Global Transcriptomic Profiling of Pulmonary Gene Expression in an Experimental Murine Model of Rickettsia conorii Infection
Abstract
:1. Introduction
2. Material and Methods
2.1. Mammalian Cell Culture and Preparation of Rickettsia conorii Stocks
2.2. Mouse Model of Infection
2.3. Purification of Total RNA, Complementary DNA Library Preparation, and RNA Sequencing
2.4. Mapping of Deep Sequencing Data and Ingenuity Pathway Analysis
2.5. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
2.6. Statistical Analysis
3. Results
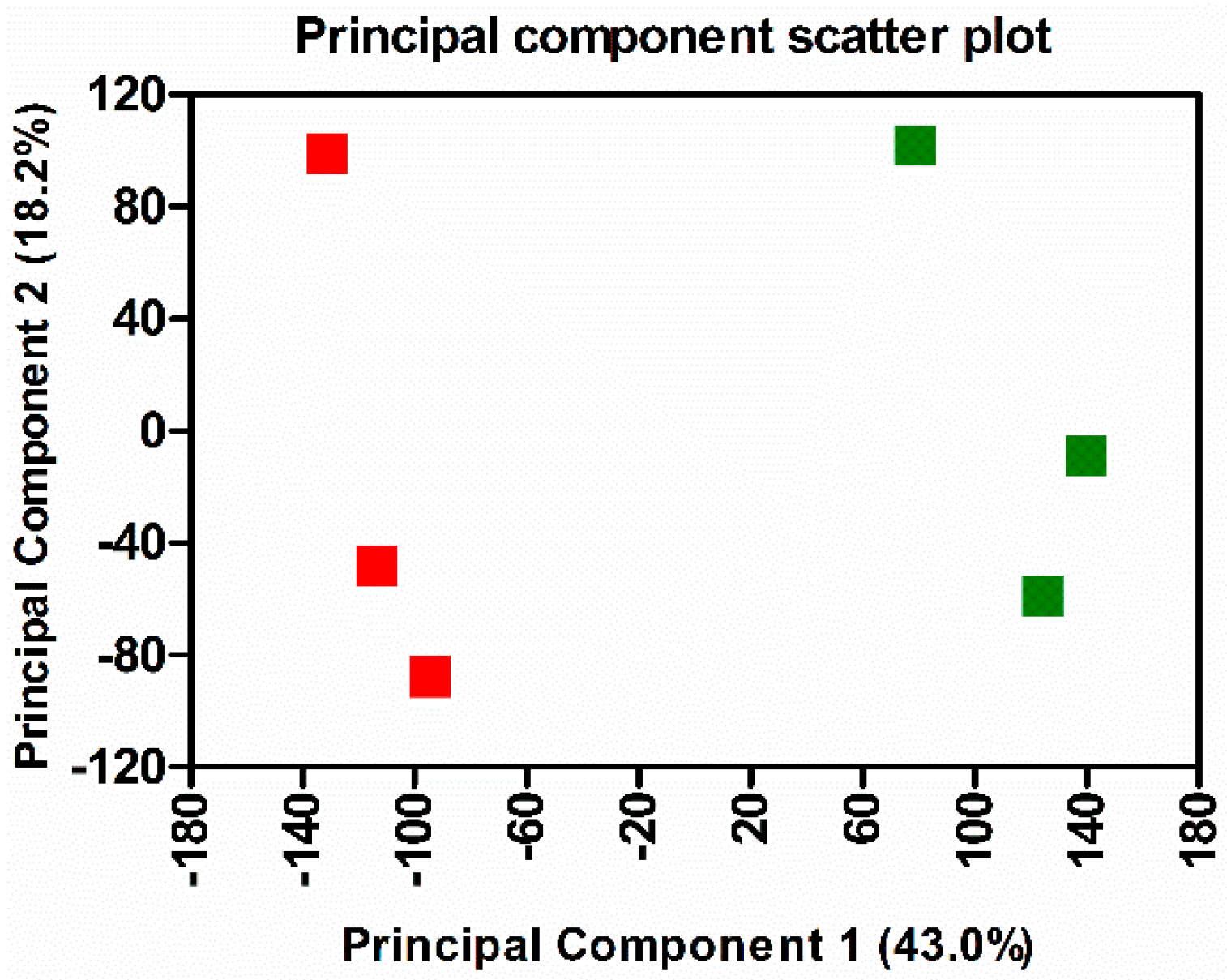
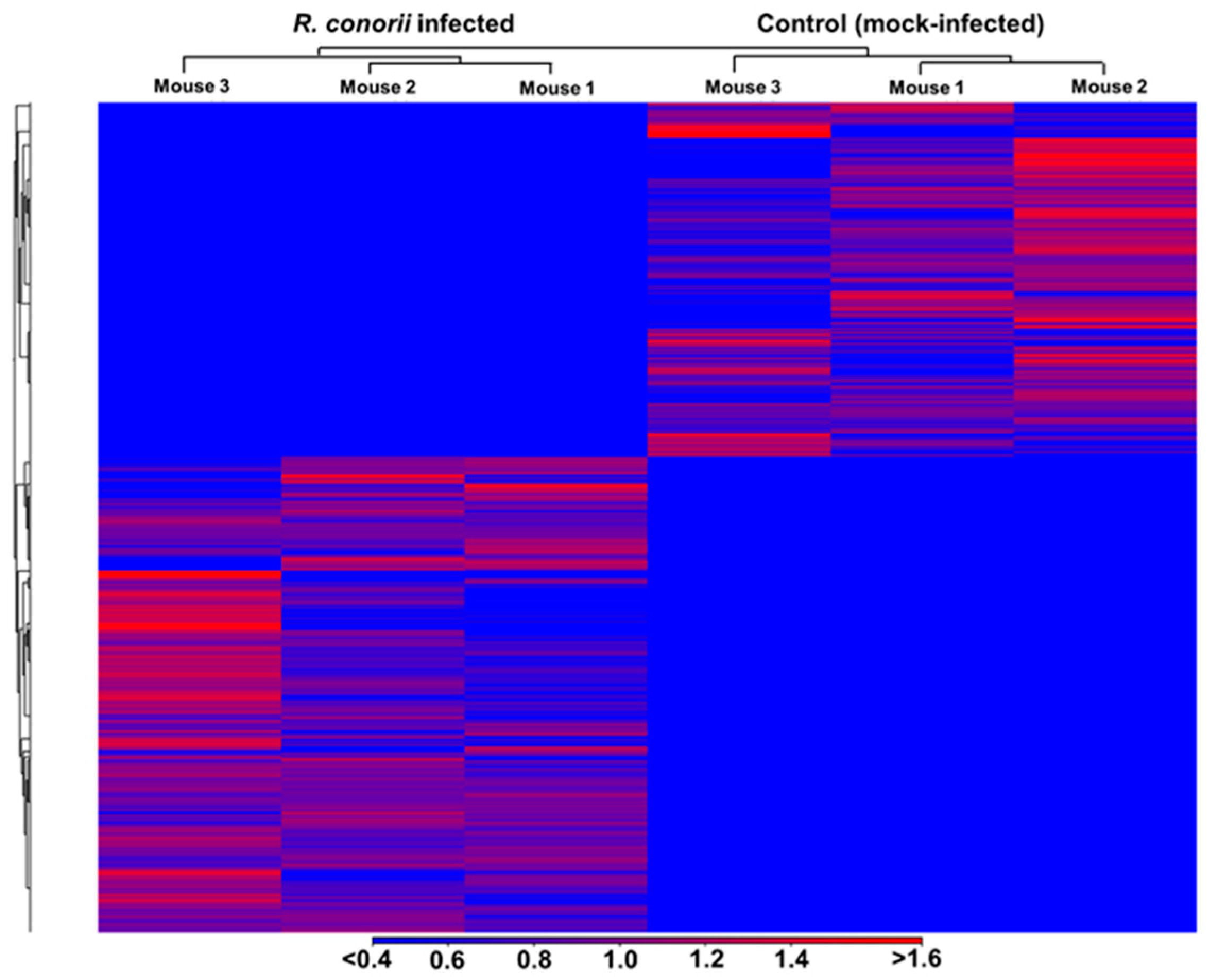
3.1. Transcriptome Sequencing, Assembly, and Analysis of Mouse Lungs During In Vivo R. conorii Infection
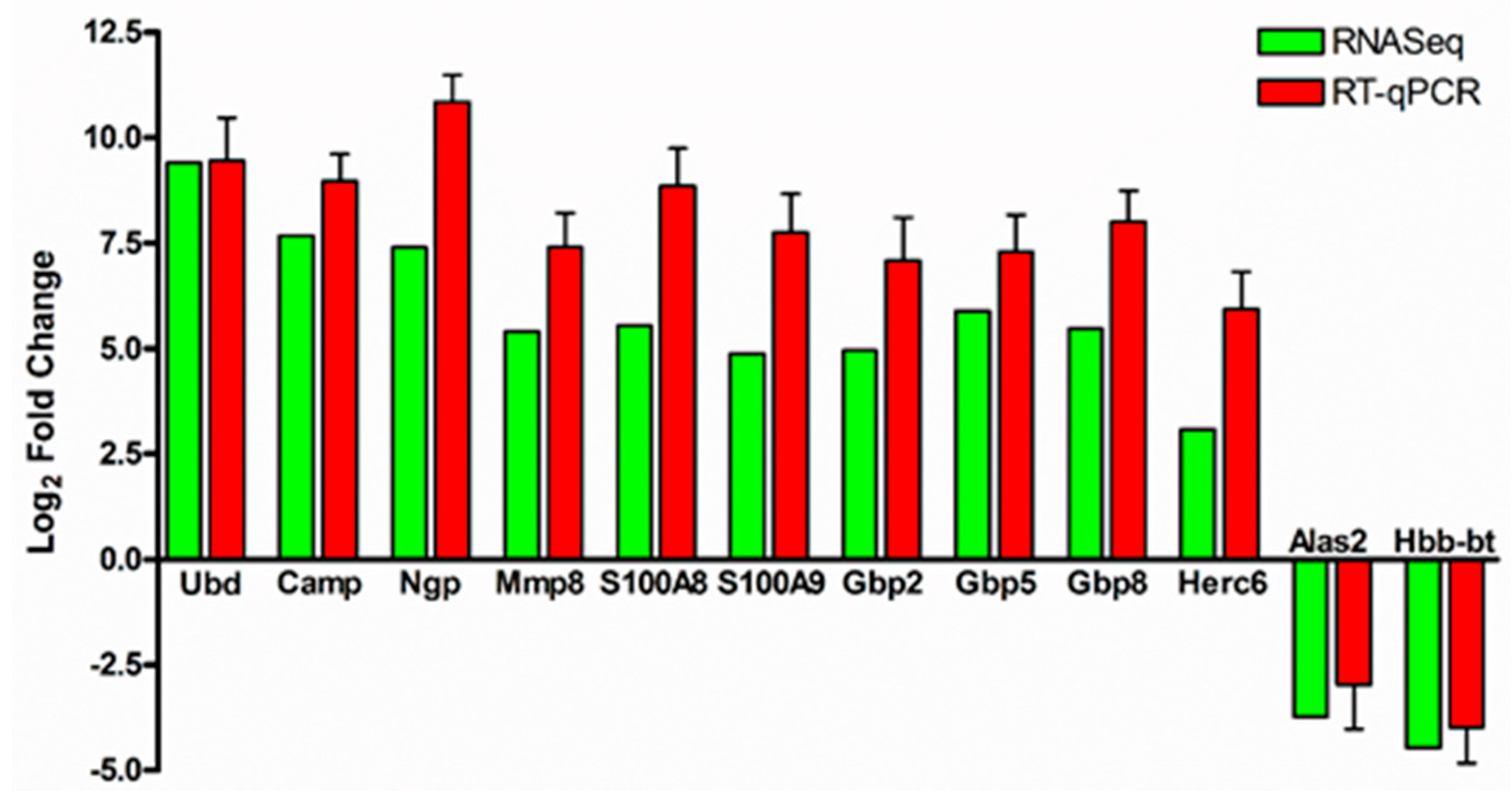
3.2. Validation of RNA-Sequencing Data by Quantitative Real-Time PCR
3.3. Functional Annotation and Classification of Transcriptome Data
3.4. Signatures of Inflammation and Tissue Damage
3.5. Antimicrobial Peptides
3.6. Activation or Inhibition of Upstream Signaling Regulators
3.7. Canonical Pathways
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sahni, A.; Fang, R.; Sahni, S.K.; Walker, D.H. Pathogenesis of rickettsial diseases: Pathogenic and immune mechanisms of an endotheliotropic infection. Annu. Rev. Pathol. 2019, 14, 127–152. [Google Scholar] [CrossRef]
- Parola, P.; Paddock, C.D.; Raoult, D. Tick-borne rickettsioses around the world: Emerging diseases challenging old concepts. Clin. Microbiol. Rev. 2005, 18, 719–756. [Google Scholar] [CrossRef] [PubMed]
- Parola, P.; Socolovschi, C.; Jeanjean, L.; Bitam, I.; Fournier, P.E.; Sotto, A.; Labauge, P.; Raoult, D. Warmer weather linked to tick attack and emergence of severe rickettsioses. PLoS Negl. Trop. Dis. 2008, 2, e338. [Google Scholar] [CrossRef] [PubMed]
- Parola, P.; Paddock, C.D.; Socolovschi, C.; Labruna, M.B.; Mediannikov, O.; Kernif, T.; Abdad, M.Y.; Stenos, J.; Bitam, I.; Fournier, P.E.; et al. Update on tick-borne rickettsioses around the world: A geographic approach. Clin. Microbiol. Rev. 2013, 26, 657–702. [Google Scholar] [CrossRef] [PubMed]
- Sahni, S.K.; Narra, H.P.; Sahni, A.; Walker, D.H. Recent molecular insights into rickettsial pathogenesis and immunity. Future Microbiol. 2013, 8, 1265–1288. [Google Scholar] [CrossRef] [PubMed]
- Rydkina, E.; Sahni, S.K.; Santucci, L.A.; Turpin, L.C.; Baggs, R.B.; Silverman, D.J. Selective modulation of antioxidant enzyme activities in host tissues during Rickettsia conorii infection. Microb. Pathog. 2004, 36, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Rydkina, E.; Silverman, D.J.; Sahni, S.K. Similarities and differences in host cell signaling following infection with different Rickettsia species. Ann. N. Y. Acad. Sci. 2005, 1063, 203–206. [Google Scholar] [CrossRef]
