Overexpression of Two Members of D7 Salivary Genes Family is Associated with Pyrethroid Resistance in the Malaria Vector Anopheles Funestus s.s. but Not in Anopheles Gambiae in Cameroon
Abstract
:1. Introduction
2. Methods
2.1. Study Area and Mosquito Collection
2.2. Molecular Species Identification
2.3. Plasmodium sporozoite Infection Rate
2.4. Insecticide Susceptibility Assays
2.5. Expression Profile of D7 Salivary Genes Using Real-Time Quantitative PCR
2.6. Sequencing of D7r3 and D7r4 Genomic DNA from Alive and Dead An. funestus Mosquitoes
2.7. Genotyping of Resistance Molecular Markers in An. funestus s.s. and in An. gambiae
3. Results
3.1. Mosquito Species Composition
3.2. Plasmodium Infection
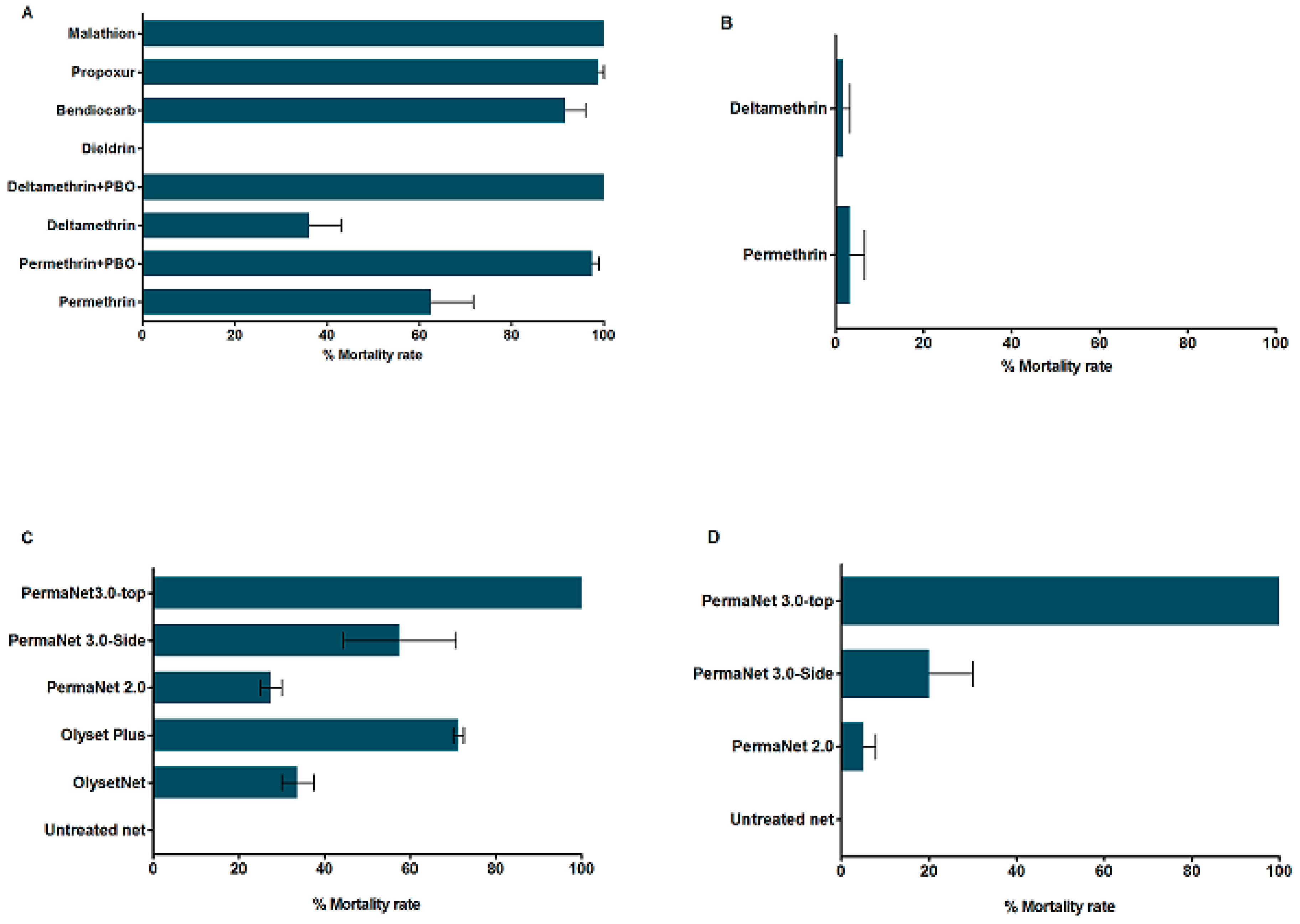
3.3. Insecticide Susceptibility Bioassays
3.3.1. Insecticide Susceptibility in An. funestus s.s.
3.3.2. Insecticide Susceptibility in An. gambiae
3.3.3. Synergist Assays for Pyrethroid Resistance in An. funestus s.s.
3.3.4. Insecticide Treated bed nets Efficacy Assessment
3.4. Expression on D7 Salivary Genes Family in An. funestus and An. gambiae Mosquitoes
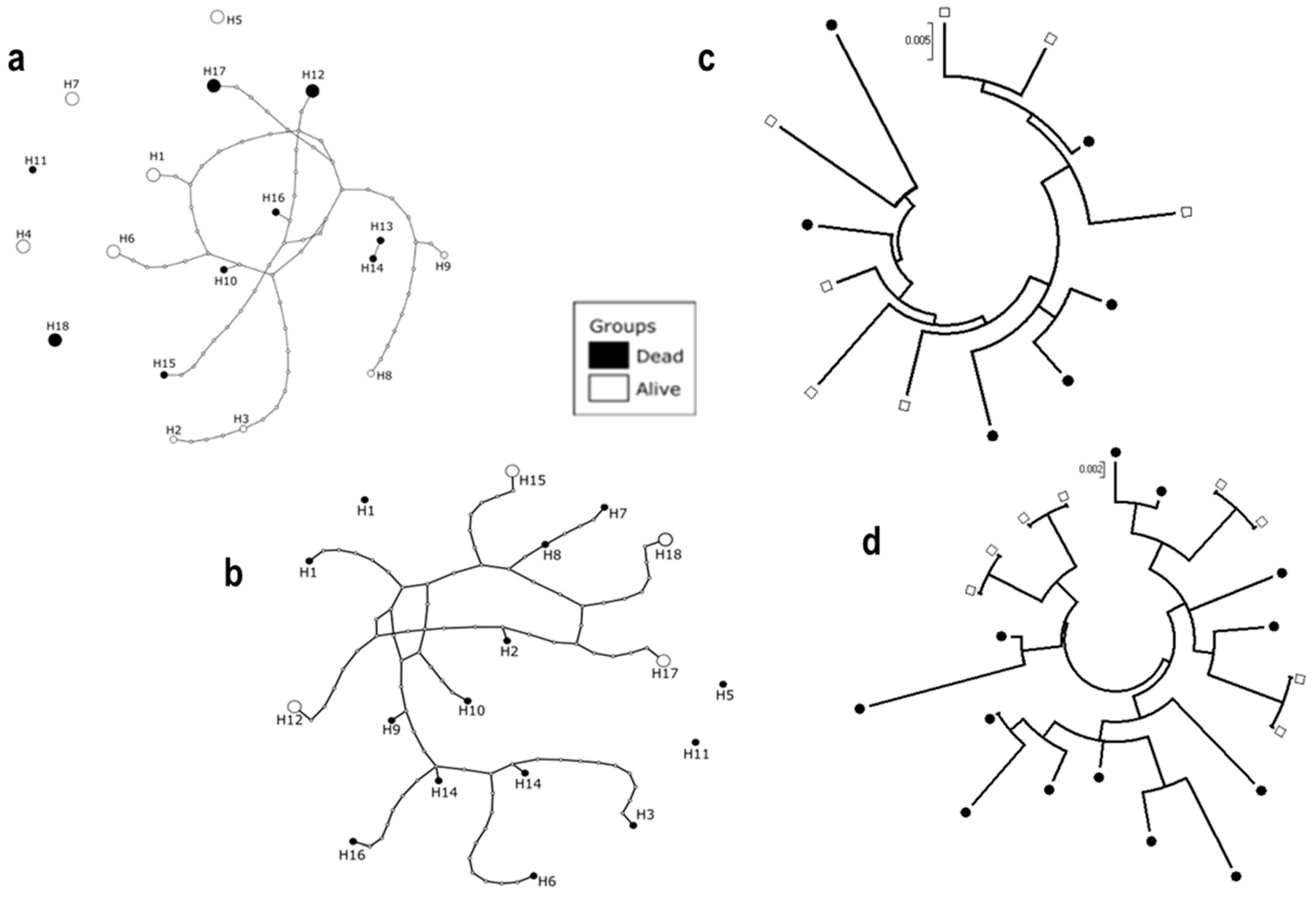
3.5. Sequencing of D7r3 and D7r4 Genomic DNA from An. funestus Mosquitoes
3.6. Molecular Basis of the Insecticide Resistance in Field Malaria Vector Populations
3.6.1. RDL-A296S and L119F-Gste2 mutations detection in An. funestus s.s.
3.6.2. L1014F mutations in An. Gambiae
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- WHO. World Malaria Report 2018; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- Bhatt, S.; Weiss, D.; Cameron, E.; Bisanzio, D.; Mappin, B.; Dalrymple, U.; Battle, K.; Moyes, C.; Henry, A.; Eckhoff, P. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature 2015, 526, 207. [Google Scholar] [CrossRef]
- Calvo, E.; Mans, B.J.; Andersen, J.F.; Ribeiro, J.M. Function and evolution of a mosquito salivary protein family. J. Biol. Chem. 2006, 281, 1935–1942. [Google Scholar] [CrossRef] [PubMed]
- Francischetti, I.M.; Valenzuela, J.G.; Pham, V.M.; Garfield, M.K.; Ribeiro, J.M. Toward a catalog for the transcripts and proteins (sialome) from the salivary gland of the malaria vector Anopheles gambiae. J. Exp. Biol. 2002, 205, 2429–2451. [Google Scholar] [PubMed]
