units of chromosome structure: Evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 1998, 140, 1285–1295. [Google Scholar] [CrossRef] [PubMed]
- Cremer, T.; Cremer, C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001, 2, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Takebayashi, S.I.; Ogata, M.; Okumura, K. Anatomy of mammalian replication domains. Genes 2017, 8, 110. [Google Scholar] [CrossRef]
- Löb, D.; Lengert, N.; Chagin, V.O.; Reinhart, M.; Casas-Delucchi, C.S.; Cardoso, M.C.; Drossel, B. 3D replicon distributions arise from stochastic initiation and domino-like DNA replication progression. Nat. Commun. 2016, 7, 11207. [Google Scholar] [CrossRef]
- Chagin, V.O.; Casas-Delucchi, C.S.; Reinhart, M.; Schermelleh, L.; Markaki, Y.; Maiser, A.; Bolius, J.J.; Bensimon, A.; Fillies, M.; Domaing, P.; et al. 4D Visualization of replication foci in mammalian cells corresponding to individual replicons. Nat. Commun. 2016, 7, 11231. [Google Scholar] [CrossRef]
- Dekker, J.; Belmont, A.S.; Guttman, M.; Leshyk, V.O.; Lis, J.T.; Lomvardas, S.; Mirny, L.A.; O’Shea, C.C.; Park, P.J.; Ren, B.; et al. The 4D nucleome project. Nature 2017, 549, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Marti-Renom, M.A.; Almouzni, G.; Bickmore, W.A.; Bystricky, K.; Cavalli, G.; Fraser, P.; Gasser, S.M.; Giorgetti, L.; Heard, E.; Nicodemi, M.; et al. Challenges and guidelines toward 4D nucleome data and model standards. Nat. Genet. 2018, 50, 1352–1358. [Google Scholar] [CrossRef] [PubMed]
- Yaffe, E.; Farkash-Amar, S.; Polten, A.; Yakhini, Z.; Tanay, A.; Simon, I. Comparative analysis of DNA replication timing reveals conserved large-scale chromosomal architecture. PLoS Genet. 2010, 6, e1001011. [Google Scholar] [CrossRef] [PubMed]
- Moindrot, B.; Audit, B.; Klous, P.; Baker, A.; Thermes, C.; De Laat, W.; Bouvet, P.; Mongelard, F.; Arneodo, A. 3D chromatin conformation correlates with replication timing and is conserved in resting cells. Nucleic Acids Res. 2012, 40, 9470–9481. [Google Scholar] [CrossRef] [PubMed]
- Dimitrova, D.S.; Gilbert, D.M. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol. Cell 1999, 4, 983–993. [Google Scholar] [CrossRef]
- Besnard, E.; Babled, A.; Lapasset, L.; Milhavet, O.; Parrinello, H.; Dantec, C.; Marin, J.M.; Lemaitre, J.M. Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat. Struct. Mol. Biol. 2012, 19, 837–844. [Google Scholar] [CrossRef] [PubMed]
- Cayrou, C.; Coulombe, P.; Vigneron, A.; Stanojcic, S.; Ganier, O.; Peiffer, I.; Rivals, E.; Puy, A.; Laurent-Chabalier, S.; Desprat, R.; et al. Genome-scale analysis of metazoan replication origins reveals their organization in specific but flexible sites defined by conserved features. Genome Res. 2011, 21, 1438–1449. [Google Scholar] [CrossRef]
- Cayrou, C.; Ballester, B.; Peiffer, I.; Fenouil, R.; Coulombe, P.; Andrau, J.C.; Van Helden, J.; Méchali, M. The chromatin environment shapes DNA replication origin organization and defines origin classes. Genome Res. 2015, 25, 1873–1885. [Google Scholar] [CrossRef]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef]
- Nozaki, T.; Imai, R.; Tanbo, M.; Nagashima, R.; Tamura, S.; Tani, T.; Joti, Y.; Tomita, M.; Hibino, K.; Kanemaki, M.T.; et al. Dynamic organization of chromatin domains revealed by super-resolution live-cell imaging. Mol. Cell 2017, 67, 282–293. [Google Scholar] [CrossRef]