- Rydkina, E.; Sahni, A.; Baggs, R.B.; Silverman, D.J.; Sahni, S.K. Infection of human endothelial cells with spotted fever group rickettsiae stimulates cyclooxygenase 2 expression and release of vasoactive prostaglandins. Infect. Immun. 2006, 74, 5067–5074. [Google Scholar] [CrossRef]
- Rydkina, E.; Sahni, A.; Silverman, D.J.; Sahni, S.K. Comparative analysis of host-cell signalling mechanisms activated in response to infection with Rickettsia conorii and Rickettsia typhi. J. Med. Microbiol. 2007, 56, 896–906. [Google Scholar] [CrossRef]
- Colonne, P.M.; Eremeeva, M.E.; Sahni, S.K. Beta interferon-mediated activation of signal transducer and activator of transcription protein 1 interferes with Rickettsia conorii replication in human endothelial cells. Infect. Immun. 2011, 79, 3733–3743. [Google Scholar] [CrossRef]
- Colonne, P.M.; Sahni, A.; Sahni, S.K. Rickettsia conorii infection stimulates the expression of ISG15 and ISG15 protease UBP43 in human microvascular endothelial cells. Biochem. Biophys. Res. Commun. 2011, 416, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Osterloh, A. Immune response against rickettsiae: Lessons from murine infection models. Med. Microbiol. Immunol. 2017, 206, 403–417. [Google Scholar] [CrossRef] [PubMed]
- Riley, S.P.; Cardwell, M.M.; Chan, Y.G.; Pruneau, L.; Del Piero, F.; Martinez, J.J. Failure of a heterologous recombinant Sca5/OmpB protein-based vaccine to elicit effective protective immunity against Rickettsia rickettsii infections in C3H/HeN mice. Pathog. Dis. 2015, 73, ftv101. [Google Scholar] [CrossRef] [PubMed]
- Eisemann, C.S.; Osterman, J.V. Proteins of typhus and spotted fever group rickettsiae. Infect. Immun. 1976, 14, 155–162. [Google Scholar] [PubMed]
- Ellison, D.W.; Clark, T.R.; Sturdevant, D.E.; Virtaneva, K.; Porcella, S.F.; Hackstadt, T. Genomic comparison of virulent Rickettsia rickettsii Sheila Smith and avirulent Rickettsia rickettsii Iowa. Infect. Immun. 2008, 76, 542–550. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.M.; Wen, J.; Walker, D.H. Rickettsia australis infection: A murine model of a highly invasive vasculopathic rickettsiosis. Am. J. Pathol. 1993, 142, 1471–1482. [Google Scholar] [PubMed]