- Jones, C.M.; Haji, K.A.; Khatib, B.O.; Bagi, J.; Mcha, J.; Devine, G.J.; Daley, M.; Kabula, B.; Ali, A.S.; Majambere, S. The dynamics of pyrethroid resistance in Anopheles arabiensis from Zanzibar and an assessment of the underlying genetic basis. Parasites Vectors 2013, 6, 343. [Google Scholar] [CrossRef]
- Riveron, J.M.; Ibrahim, S.S.; Chanda, E.; Mzilahowa, T.; Cuamba, N.; Irving, H.; Barnes, K.G.; Ndula, M.; Wondji, C.S. The highly polymorphic CYP6M7 cytochrome P450 gene partners with the directionally selected CYP6P9a and CYP6P9b genes to expand the pyrethroid resistance front in the malaria vector Anopheles funestus in Africa. BMC Genom. 2014, 15, 817. [Google Scholar] [CrossRef]
- Riveron, J.M.; Yunta, C.; Ibrahim, S.S.; Djouaka, R.; Irving, H.; Menze, B.D.; Ismail, H.M.; Hemingway, J.; Ranson, H.; Albert, A. A single mutation in the GSTe2 gene allows tracking of metabolically based insecticide resistance in a major malaria vector. Genome Biol. 2014, 15, R27. [Google Scholar] [CrossRef]
- Toé, K.H.; N’Falé, S.; Dabiré, R.K.; Ranson, H.; Jones, C.M. The recent escalation in strength of pyrethroid resistance in Anopheles coluzzi in West Africa is linked to increased expression of multiple gene families. BMC Genom. 2015, 16, 1. [Google Scholar] [CrossRef] [PubMed]
- Wilding, C.; Weetman, D.; Rippon, E.; Steen, K.; Mawejje, H.; Barsukov, I.; Donnelly, M. Parallel evolution or purifying selection, not introgression, explains similarity in the pyrethroid detoxification linked GSTE4 of Anopheles gambiae and An. arabiensis. Mol. Genet. Genom. 2015, 290, 201–215. [Google Scholar] [CrossRef] [PubMed]
- Samb, B.; Konate, L.; Irving, H.; Riveron, J.M.; Dia, I.; Faye, O.; Wondji, C.S. Investigating molecular basis of lambda-cyhalothrin resistance in an Anopheles funestus population from Senegal. Parasites Vectors 2016, 9, 449. [Google Scholar] [CrossRef] [PubMed]
- Arca, B.; Lombardo, F.; Lanfrancotti, A.; Spanos, L.; Veneri, M.; Louis, C.; Coluzzi, M. A cluster of four D7-related genes is expressed in the salivary glands of the African malaria vector Anopheles gambiae. Insect Mol. Biol. 2002, 11, 47–55. [Google Scholar] [CrossRef]