- Xiang, W.; Julia Roberti, M.; Hériché, J.K.; Huet, S.; Alexander, S.; Ellenberg, J. Correlative live and super-resolution imaging reveals the dynamic structure of replication domains. J. Cell Biol. 2018, 217, 1973–1984. [Google Scholar] [CrossRef] [PubMed]
- Solovei, I.; Thanisch, K.; Feodorova, Y. How to rule the nucleus: Divide et impera. Curr. Opin. Cell Biol. 2016, 40, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012, 485, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Nora, E.P.; Lajoie, B.R.; Schulz, E.G.; Giorgetti, L.; Okamoto, I.; Servant, N.; Piolot, T.; Van Berkum, N.L.; Meisig, J.; Sedat, J.; et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 2012, 485, 381–385. [Google Scholar] [CrossRef]
- Ragoczy, T.; Telling, A.; Scalzo, D.; Kooperberg, C.; Groudine, M. Functional redundancy in the nuclear compartmentalization of the Late-Replicating genome. Nucleus 2014, 5, 626–635. [Google Scholar] [CrossRef] [PubMed]
- Vieux-Rochas, M.; Fabre, P.J.; Leleu, M.; Duboule, D.; Noordermeer, D. Clustering of mammalian HOX genes with other H3K27me3 targets within an active nuclear domain. Proc. Natl. Acad. Sci. USA 2015, 112, 4672–4677. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Singh, P.B.; Gilbert, D.M. Uncoupling global and fine-tuning replication timing determinants for mouse pericentric heterochromatin. J. Cell Biol. 2006, 174, 185–194. [Google Scholar] [CrossRef]
- Walter, J.; Schermelleh, L.; Cremer, M.; Tashiro, S.; Cremer, T. Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. J. Cell Biol. 2003, 160, 685–697. [Google Scholar] [CrossRef]
- Thomson, I.; Gilchrist, S.; Bickmore, W.A.; Chubb, J.R. The radial positioning of chromatin is not inherited through mitosis but is established de novo in early G1. Curr. Biol. 2004, 14, 166–172. [Google Scholar] [CrossRef]
- Dileep, V.; Ay, F.; Sima, J.; Vera, D.L.; Noble, W.S.; Gilbert, D.M. Topologically associating domains and their long-range contacts are established during early G1 coincident with the establishment of the replication timing program. Genome Res. 2015, 25, 1104–1113. [Google Scholar] [CrossRef]
- Nora, E.P.; Goloborodko, A.; Valton, A.L.; Gibcus, J.H.; Uebersohn, A.; Abdennur, N.; Dekker, J.; Mirny, L.A.; Bruneau, B.G. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 2017, 169, 930–944. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.S.P.; Huang, S.C.; Glenn St Hilaire, B.; Engreitz, J.M.; Perez, E.M.; Kieffer-Kwon, K.R.; Sanborn, A.L.; Johnstone, S.E.; Bascom, G.D.; Bochkov, I.D.; et al. Cohesin loss eliminates all loop domains. Cell 2017, 171, 305–320. [Google Scholar] [CrossRef] [PubMed]
- Sima, J.; Chakraborty, A.; Dileep, V.; Michalski, M.; Klein, K.N.; Holcomb, N.P.; Turner, J.L.; Paulsen, M.T.; Rivera-Mulia, J.C.; Trevilla-Garcia, C.; et al. Identifying cis elements for spatiotemporal control of mammalian DNA replication. Cell 2019, 176, 816–830. [Google Scholar] [CrossRef]
- Nagano, T.; Lubling, Y.; Várnai, C.; Dudley, C.; Leung, W.; Baran, Y.; Mendelson Cohen, N.; Wingett, S.; Fraser, P.; Tanay, A. Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature 2017, 547, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Takebayashi, S.; Dileep, V.; Ryba, T.; Dennis, J.H.; Gilbert, D.M. Chromatin-interaction compartment switch at developmentally regulated chromosomal domains reveals an unusual principle of chromatin folding. Proc. Natl. Acad. Sci. USA 2012, 109, 12574–12579. [Google Scholar] [CrossRef] [PubMed]