- De Sousa, R.; Ismail, N.; Nobrega, S.D.; Franca, A.; Amaro, M.; Anes, M.; Pocas, J.; Coelho, R.; Torgal, J.; Bacellar, F.; et al. Intralesional expression of mRNA of interferon-γ, tumor necrosis factor-α, interleukin-10, nitric oxide synthase, indoleamine-2,3-dioxygenase, and RANTES is a major immune effector in Mediterranean spotted fever rickettsiosis. J. Infect. Dis. 2007, 196, 770–781. [Google Scholar] [CrossRef]
- Walker, D.H.; Popov, V.L.; Wen, J.; Feng, H.M. Rickettsia conorii infection of C3H/HeN mice. A model of endothelial-target rickettsiosis. Lab. Investig. J. Tech. Methods Pathol. 1994, 70, 358–368. [Google Scholar]
- Sahni, S.K.; Kiriakidi, S.; Colonne, M.P.; Sahni, A.; Silverman, D.J. Selective activation of signal transducer and activator of transcription (STAT) proteins STAT1 and STAT3 in human endothelial cells infected with Rickettsia rickettsii. Clin. Microbiol. Infect. 2009, 15 (Suppl. 2), 303–304. [Google Scholar] [CrossRef]
- Rydkina, E.; Turpin, L.C.; Sahni, A.; Sahni, S.K. Regulation of inducible heme oxygenase and cyclooxygenase isozymes in a mouse model of spotted fever group rickettsiosis. Microb. Pathog. 2012, 53, 28–36. [Google Scholar] [CrossRef]
- Colonne, P.M.; Sahni, A.; Sahni, S.K. Suppressor of cytokine signalling protein SOCS1 and UBP43 regulate the expression of type I interferon-stimulated genes in human microvascular endothelial cells infected with Rickettsia conorii. J. Med. Microbiol. 2013, 62, 968–979. [Google Scholar] [CrossRef] [PubMed]
- Sahni, A.; Patel, J.; Narra, H.P.; Schroeder, C.L.C.; Walker, D.H.; Sahni, S.K. Fibroblast growth factor receptor-1 mediates internalization of pathogenic spotted fever rickettsiae into host endothelium. PLoS ONE 2017, 12, e0183181. [Google Scholar] [CrossRef] [PubMed]
- Caro-Gomez, E.; Gazi, M.; Cespedes, M.A.; Goez, Y.; Teixeira, B.; Valbuena, G. Phenotype of the anti-Rickettsia CD8+ T cell response suggests cellular correlates of protection for the assessment of novel antigens. Vaccine 2014, 32, 4960–4967. [Google Scholar] [CrossRef] [PubMed]
- Fang, R.; Ismail, N.; Walker, D.H. Contribution of NK cells to the innate phase of host protection against an intracellular bacterium targeting systemic endothelium. Am. J. Pathol. 2012, 181, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Marth, C.D.; Young, N.D.; Glenton, L.Y.; Noden, D.M.; Browning, G.F.; Krekeler, N. Deep sequencing of the uterine immune response to bacteria during the equine oestrous cycle. BMC Genom. 2015, 16, 934. [Google Scholar] [CrossRef] [PubMed]