- Arcà, B.; Lombardo, F.; Valenzuela, J.G.; Francischetti, I.M.; Marinotti, O.; Coluzzi, M.; Ribeiro, J.M. An updated catalogue of salivary gland transcripts in the adult female mosquito, Anopheles gambiae. J. Exp. Biol. 2005, 208, 3971–3986. [Google Scholar] [CrossRef]
- Lanfrancotti, A.; Lombardo, F.; Santolamazza, F.; Veneri, M.; Castrignanò, T.; Coluzzi, M.; Arcà, B. Novel cDNAs encoding salivary proteins from the malaria vector Anopheles gambiae. FEBS Lett. 2002, 517, 67–71. [Google Scholar] [CrossRef]
- Mans, B.J.; Calvo, E.; Ribeiro, J.M.; Andersen, J.F. The crystal structure of D7r4, a salivary biogenic amine-binding protein from the malaria mosquito Anopheles gambiae. J. Biol. Chem. 2007, 282, 36626–36633. [Google Scholar] [CrossRef] [PubMed]
- Gillies, M.; Coetzee, M. A supplement to the Anophelinae of Africa south of the Sahara (Afrotropical region). Publ. S. Afr. Inst. Med. Res. 1987, 55, 1–143. [Google Scholar]
- Morgan, J.C.; Irving, H.; Okedi, L.M.; Steven, A.; Wondji, C.S. Pyrethroid resistance in an Anopheles funestus population from Uganda. PLoS ONE 2010, 5, e11872. [Google Scholar] [CrossRef]
- Livak, K.J. Organization and mapping of a sequence on the Drosophila melanogaster X and Y chromosomes that is transcribed during spermatogenesis. Genetics 1984, 107, 611–634. [Google Scholar] [PubMed]
- Koekemoer, L.; Kamau, L.; Hunt, R.; Coetzee, M. A cocktail polymerase chain reaction assay to identify members of the Anopheles funestus (Diptera: Culicidae) group. Am. J. Trop. Med. Hyg. 2002, 66, 804–811. [Google Scholar] [CrossRef]
- Santolamazza, F.; Mancini, E.; Simard, F.; Qi, Y.; Tu, Z.; della Torre, A. Insertion polymorphisms of SINE200 retrotransposons within speciation islands of Anopheles gambiae molecular forms. Malar. J. 2008, 7, 163. [Google Scholar] [CrossRef]
- Bass, C.; Nikou, D.; Blagborough, A.M.; Vontas, J.; Sinden, R.E.; Williamson, M.S.; Field, L.M. PCR-based detection of Plasmodium in Anopheles mosquitoes: A comparison of a new high-throughput assay with existing methods. Malar. J. 2008, 7, 177. [Google Scholar] [CrossRef]
- WHO. Test Procedures for Insecticide Resistance Monitoring in Malaria Vector Mosquitoes, 2nd ed.; WHO: Geneva, Switzerland, 2018. [Google Scholar]
- WHO. Test Procedures for Insecticide Resistance Monitoring in Malaria Vector Mosquitoes; WHO: Geneva, Switzerland, 2013. [Google Scholar]