- Hassan-Zadeh, V.; Chilaka, S.; Cadoret, J.C.; Ma, M.K.W.; Boggetto, N.; West, A.G.; Prioleau, M.N. USF binding sequences from the HS4 insulator element impose early replication timing on a vertebrate replicator. PLoS Biol. 2012, 10, e1001277. [Google Scholar] [CrossRef] [PubMed]
- Shang, W.H.; Hori, T.; Martins, N.M.C.; Toyoda, A.; Misu, S.; Monma, N.; Hiratani, I.; Maeshima, K.; Ikeo, K.; Fujiyama, A.; et al. Chromosome engineering allows the efficient isolation of vertebrate neocentromeres. Dev. Cell 2013, 24, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Siefert, J.C.; Georgescu, C.; Wren, J.D.; Koren, A.; Sansam, C.L. DNA replication timing during development anticipates transcriptional programs and parallels enhancer activation. Genome Res. 2017, 27, 1406–1416. [Google Scholar] [CrossRef] [PubMed]
- Pourkarimi, E.; Bellush, J.M.; Whitehouse, I. Spatiotemporal coupling and decoupling of gene transcription with DNA replication origins during embryogenesis in C. elegans. eLife 2016, 5, 1–12. [Google Scholar] [CrossRef]
- Wear, E.E.; Song, J.; Zynda, G.; LeBlanc, C.; Lee, T.-J.; Mickelson-Young, L.; Concia, L.; Mulvaney, P.; Szymanski, E.S.; Allen, G.C.; et al. Genomic analysis of the DNA replication timing program during mitotic S phase in maize (Zea mays L.) root tips. Plant Cell 2017, 29, 2126–2149. [Google Scholar] [CrossRef]
- Concia, L.; Brooks, A.M.; Wheeler, E.; Zynda, G.; Wear, E.E.; LeBlanc, C.; Song, J.; Lee, T.-J.; Pascuzzi, P.E.; Martienssen, R.; et al. Genome-wide analysis of the Arabidopsis thaliana replication timing program. Plant Physiol. 2018, 176, 2166–2185. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, R.L.; Penke, T.J.R.; Strahl, B.D.; Matera, A.G.; McKay, D.J.; MacAlpine, D.M.; Duronio, R.J. Chromatin conformation and transcriptional activity are permissive regulators of DNA replication initiation in Drosophila. Genome Res. 2018, 28, 1688–1700. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, M.; Malla, S.; Blythe, M.J.; Nieduszynski, C.A.; Allers, T. Accelerated growth in the absence of DNA replication origins. Nature 2013, 503, 544–547. [Google Scholar] [CrossRef] [PubMed]
- Koren, A.; Tsai, H.; Tirosh, I.; Burrack, L.S.; Barkai, N.; Berman, J. Epigenetically-inherited centromere and neocentromere DNA replicates earliest in S-phase. PLoS Genet. 2010, 6, e1001068. [Google Scholar] [CrossRef] [PubMed]
- Hyrien, O.; Maric, C.; Méchali, M. Transition in specification of embryonic metazoan DNA replication origins. Science 1995, 270, 994–997. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, T.; Sawado, T.; Yamaguchi, M.; Shinomiya, T. Specification of regions of DNA replication initiation during embryogenesis in the 65-kilobase DNApolalpha-dE2F locus of Drosophila melanogaster. Mol. Cell. Biol. 1999, 19, 547–555. [Google Scholar] [CrossRef] [PubMed]
- Tazumi, A.; Fukuura, M.; Nakato, R.; Kishimoto, A.; Takenaka, T.; Ogawa, S.; Song, J.H.; Takahashi, T.S.; Nakagawa, T.; Shirahige, K.; et al. Telomere-binding protein Taz1 controls global replication timing through its localization near late replication origins in fission yeast. Genes Dev. 2012, 26, 2050–2062. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, S.; Kido, S.; Handa, T.; Ogawa, H.; Asakawa, H.; Takahashi, T.S.; Nakagawa, T.; Hiraoka, Y.; Masukata, H. Shelterin promotes tethering of late replication origins to telomeres for replication-timing control. EMBO J. 2018, 37, e98997. [Google Scholar] [CrossRef] [PubMed]