- Langelier, C.; Kalantar, K.L.; Moazed, F.; Wilson, M.R.; Crawford, E.D.; Deiss, T.; Belzer, A.; Bolourchi, S.; Caldera, S.; Fung, M.; et al. Integrating host response and unbiased microbe detection for lower respiratory tract infection diagnosis in critically ill adults. Proc. Natl. Acad. Sci. USA 2018, 115, E12353–E12362. [Google Scholar] [CrossRef] [PubMed]
- Xiang, L.X.; He, D.; Dong, W.R.; Zhang, Y.W.; Shao, J.Z. Deep sequencing-based transcriptome profiling analysis of bacteria-challenged Lateolabrax japonicus reveals insight into the immune-relevant genes in marine fish. BMC Genom. 2010, 11, 472. [Google Scholar] [CrossRef]
- Narra, H.P.; Schroeder, C.L.; Sahni, A.; Rojas, M.; Khanipov, K.; Fofanov, Y.; Sahni, S.K. Small regulatory RNAs of Rickettsia conorii. Sci. Rep. 2016, 6, 36728. [Google Scholar] [CrossRef]
- Rydkina, E.; Turpin, L.C.; Sahni, S.K. Rickettsia rickettsii infection of human macrovascular and microvascular endothelial cells reveals activation of both common and cell type-specific host response mechanisms. Infect. Immun. 2010, 78, 2599–2606. [Google Scholar] [CrossRef]
- Chowdhury, I.H.; Narra, H.P.; Sahni, A.; Khanipov, K.; Fofanov, Y.; Sahni, S.K. Enhancer associated long non-coding RNA transcription and gene regulation in experimental models of rickettsial infection. Front. Immunol. 2018, 9, 3014. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Azad, A.F.; Beard, C.B. Rickettsial pathogens and their arthropod vectors. Emerg. Infect. Dis. 1998, 4, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Raoult, D. New rickettsial pathogens. Int. J. Clin. Pract. Suppl. 2000, 115, 79–87. [Google Scholar]
- Sahni, A.; Narra, H.P.; Walker, D.H.; Sahni, S.K. Endothelial activation and injury: The mechanisms of rickettsial vasculitis. In Vascular Responses to Pathogens; Gavins, F., Stokes, K.Y., Eds.; Elsevier: Atlanta, GA, USA, 2016; pp. 111–122. [Google Scholar]
- Riley, S.P.; Pruneau, L.; Martinez, J.J. Evaluation of changes to the Rickettsia rickettsii transcriptome during mammalian infection. PLoS ONE 2017, 12, e0182290. [Google Scholar] [CrossRef] [PubMed]
- Herath, H.; Jayasundara, J.; Senadhira, S.D.N.; Kularatne, S.A.M.; Kularatne, W.K.S. Spotted fever rickettsioses causing myocarditis and ARDS: A case from Sri Lanka. BMC Infect. Dis. 2018, 18, 705. [Google Scholar] [CrossRef] [PubMed]
- Snyder, A.; Alsauskas, Z.; Gong, P.; Rosenstiel, P.E.; Klotman, M.E.; Klotman, P.E.; Ross, M.J. FAT10: A novel mediator of Vpr-induced apoptosis in human immunodeficiency virus-associated nephropathy. J. Virol. 2009, 83, 11983–11988. [Google Scholar] [CrossRef] [PubMed]
- Oliva, J.; French, S.W. UBD (ubiquitin D). Atlas Genet. Cytogenet. Oncol. Haematol. 2012, 16, 289–292. [Google Scholar] [CrossRef]