- Djouaka, R.; Irving, H.; Tukur, Z.; Wondji, C.S. Exploring mechanisms of multiple insecticide resistance in a population of the malaria vector Anopheles funestus in Benin. PLoS ONE 2011, 6, e27760. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Torres, D.; Chandre, F.; Williamson, M.; Darriet, F.; Berge, J.B.; Devonshire, A.L.; Guillet, P.; Pasteur, N.; Pauron, D. Molecular characterization of pyrethroid knockdown resistance (kdr) in the major malaria vector Anopheles gambiae ss. Insect Mol. Biol. 1998, 7, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Djouaka, R.; Riveron, J.M.; Yessoufou, A.; Tchigossou, G.; Akoton, R.; Irving, H.; Djegbe, I.; Moutairou, K.; Adeoti, R.; Tamò, M. Multiple insecticide resistance in an infected population of the malaria vector Anopheles funestus in Benin. Parasites Vectors 2016, 9, 453. [Google Scholar] [CrossRef]
- Tchouakui, M.; Riveron, J.M.; Djonabaye, D.; Tchapga, W.; Irving, H.; Takam, P.S.; Njiokou, F.; Wondji, C.S. Fitness Costs of the Glutathione S-Transferase Epsilon 2 (L119F-GSTe2) Mediated Metabolic Resistance to Insecticides in the Major African Malaria Vector Anopheles Funestus. Genes 2018, 9, 645. [Google Scholar] [CrossRef] [PubMed]
- Menze, B.D.; Wondji, M.J.; Tchapga, W.; Tchoupo, M.; Riveron, J.M.; Wondji, C.S. Bionomics and insecticides resistance profiling of malaria vectors at a selected site for experimental hut trials in central Cameroon. Malar. J. 2018, 17, 317. [Google Scholar] [CrossRef]
- Rivero, A.; Vezilier, J.; Weill, M.; Read, A.F.; Gandon, S. Insecticide control of vector-borne diseases: When is insecticide resistance a problem? PLoS Pathog. 2010, 6, e1001000. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, J.M.; Francischetti, I.M. Role of arthropod saliva in blood feeding: Sialome and post-sialome perspectives. Annu. Rev. Entomol. 2003, 48, 73–88. [Google Scholar] [CrossRef] [PubMed]
- Cohuet, A.; Simard, F.; Wondji, C.S.; Antonio-Nkondjio, C.; Awono-Ambene, P.; Fontenille, D. High malaria transmission intensity due to Anopheles funestus (Diptera: Culicidae) in a village of savannah–forest transition area in Cameroon. J. Med. Entomol. 2004, 41, 901–905. [Google Scholar] [CrossRef]