- Hayano, M.; Kanoh, Y.; Matsumoto, S.; Renard-Guillet, C.; Shirahige, K.; Masai, H. Rif1 is a global regulator of timing of replication origin firing in fission yeast. Genes Dev. 2012, 26, 137–150. [Google Scholar] [CrossRef]
- Peace, J.M.; Ter-Zakarian, A.; Aparicio, O.M. Rif1 regulates initiation timing of late replication origins throughout the S. cerevisiae genome. PLoS ONE 2014, 9, e98501. [Google Scholar] [CrossRef]
- Knott, S.R.V.; Peace, J.M.; Ostrow, A.Z.; Gan, Y.; Rex, A.E.; Viggiani, C.J.; Tavaré, S.; Aparicio, O.M. Forkhead transcription factors establish origin timing and long-range clustering in S. cerevisiae. Cell 2012, 148, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Ostrow, A.Z.; Kalhor, R.; Gan, Y.; Villwock, S.K.; Linke, C.; Barberis, M.; Chen, L.; Aparicio, O.M. Conserved forkhead dimerization motif controls DNA replication timing and spatial organization of chromosomes in S. cerevisiae. Proc. Natl. Acad. Sci. USA 2017, 114, E2411–E2419. [Google Scholar] [CrossRef] [PubMed]
- Dileep, V.; Rivera-Mulia, J.C.; Sima, J.; Gilbert, D.M. Large-scale chromatin structure-function relationships during the cell cycle and development: Insights from replication timing. Cold Spring Harb. Symp. Quant. Biol. 2015, 80, 53–63. [Google Scholar] [CrossRef]
- Yamazaki, S.; Ishii, A.; Kanoh, Y.; Oda, M.; Nishito, Y.; Masai, H. Rif1 regulates the replication timing domains on the human genome. EMBO J. 2012, 31, 3667–3677. [Google Scholar] [CrossRef] [PubMed]
- Cornacchia, D.; Dileep, V.; Quivy, J.P.; Foti, R.; Tili, F.; Santarella-Mellwig, R.; Antony, C.; Almouzni, G.; Gilbert, D.M.; Buonomo, S.B.C. Mouse Rif1 is a key regulator of the replication-timing programme in mammalian cells. EMBO J. 2012, 31, 3678–3690. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, H.F.; Azuara, V.; Amoils, S.; Spivakov, M.; Terry, A.; Nesterova, T.; Cobb, B.S.; Ramsahoye, B.; Merkenschlager, M.; Fisher, A.G. The impact of chromatin modifiers on the timing of locus replication in mouse embryonic stem cells. Genome Biol. 2007, 8, 1–13. [Google Scholar] [CrossRef]
- Li, J.; Santoro, R.; Koberna, K.; Grummt, I. The chromatin remodeling complex NoRC controls replication timing of rRNA genes. EMBO J. 2005, 24, 120–127. [Google Scholar] [CrossRef]
- Takebayashi, S.I.; Lei, I.; Ryba, T.; Sasaki, T.; Dileep, V.; Battaglia, D.; Gao, X.; Fang, P.; Fan, Y.; Esteban, M.A.; et al. Murine esBAF chromatin remodeling complex subunits BAF250a and Brg1 are necessary to maintain and reprogram pluripotency-specific replication timing of select replication domains. Epigenetics Chromatin 2013, 6, 1–12. [Google Scholar] [CrossRef]
- Schwaiger, M.; Stadler, M.B.; Bell, O.; Kohler, H.; Oakeley, E.J.; Schübeler, D. Chromatin state marks cell-type- and gender-specific replication of the Drosophila genome. Genes Dev. 2009, 23, 589–601. [Google Scholar] [CrossRef]
- Sansam, C.G.; Pietrzak, K.; Majchrzycka, B.; Kerlin, M.A.; Chen, J.; Rankin, S.; Sansam, C.L. A mechanism for epigenetic control of DNA replication. Genes Dev. 2018, 32, 224–229. [Google Scholar] [CrossRef]