- Xhindoli, D.; Pacor, S.; Benincasa, M.; Scocchi, M.; Gennaro, R.; Tossi, A. The human cathelicidin LL-37—A pore-forming antibacterial peptide and host-cell modulator. Biochim. Biophys. Acta 2016, 1858, 546–566. [Google Scholar] [CrossRef]
- Rivas-Santiago, B.; Hernandez-Pando, R.; Carranza, C.; Juarez, E.; Contreras, J.L.; Aguilar-Leon, D.; Torres, M.; Sada, E. Expression of cathelicidin LL-37 during Mycobacterium tuberculosis infection in human alveolar macrophages, monocytes, neutrophils, and epithelial cells. Infect. Immun. 2008, 76, 935–941. [Google Scholar] [CrossRef]
- Soong, L.; Wang, H.; Shelite, T.R.; Liang, Y.; Mendell, N.L.; Sun, J.; Gong, B.; Valbuena, G.A.; Bouyer, D.H.; Walker, D.H. Strong type 1, but impaired type 2, immune responses contribute to Orientia tsutsugamushi-induced pathology in mice. PLoS Negl. Trop. Dis. 2014, 8, e3191. [Google Scholar] [CrossRef]
- Papp, S.; Moderzynski, K.; Rauch, J.; Heine, L.; Kuehl, S.; Richardt, U.; Mueller, H.; Fleischer, B.; Osterloh, A. Liver necrosis and lethal systemic inflammation in a murine model of Rickettsia typhi infection: Role of neutrophils, macrophages and NK cells. PLoS Negl. Trop. Dis. 2016, 10, e0004935. [Google Scholar] [CrossRef] [PubMed]
- Moderzynski, K.; Papp, S.; Rauch, J.; Heine, L.; Kuehl, S.; Richardt, U.; Fleischer, B.; Osterloh, A. CD4+ T cells are as protective as CD8+ T cells against Rickettsia typhi infection by activating macrophage bactericidal activity. PLoS Negl. Trop. Dis. 2016, 10, e0005089. [Google Scholar] [CrossRef] [PubMed]
- Walker, D.H.; Dumler, J.S. The role of CD8 T lymphocytes in rickettsial infections. Semin. Immunopathol. 2015, 37, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Popov, V.L.; Yuoh, G.; Walker, D.H. Role of T lymphocyte subsets in immunity to spotted fever group Rickettsiae. J. Immunol. 1997, 158, 5314–5320. [Google Scholar] [PubMed]
- Wandel, M.P.; Pathe, C.; Werner, E.I.; Ellison, C.J.; Boyle, K.B.; von der Malsburg, A.; Rohde, J.; Randow, F. GBPs inhibit motility of Shigella flexneri but are targeted for degradation by the bacterial ubiquitin ligase IpaH9.8. Cell Host Microbe 2017, 22, 507–518.e5. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Jiang, W.; Yu, Q.; Liu, W.; Zhou, P.; Li, J.; Xu, J.; Xu, B.; Wang, F.; Shao, F. Ubiquitination and degradation of GBPs by a Shigella effector to suppress host defence. Nature 2017, 551, 378–383. [Google Scholar] [CrossRef] [PubMed]