- Menze, B.D.; Riveron, J.M.; Ibrahim, S.S.; Irving, H.; Antonio-Nkondjio, C.; Awono-Ambene, P.H.; Wondji, C.S. Multiple insecticide resistance in the malaria vector Anopheles funestus from Northern Cameroon is mediated by metabolic resistance alongside potential target site insensitivity mutations. PLoS ONE 2016, 11, e0163261. [Google Scholar] [CrossRef]
- Ndo, C.; Kopya, E.; Menze-Djantio, B.; Toto, J.C.; Awono-Ambene, P.; Lycett, G.; Wondji, C.S. High susceptibility of wild Anopheles funestus to infection with natural Plasmodium falciparum gametocytes using membrane feeding assays. Parasites Vectors 2016, 9, 341. [Google Scholar] [CrossRef]
- Kamdem, C.; Fossog, B.T.; Simard, F.; Etouna, J.; Ndo, C.; Kengne, P.; Boussès, P.; Etoa, F.-X.; Awono-Ambene, P.; Fontenille, D. Anthropogenic habitat disturbance and ecological divergence between incipient species of the malaria mosquito Anopheles gambiae. PLoS ONE 2012, 7, e39453. [Google Scholar] [CrossRef]
- Chouaïbou, M.; Etang, J.; Brévault, T.; Nwane, P.; Hinzoumbé, C.K.; Mimpfoundi, R.; Simard, F. Dynamics of insecticide resistance in the malaria vector Anopheles gambiae sl from an area of extensive cotton cultivation in Northern Cameroon. Trop. Med. Int. Health 2008, 13, 476–486. [Google Scholar] [CrossRef] [PubMed]
- Mueller, P.; Chouaibou, M.; Pignatelli, P.; Etang, J.; Walker, E.D.; Donnelly, M.J.; Simard, F.; Ranson, H. Pyrethroid tolerance is associated with elevated expression of antioxidants and agricultural practice in Anopheles arabiensis sampled from an area of cotton fields in Northern Cameroon. Mol. Ecol. 2008, 17, 1145–1155. [Google Scholar] [CrossRef]
- Ndo, C.; Kopya, E.; Donbou, M.A.; Njiokou, F.; Awono-Ambene, P.; Wondji, C. Elevated Plasmodium infection rates and high pyrethroid resistance in major malaria vectors in a forested area of Cameroon highlight challenges of malaria control. Parasites Vectors 2018, 11, 157. [Google Scholar] [CrossRef]
- Antonio-Nkondjio, C.; Fossog, B.T.; Ndo, C.; Djantio, B.M.; Togouet, S.Z.; Awono-Ambene, P.; Costantini, C.; Wondji, C.S.; Ranson, H. Anopheles gambiae distribution and insecticide resistance in the cities of Douala and Yaounde (Cameroon): Influence of urban agriculture and pollution. Malar. J. 2011, 10, 154. [Google Scholar] [CrossRef] [PubMed]