- Hiraga, S.I.; Alvino, G.M.; Chang, F.; Lian, H.Y.; Sridhar, A.; Kubota, T.; Brewer, B.J.; Weinreich, M.; Raghuraman, M.K.; Donaldson, A.D. Rif1 controls DNA replication by directing Protein Phosphatase 1 to reverse Cdc7- mediated phosphorylation of the MCM complex. Genes Dev. 2014, 28, 372–383. [Google Scholar] [CrossRef] [PubMed]
- Hiraga, S.; Ly, T.; Garzón, J.; Hořejší, Z.; Ohkubo, Y.; Endo, A.; Obuse, C.; Boulton, S.J.; Lamond, A.I.; Donaldson, A.D. Human RIF1 and protein phosphatase 1 stimulate DNA replication origin licensing but suppress origin activation. EMBO Rep. 2017, 18, 403–419. [Google Scholar] [CrossRef] [PubMed]
- Foti, R.; Gnan, S.; Cornacchia, D.; Dileep, V.; Bulut-Karslioglu, A.; Diehl, S.; Buness, A.; Klein, F.A.; Huber, W.; Johnstone, E.; et al. Nuclear architecture organized by Rif1 underpins the replication-timing program. Mol. Cell 2016, 61, 260–273. [Google Scholar] [CrossRef] [PubMed]
- Kanoh, Y.; Matsumoto, S.; Fukatsu, R.; Kakusho, N.; Kono, N.; Renard-Guillet, C.; Masuda, K.; Iida, K.; Nagasawa, K.; Shirahige, K.; et al. Rif1 binds to G quadruplexes and suppresses replication over long distances. Nat. Struct. Mol. Biol. 2015, 22, 889–897. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, K.; Yoshizawa-Sugata, N.; Masai, H. Oligomer formation and G-quadruplex binding by purified murine Rif1 protein, a key organizer of higher-order chromatin architecture. J. Biol. Chem. 2018, 293, 3607–3624. [Google Scholar] [CrossRef] [PubMed]
- Davé, A.; Cooley, C.; Garg, M.; Bianchi, A. Protein Phosphatase 1 recruitment by Rif1 regulates DNA replication origin firing by counteracting DDK activity. Cell Rep. 2014, 7, 53–61. [Google Scholar] [CrossRef]
- Mattarocci, S.; Shyian, M.; Lemmens, L.; Damay, P.; Altintas, D.M.; Shi, T.; Bartholomew, C.R.; Thomä, N.H.; Hardy, C.F.J.; Shore, D. Rif1 Controls DNA replication timing in yeast through the PP1 Phosphatase Glc7. Cell Rep. 2014, 7, 62–69. [Google Scholar] [CrossRef]
- Tanaka, S.; Araki, H. Helicase activation and establishment of replication forks at chromosomal origins of replication. Cold Spring Harb. Perspect. Biol. 2013, 5, a010371. [Google Scholar] [CrossRef]
- Ferguson, B.M.; Fangman, W.L. A position effect on the time of replication origin activation in yeast. Cell 1992, 68, 333–339. [Google Scholar] [CrossRef]
- Stevenson, J.B.; Gottschling, D.E. Telomeric chromatin modulates replication timing near chromosome ends. Genes Dev. 1999, 13, 146–151. [Google Scholar] [CrossRef]
- Zappulla, D.C.; Sternglanz, R.; Leatherwood, J. Control of replication timing by a transcriptional silencer. Curr. Biol. 2002, 12, 869–875. [Google Scholar] [CrossRef]
- Cosgrove, A.J.; Nieduszynski, C.A.; Donaldson, A.D. Ku complex controls the replication time of DNA in telomere regions. Genes Dev. 2002, 16, 2485–2490. [Google Scholar] [CrossRef] [PubMed]
- Lian, H.-Y.; Robertson, E.D.; Hiraga, S.; Alvino, G.M.; Collingwood, D.; McCune, H.J.; Sridhar, A.; Brewer, B.J.; Raghuraman, M.K.; Donaldson, A.D. The effect of Ku on telomere replication time is mediated by telomere length but is independent of histone tail acetylation. Mol. Biol. Cell 2011, 22, 1753–1765. [Google Scholar] [CrossRef]
- Heun, P.; Laroche, T.; Raghuraman, M.K.; Gasser, S.M. The positioning and dynamics of origins of replication in the budding yeast nucleus. J. Cell Biol. 2001, 152, 385–400. [Google Scholar] [CrossRef]