- Man, S.M.; Karki, R.; Malireddi, R.K.; Neale, G.; Vogel, P.; Yamamoto, M.; Lamkanfi, M.; Kanneganti, T.D. The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat. Immunol. 2015, 16, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Meunier, E.; Wallet, P.; Dreier, R.F.; Costanzo, S.; Anton, L.; Ruhl, S.; Dussurgey, S.; Dick, M.S.; Kistner, A.; Rigard, M.; et al. Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat. Immunol. 2015, 16, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Smalley, C.; Bechelli, J.; Rockx-Brouwer, D.; Saito, T.; Azar, S.R.; Ismail, N.; Walker, D.H.; Fang, R. Rickettsia australis activates inflammasome in human and murine macrophages. PLoS ONE 2016, 11, e0157231. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, N.; Bahnan, W.; Wiley, D.J.; Barber, G.; Fields, K.A.; Schesser, K. Eukaryotic initiation factor 2 (eIF2) signaling regulates proinflammatory cytokine expression and bacterial invasion. J. Biol. Chem. 2012, 287, 28738–28744. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.G.; Francis, C.W.; Silverman, D.J.; Sahni, S.K. Nuclear factor κB protects against host cell apoptosis during Rickettsia rickettsii infection by inhibiting activation of apical and effector caspases and maintaining mitochondrial integrity. Infect. Immun. 2003, 71, 4127–4136. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.G.; Francis, C.W.; Silverman, D.J.; Sahni, S.K. NF-κB activation suppresses host cell apoptosis during Rickettsia rickettsii infection via regulatory effects on intracellular localization or levels of apoptogenic and anti-apoptotic proteins. FEMS Microbiol. Lett. 2004, 234, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Valbuena, G.; Walker, D.H.; Gazi, M.; Hidalgo, M.; DeSousa, R.; Oteo, J.A.; Goez, Y.; Brasier, A.R. Endothelial cell proteomic response to Rickettsia conorii infection reveals activation of the Janus Kinase (JAK)-Signal Transducer and Activator of Transcription (STAT)-Inferferon Stimulated Gene (ISG)15 pathway and reprogramming plasma membrane integrin/cadherin signaling. Mol. Cell. Proteom. 2016, 15, 289–304. [Google Scholar] [CrossRef]
- Bechelli, J.; Vergara, L.; Smalley, C.; Buzhdygan, T.P.; Bender, S.; Zhang, W.; Liu, Y.; Popov, V.L.; Wang, J.; Garg, N.; et al. Atg5 supports Rickettsia australis infection in macrophages in vitro and in vivo. Infect. Immun. 2019. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, T.; Kishi, M.; Ogawa, M. Restriction of the growth of a nonpathogenic spotted fever group Rickettsia. FEMS Immunol. Med. Microbiol. 2012, 64, 42–47. [Google Scholar] [CrossRef] [PubMed]
| Symbol | Gene Name | p-Value | FDR (q-Value) | FC | Log2 FC |
|---|---|---|---|---|---|