- Nwane, P.; Etang, J.; Chouaїbou, M.; Toto, J.C.; Koffi, A.; Mimpfoundi, R.; Simard, F. Multiple insecticide resistance mechanisms in Anopheles gambiae sl populations from Cameroon, Central Africa. Parasites Vectors 2013, 6, 41. [Google Scholar] [CrossRef] [PubMed]
- Tene Fossog, B.; Ayala, D.; Acevedo, P.; Kengne, P.; Ngomo Abeso Mebuy, I.; Makanga, B.; Magnus, J.; Awono-Ambene, P.; Njiokou, F.; Pombi, M. Habitat segregation and ecological character displacement in cryptic African malaria mosquitoes. Evol. Appl. 2015, 8, 326–345. [Google Scholar] [CrossRef]
- Isaacs, A.T.; Mawejje, H.D.; Tomlinson, S.; Rigden, D.J.; Donnelly, M.J. Genome-wide transcriptional analyses in Anopheles mosquitoes reveal an unexpected association between salivary gland gene expression and insecticide resistance. BMC Genom. 2018, 19, 225. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, B.J.; Bibby, J.; Pignatelli, P.; Muangnoicharoen, S.; O’Neill, P.M.; Lian, L.-Y.; Müller, P.; Nikou, D.; Steven, A.; Hemingway, J. Cytochrome P450 6M2 from the malaria vector Anopheles gambiae metabolizes pyrethroids: Sequential metabolism of deltamethrin revealed. Insect Biochem. Mol. Biol. 2011, 41, 492–502. [Google Scholar] [CrossRef]
- Mitchell, S.N.; Stevenson, B.J.; Müller, P.; Wilding, C.S.; Egyir-Yawson, A.; Field, S.G.; Hemingway, J.; Paine, M.J.; Ranson, H.; Donnelly, M.J. Identification and validation of a gene causing cross-resistance between insecticide classes in Anopheles gambiae from Ghana. Proc. Natl. Acad. Sci. USA 2012, 109, 6147–6152. [Google Scholar] [CrossRef]
- Ibrahim, S.S.; Ndula, M.; Riveron, J.M.; Irving, H.; Wondji, C.S. The P450 CYP 6Z1 confers carbamate/pyrethroid cross-resistance in a major African malaria vector beside a novel carbamate-insensitive N485I acetylcholinesterase-1 mutation. Mol. Ecol. 2016, 25, 3436–3452. [Google Scholar] [CrossRef]
- Thomsen, E.K.; Strode, C.; Hemmings, K.; Hughes, A.J.; Chanda, E.; Musapa, M.; Kamuliwo, M.; Phiri, F.N.; Muzia, L.; Chanda, J. Underpinning sustainable vector control through informed insecticide resistance management. PLoS ONE 2014, 9, e99822. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, H.; Wilding, C.S.; Nardini, L.; Pignatelli, P.; Koekemoer, L.L.; Ranson, H.; Coetzee, M. Insecticide resistance in Anopheles arabiensis in Sudan: Temporal trends and underlying mechanisms. Parasites Vectors 2014, 7, 213. [Google Scholar] [CrossRef] [PubMed]