- Pohl, T.J.; Brewer, B.J.; Raghuraman, M.K. Functional centromeres determine the activation time of pericentric origins of DNA replication in Saccharomyces cerevisiae. PLoS Genet. 2012, 8, e1002677. [Google Scholar] [CrossRef]
- Kim, S.-M.; Dubey, D.D.; Huberman, J.A. Early-replicating heterochromatin. Genes Dev. 2003, 17, 330–335. [Google Scholar] [CrossRef]
- Hayashi, M.T.; Takahashi, T.S.; Nakagawa, T.; Nakayama, J.I.; Masukata, H. The heterochromatin protein Swi6/HP1 activates replication origins at the pericentromeric region and silent mating-type locus. Nat. Cell Biol. 2009, 11, 357–362. [Google Scholar] [CrossRef]
- Goren, A.; Tabib, A.; Hecht, M.; Cedar, H. DNA replication timing of the human β-globin domain is controlled by histone modification at the origin. Genes Dev. 2008, 22, 1319–1324. [Google Scholar] [CrossRef]
- Therizols, P.; Illingworth, R.S.; Courilleau, C.; Boyle, S.; Wood, A.J.; Bickmore, W.A. Chromatin decondensation is sufficient to alter nuclear organization in embryonic stem cells. Science 2014, 346, 1238–1242. [Google Scholar] [CrossRef]
- Koren, A.; Handsaker, R.E.; Kamitaki, N.; Karlić, R.; Ghosh, S.; Polak, P.; Eggan, K.; McCarroll, S.A. Genetic variation in human DNA replication timing. Cell 2014, 159, 1015–1026. [Google Scholar] [CrossRef]
- Platt, E.J.; Smith, L.; Thayer, M.J. L1 retrotransposon antisense RNA within ASAR lncRNAs controls chromosome-wide replication timing. J. Cell Biol. 2018, 217, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Li, F.; Murphy, C.S.; Davidson, M.W.; Gilbert, D.M. G2 phase chromatin lacks determinants of replication timing. J. Cell Biol. 2010, 189, 967–980. [Google Scholar] [CrossRef]
- Ryba, T.; Battaglia, D.; Pope, B.D.; Hiratani, I.; Gilbert, D.M. Genome-scale analysis of replication timing: From bench to bioinformatics. Nat. Protoc. 2011, 6, 870–895. [Google Scholar] [CrossRef]
- Takebayashi, S.; Ogata, S.; Ogata, M.; Okumura, K. Mapping mammalian replication domains using the ion torrent semiconductor sequencing platform. Biosci. Biotechnol. Biochem. 2018, 82, 2098–2100. [Google Scholar] [CrossRef] [PubMed]
- Müller, C.A.; Hawkins, M.; Retkute, R.; Malla, S.; Wilson, R.; Blythe, M.J.; Nakato, R.; Komata, M.; Shirahige, K.; De Moura, A.P.S.; et al. The dynamics of genome replication using deep sequencing. Nucleic Acids Res. 2013, 42, e3. [Google Scholar] [CrossRef] [PubMed]
- Van Der Aa, N.; Cheng, J.; Mateiu, L.; Esteki, M.Z.; Kumar, P.; Dimitriadou, E.; Vanneste, E.; Moreau, Y.; Vermeesch, J.R.; Voet, T. Genome-wide copy number profiling of single cells in S-phase reveals DNA-replication domains. Nucleic Acids Res. 2013, 41, e66. [Google Scholar] [CrossRef]
- Buenrostro, J.D.; Wu, B.; Litzenburger, U.M.; Ruff, D.; Gonzales, M.L.; Snyder, M.P.; Chang, H.Y.; Greenleaf, W.J. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 2015, 523, 486–490. [Google Scholar] [CrossRef] [PubMed]
- Risca, V.I.; Greenleaf, W.J. Unraveling the 3D genome: Genomics tools for multiscale exploration. Trends Genet. 2015, 31, 357–372. [Google Scholar] [CrossRef]
- Kelsey, G.; Stegle, O.; Reik, W. Single-cell epigenomics: Recording the past and predicting the future. Science 2017, 358, 69–75. [Google Scholar] [CrossRef]
- Hu, Y.; An, Q.; Sheu, K.; Trejo, B.; Fan, S.; Guo, Y. Single cell multi-omics technology: Methodology and application. Front. Cell Dev. Biol. 2018, 6, 28. [Google Scholar] [CrossRef]