| Up-regulated molecules | |||||
| Ubd | Ubiquitin D | 0 | 0 | 679.8 | 9.4 |
| Acod1 | Aconitate decarboxylase 1 | 0 | 0 | 433.9 | 8.8 |
| Cxcl9 | Chemokine (C-X-C motif) ligand 9 | 0 | 0 | 405.4 | 8.7 |
| Ido1 | Indoleamine 2,3-dioxygenase 1 | 0 | 0 | 332.3 | 8.4 |
| Cxcl10 | C-X-C motif chemokine ligand 10 | 0 | 0 | 331.6 | 8.4 |
| Ly6a | Lymphocyte antigen 6 complex, locus A | 0 | 0 | 223.2 | 7.8 |
| Camp | Cathelicidin antimicrobial peptide | 0 | 0 | 204 | 7.7 |
| Ngp | Neutrophilic granule protein | 0 | 0 | 168 | 7.4 |
| Gzmk | Granzyme K | 0 | 0 | 135.7 | 7.1 |
| Gbp6 | Guanylate binding protein family member 6 | 0 | 0 | 127.8 | 7 |
| Ifn-γ | Interferon γ | 0 | 0 | 119.8 | 6.9 |
| Serpina3g | Serine (or cysteine) peptidase inhibitor, clade A, member 3G | 0 | 0 | 117.5 | 6.9 |
| Gzmb | Granzyme B | 0 | 0 | 103.9 | 6.7 |
| Gzma | Granzyme A | 0 | 0 | 90.4 | 6.5 |
| Klrg1 | Killer cell lectin like receptor G1 | 0 | 0 | 87.7 | 6.5 |
| Ccl7 | Chemokine (C-C motif) ligand 7 | 0 | 0 | 83.9 | 6.4 |
| Ccl2 | Chemokine (C-C motif) ligand 2 | 0 | 0 | 74.8 | 6.2 |
| Tgtp1/Tgtp2 | T cell specific GTPase 1 | 0 | 0 | 73.4 | 6.2 |
| Gbp5 | Guanylate binding protein 5 | 0 | 0 | 59.1 | 5.9 |
| Xcl1 | X-C motif chemokine ligand 1 | 0 | 0 | 52.6 | 5.7 |
| Calhm6 | Calcium homeostasis modulator family member 6 | 0 | 0 | 52.2 | 5.7 |
| Down-regulated molecules | |||||
| Hbb-bt | Hemoglobin subunit β | 0 | 0 | −25.2 | −4.7 |
| Hba1/Hba2 | Hemoglobin subunit α 2 | 0 | 0 | −22 | −4.5 |
| Alas2 | 5’-aminolevulinate synthase 2 | 0 | 0 | −13.3 | −3.7 |
| Cyp26b1 | Cytochrome P450 family 26 subfamily B member 1 | 3 × 10−7 | 0.000005 | −10.2 | −3.3 |
| Alb | Albumin | 0.000546 | 0.00461 | −9.2 | −3.2 |
| Hey1 | Hes related family bHLH transcription factor with YRPW motif 1 | 0 | 0 | −6.9 | −2.8 |
| Aplnr | Apelin receptor | 0 | 0 | −6.6 | −2.7 |
| Nr1d1 | Nuclear receptor subfamily 1 group D member 1 | 0 | 0 | −5.9 | −2.6 |
| Ptprb | Protein tyrosine phosphatase, receptor type B | 0 | 0 | −5.8 | −2.5 |
| Dll4 | Delta like canonical Notch ligand 4 | 0 | 0 | −5.7 | −2.5 |
| Efnb2 | Ephrin B2 | 0 | 0 | −5.1 | −2.3 |
| Ltbp4 | Latent transforming growth factor β binding protein 4 | 0 | 0 | −5 | −2.3 |
| Hmcn1 | Hemicentin 1 | 0 | 0 | −4.9 | −2.3 |
| Angptl2 | Angiopoietin like 2 | 0 | 0 | −4.7 | −2.2 |
| Ednrb | Endothelin receptor type B | 0 | 0 | −4.1 | −2 |
| Spock2 | SPARC/osteonectin, cwcv and kazal like domains proteoglycan 2 | 0 | 0 | −4.1 | −2 |
| Sh3pxd2a | SH3 and PX domains 2A | 0 | 0 | −3.7 | −1.9 |