- Djegbe, I.; Cornelie, S.; Rossignol, M.; Demettre, E.; Seveno, M.; Remoue, F.; Corbel, V. Differential expression of salivary proteins between susceptible and insecticide-resistant mosquitoes of Culex quinquefasciatus. PLoS ONE 2011, 6, e17496. [Google Scholar] [CrossRef] [PubMed]
- Cornelie, S.; Rossignol, M.; Seveno, M.; Demettre, E.; Mouchet, F.; Djègbè, I.; Marin, P.; Chandre, F.; Corbel, V.; Remoué, F. Salivary gland proteome analysis reveals modulation of anopheline unique proteins in insensitive acetylcholinesterase resistant Anopheles gambiae mosquitoes. PLoS ONE 2014, 9, e103816. [Google Scholar] [CrossRef] [PubMed]
- Djègbè, I.; Boussari, O.; Sidick, A.; Martin, T.; Ranson, H.; Chandre, F.; Akogbéto, M.; Corbel, V. Dynamics of insecticide resistance in malaria vectors in Benin: First evidence of the presence of L1014S kdr mutation in Anopheles gambiae from West Africa. Malar. J. 2011, 10, 261. [Google Scholar] [CrossRef] [PubMed]

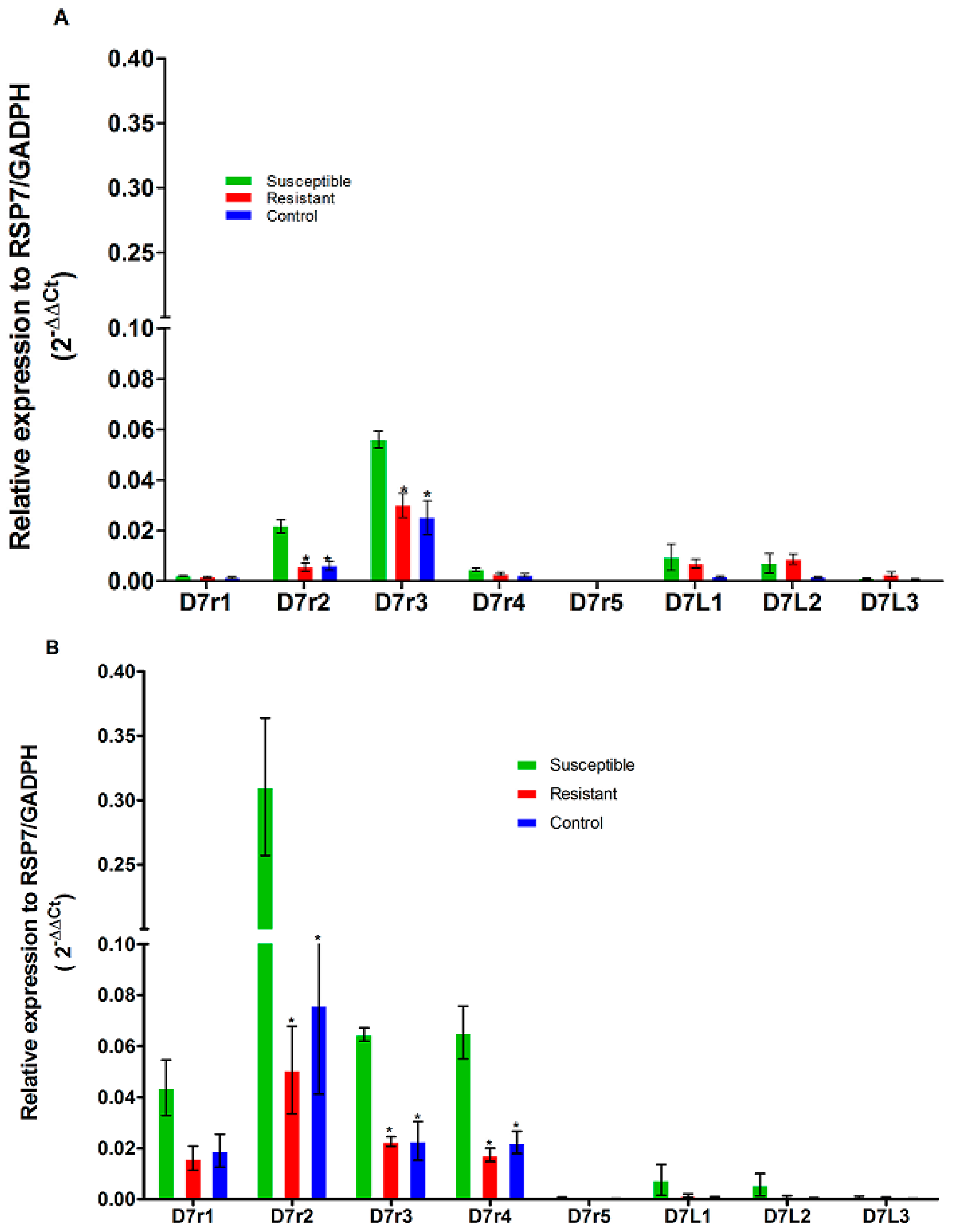
| Species | Gene | Fold Change Resistant vs. Susceptible | Log2FC | p-Value | Fold Change Control vs. Susceptible | Log2FC | p-Value |
|---|---|---|---|---|---|---|---|
| An. funestus | D7r2 | 6.66 | 2.73 | NS | 2.24 | 1.16 | NS |
| D7r3 | 11.99 | 3.58 | 0.04 | 4.44 | 2.15 | 0.006 | |
| D7r4 | 6.24 | 2.64 | NS | 4.012 | 2 | 0.02 | |
| An. gambiae | D7r1 | 0.744 | −0.425 | NS | 0.659 | −0.60 | NS |
| D7r2 | 0.257 | −1.95 | 0.007 | 0.27 | −1.84 | 0.008 | |
| D7r3 | 0.535 | −0.90 | 0.01 | 0.44 | −1.15 | 0.01 |
| Species | Gene | Fold Change Resistant vs. Susceptible | Log2FC | p-Value | Fold Change Control vs. Susceptible | Log2FC | p-Value |
|---|---|---|---|---|---|---|---|
| An. funestus | D7r2 | 2.2 | 1.1 | NS | 4.19 | 2.06 | 0.0002 |
| D7r3 | 9.88 | 3.304 | 0.001 | 8.404 | 3.07 | <0.0001 | |
| D7r4 | 4.45 | 2.15 | 0.026 | 6.76 | 2.75 | 0.04 | |
| An. gambiae | D7r1 | 0.36 | −1.44 | NS | 0.434 | −1.20 | NS |
| D7r2 | 0.163 | −2.61 | 0.001 | 0.245 | −2.02 | 0.02 | |
| D7r3 | 0.349 | −1.51 | 0.0002 | 0.353 | −1.5 | 0.006 | |