- Shema, E.; Bernstein, B.E.; Buenrostro, J.D. Single-cell and single-molecule epigenomics to uncover genome regulation at unprecedented resolution. Nat. Genet. 2019, 51, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Stevens, T.J.; Lando, D.; Basu, S.; Liam, P.; Cao, Y.; Lee, S.F.; Leeb, M.; Wohlfahrt, K.J.; Boucher, W.; Shaughnessy-kirwan, A.O.; et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 2017, 544, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Flyamer, I.M.; Gassler, J.; Imakaev, M.; Ulyanov, S.V.; Abdennur, N.; Razin, S.V.; Mirny, L.; Tachibana-Konwalski, K. Single-cell Hi-C reveals unique chromatin reorganization at oocyte-tozygote transition. Nature 2017, 544, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Kind, J.; Pagie, L.; De Vries, S.S.; Nahidiazar, L.; Dey, S.S.; Bienko, M.; Zhan, Y.; Lajoie, B.; De Graaf, C.A.; Amendola, M.; et al. Genome-wide maps of nuclear lamina interactions in single Human cells. Cell 2015, 163, 134–147. [Google Scholar] [CrossRef]
- Baslan, T.; Kendall, J.; Ward, B.; Cox, H.; Leotta, A.; Rodgers, L.; Riggs, M.; D’Italia, S.; Sun, G.; Yong, M.; et al. Optimizing sparse sequencing of single cells for highly multiplex copy number profiling. Genome Res. 2015, 125, 714–724. [Google Scholar] [CrossRef] [PubMed]
- Courbet, S.; Gay, S.; Arnoult, N.; Wronka, G.; Anglana, M.; Brison, O.; Debatisse, M. Replication fork movement sets chromatin loop size and origin choice in mammalian cells. Nature 2008, 455, 557–560. [Google Scholar] [CrossRef]
- Letessier, A.; Millot, G.A.; Koundrioukoff, S.; Lachagès, A.-M.; Vogt, N.; Hansen, R.S.; Malfoy, B.; Brison, O.; Debatisse, M. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature 2011, 470, 120–123. [Google Scholar] [CrossRef] [PubMed]
- Koren, A.; McCarroll, S.A. Random replication of the inactive X chromosome. Genome Res. 2014, 24, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Mulia, J.C.; Dimond, A.; Vera, D.; Trevilla-Garcia, C.; Sasaki, T.; Zimmerman, J.; Dupont, C.; Gribnau, J.; Fraser, P.; Gilbert, D.M. Allele-specific control of replication timing and genome organization during development. Genome Res. 2018, 28, 800–811. [Google Scholar] [CrossRef] [PubMed]
- Weinberger, L.; Ayyash, M.; Novershtern, N.; Hanna, J.H. Dynamic stem cell states: Naive to primed pluripotency in rodents and humans. Nat. Rev. Mol. Cell Biol. 2016, 17, 155–169. [Google Scholar] [CrossRef]
- Takahashi, S.; Kobayashi, S.; Hiratani, I. Epigenetic differences between naïve and primed pluripotent stem cells. Cell. Mol. Life Sci. 2018, 75, 1191–1203. [Google Scholar] [CrossRef]
- Nagano, T.; Lubling, Y.; Stevens, T.J.; Schoenfelder, S.; Yaffe, E.; Dean, W.; Laue, E.D.; Tanay, A.; Fraser, P. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 2013, 502, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Quinodoz, S.A.; Ollikainen, N.; Tabak, B.; Palla, A.; Schmidt, J.M.; Detmar, E.; Lai, M.M.; Shishkin, A.A.; Bhat, P.; Takei, Y.; et al. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell 2018, 174, 744–757. [Google Scholar] [CrossRef] [PubMed]
- Ryba, T.; Crockett, D.K.; Battaglia, D.; Ridge, P.G.; Chang, B.H.; Wilson, A.R.; Shirley, J.W.; Lyon, E.; Buckley, Q.; Williams, M.S.; et al. Abnormal developmental control of replication-timing domains in pediatric acute lymphoblastic leukemia. Genome Res. 2012, 22, 1833–1844. [Google Scholar] [CrossRef]