| D7r4 | 0.26 | −1.91 | 0.01 | 0.34 | −1.55 | 0.01 |
| 2N | H | S | Hd | π | D | D * | |
|---|---|---|---|---|---|---|---|
| Alive | 14 | 9 | 52 | 0.956 | 0.00142 | 0.175 | 1.132 |
| Dead | 12 | 9 | 49 | 0.955 | 0.0022 | −0.53006 | 0.0895 |
| Total | 26 | 18 | 84 | 0.978 | 0.024 | −1.04 | −1.54 |
| 2N | H | S | Hd | π | D | D * | |
|---|---|---|---|---|---|---|---|
| Alive | 8 | 4 | 28 | 0.857 | 0.017 | 0.98 | 5.27 * |
| Dead | 14 | 14 | 67 | 1 | 0.022 | −0.95 | −4.19 * |
| Total | 22 | 18 | 77 | 0.983 | 0.022 | −0.98 | −2.610 * |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elanga-Ndille, E.; Nouage, L.; Binyang, A.; Assatse, T.; Tene-Fossog, B.; Tchouakui, M.; Nguete Nguiffo, D.; Irving, H.; Ndo, C.; Awono-Ambene, P.; et al. Overexpression of Two Members of D7 Salivary Genes Family is Associated with Pyrethroid Resistance in the Malaria Vector Anopheles Funestus s.s. but Not in Anopheles Gambiae in Cameroon. Genes 2019, 10, 211. https://doi.org/10.3390/genes10030211
Elanga-Ndille E, Nouage L, Binyang A, Assatse T, Tene-Fossog B, Tchouakui M, Nguete Nguiffo D, Irving H, Ndo C, Awono-Ambene P, et al. Overexpression of Two Members of D7 Salivary Genes Family is Associated with Pyrethroid Resistance in the Malaria Vector Anopheles Funestus s.s. but Not in Anopheles Gambiae in Cameroon. Genes. 2019; 10(3):211. https://doi.org/10.3390/genes10030211
Chicago/Turabian StyleElanga-Ndille, Emmanuel, Lynda Nouage, Achille Binyang, Tatiane Assatse, Billy Tene-Fossog, Magellan Tchouakui, Daniel Nguete Nguiffo, Helen Irving, Cyrille Ndo, Parfait Awono-Ambene, and et al. 2019. "Overexpression of Two Members of D7 Salivary Genes Family is Associated with Pyrethroid Resistance in the Malaria Vector Anopheles Funestus s.s. but Not in Anopheles Gambiae in Cameroon" Genes 10, no. 3: 211. https://doi.org/10.3390/genes10030211
APA StyleElanga-Ndille, E., Nouage, L., Binyang, A., Assatse, T., Tene-Fossog, B., Tchouakui, M., Nguete Nguiffo, D., Irving, H., Ndo, C., Awono-Ambene, P., & Wondji, C. S. (2019). Overexpression of Two Members of D7 Salivary Genes Family is Associated with Pyrethroid Resistance in the Malaria Vector Anopheles Funestus s.s. but Not in Anopheles Gambiae in Cameroon. Genes, 10(3), 211. https://doi.org/10.3390/genes10030211