- Evers, D.L.; Fowler, C.B.; Cunningham, B.R.; Mason, J.T.; O’Leary, T.J. The effect of formaldehyde fixation on RNA: Optimization of formaldehyde adduct removal. J. Mol. Diagn. 2011, 13, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Dey, S.S.; Kester, L.; Spanjaard, B.; Bienko, M.; van Oudenaarden, A. Integrated genome and transcriptome sequencing of the same cell. Nat. Biotechnol. 2015, 33, 285–289. [Google Scholar] [CrossRef] [PubMed]
- Macaulay, I.C.; Haerty, W.; Kumar, P.; Li, Y.I.; Hu, T.X.; Teng, M.J.; Goolam, M.; Saurat, N.; Coupland, P.; Shirley, L.M.; et al. G&T-seq: Parallel sequencing of single-cell genomes and transcriptomes. Nat. Methods 2015, 12, 519–522. [Google Scholar]
- Macaulay, I.C.; Teng, M.J.; Haerty, W.; Kumar, P.; Ponting, C.P.; Voet, T. Separation and parallel sequencing of the genomes and transcriptomes of single cells using G&T-seq. Nat. Protoc. 2016, 11, 2081–2103. [Google Scholar]
- Clark, S.J.; Argelaguet, R.; Kapourani, C.A.; Stubbs, T.M.; Lee, H.J.; Alda-Catalinas, C.; Krueger, F.; Sanguinetti, G.; Kelsey, G.; Marioni, J.C.; et al. ScNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells e. Nat. Commun. 2018, 9, 781. [Google Scholar] [CrossRef]
- Dixit, A.; Parnas, O.; Li, B.; Chen, J.; Fulco, C.P.; Jerby-Arnon, L.; Marjanovic, N.D.; Dionne, D.; Burks, T.; Raychowdhury, R.; et al. Perturb-Seq: Dissecting molecular circuits with scalable single-cell RNA Profiling of pooled genetic screens. Cell 2016, 167, 1853–1866. [Google Scholar] [CrossRef]
- Boettiger, A.N.; Bintu, B.; Moffitt, J.R.; Wang, S.; Beliveau, B.J.; Fudenberg, G.; Imakaev, M.; Mirny, L.A.; Wu, C.T.; Zhuang, X. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature 2016, 529, 418–422. [Google Scholar] [CrossRef]
- Wang, S.; Su, J.-H.; Beliveau, B.J.; Bintu, B.; Moffitt, J.R.; Wu, C.; Zhuang, X. Spatial organization of chromatin domains and compartments in single chromosomes. Science 2016, 353, 598–602. [Google Scholar] [CrossRef] [PubMed]
- Szabo, Q.; Jost, D.; Chang, J.M.; Cattoni, D.I.; Papadopoulos, G.L.; Bonev, B.; Sexton, T.; Gurgo, J.; Jacquier, C.; Nollmann, M.; et al. TADs are 3D structural units of higher-order chromosome organization in Drosophila. Sci. Adv. 2018, 4, eaar8082. [Google Scholar] [CrossRef]
- Bintu, B.; Mateo, L.J.; Su, J.H.; Sinnott-Armstrong, N.A.; Parker, M.; Kinrot, S.; Yamaya, K.; Boettiger, A.N.; Zhuang, X. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science 2018, 362, eaau1783. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Xing, D.; Tan, L.; Li, H.; Zhou, G.; Huang, L.; Xie, X.S. Single-cell whole-genome analyses by Linear Amplification via Transposon Insertion (LIANTI). Science 2017, 356, 189–194. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hiratani, I.; Takahashi, S. DNA Replication Timing Enters the Single-Cell Era. Genes 2019, 10, 221. https://doi.org/10.3390/genes10030221
Hiratani I, Takahashi S. DNA Replication Timing Enters the Single-Cell Era. Genes. 2019; 10(3):221. https://doi.org/10.3390/genes10030221
Chicago/Turabian StyleHiratani, Ichiro, and Saori Takahashi. 2019. "DNA Replication Timing Enters the Single-Cell Era" Genes 10, no. 3: 221. https://doi.org/10.3390/genes10030221
APA StyleHiratani, I., & Takahashi, S. (2019). DNA Replication Timing Enters the Single-Cell Era. Genes, 10(3), 221. https://doi.org/10.3390/genes10030221