| Ndst1 | N-deacetylase and N-sulfotransferase 1 | 0 | 0 | −3.7 | −1.9 |
| Igfbp5 | Insulin like growth factor binding protein 5 | 6.68 × 10−7 | 1.04 × 10−5 | −3.5 | −1.8 |
| Pdgfrb | Platelet derived growth factor receptor β | 0 | 0 | −3.5 | −1.8 |
| Regulator | Molecule Type | p-Value | Activation z-Score |
|---|---|---|---|
| Activated upstream regulators | |||
| Irf3 | Transcription regulator | 2.21 × 10−24 | 6.8 |
| Ifn-γ | Cytokine | 1.82 × 10−27 | 6.5 |
| Ticam1 | Other | 4.13 × 10−22 | 6.3 |
| Irf7 | Transcription regulator | 1.62 × 10−24 | 5.8 |
| Stat1 | Transcription regulator | 1.62 × 10−17 | 5.7 |
| Myd88 | Other | 8.02 × 10−20 | 5.5 |
| Cd38 | Enzyme | 1.17 × 10−8 | 5.5 |
| Samsn1 | Other | 5.86 × 10−12 | 5.4 |
| Il5 | Cytokine | 3.07 × 10−9 | 5.2 |
| Rb1 | Transcription regulator | 8.56 × 10−6 | 4.9 |
| Dock8 | Other | 5.24 × 10−11 | 4.9 |
| Sash1 | Other | 1.03 × 10−9 | 4.8 |
| Tlr4 | Transmembrane receptor | 4.04 × 10−16 | 4.7 |
| Nfatc2 | Transcription regulator | 1.88 × 10−8 | 4.6 |
| Tnf | Cytokine | 3.54 × 10−13 | 4.6 |
| Chuk | Kinase | 9.36 × 10−14 | 4.5 |
| Mavs | Other | 1.04 × 10−14 | 4.3 |
| Arhgap21 | Other | 7.33 × 10−7 | 4.1 |
| Ikbkb | Kinase | 8.43 × 10−17 | 4.1 |
| Irf5 | Transcription regulator | 2.48 × 10−9 | 4 |
| Inhibited upstream regulators | |||
| Il10ra | Transmembrane receptor | 1.85 × 10−19 | −7.4 |
| Ptger4 | G-protein coupled receptor | 9.94 × 10−16 | −6 |
| Kdm5a | Transcription regulator | 4.61 × 10−7 | −5.2 |
| Socs1 | Other | 1.19 × 10−15 | −4.6 |
| Irgm1 | Other | 2.56 × 10−10 | −4 |
| Bcl6 | Transcription regulator | 7.76 × 10−5 | −3.6 |
| Srf | Transcription regulator | 0.000721 | −3.4 |
| Nr3c1 | Ligand-dependent nuclear receptor | 7.27 × 10−8 | −3.4 |
| Gfi1 | Transcription regulator | 0.000536 | −3.3 |
| Ncstn | Peptidase | 2.48 × 10−9 | −3.3 |
| Apoe | Transporter | 5.75 × 10−6 | −2.9 |
| Por | Enzyme | 5.08 × 10−5 | −2.9 |
| Dusp1 | Phosphatase | 0.013 | −2.7 |
| C5 | Cytokine | 0.0151 | −2.6 |
| Tgfbr1 | Kinase | 0.0333 | −2.6 |
| Dicer1 | Enzyme | 0.00974 | −2.6 |
| Stat6 | Transcription regulator | 5.24 × 10−14 | −2.5 |
| Cyb5r4 | Enzyme | 0.000106 | −2.4 |
| Abcg1 | Transporter | 0.00048 | −2.4 |
| Irf9 | Transcription regulator | 2.32 × 10−8 | −2.3 |
| Canonical Pathway Name | p-Value | Differentially Expressed Genes/Total Number of Genes in the Pathway |
|---|---|---|
| EIF2 Signaling | 1.54 × 10−42 | 100/205 (48.8%) |
| Oxidative Phosphorylation | 1.29 × 10−21 | 48/96 (50.0%) |
| Mitochondrial Dysfunction | 3.29 × 10−19 | 59/153 (38.6%) |
| Antigen Presentation Pathway | 3.90 × 10−18 | 23/28 (82.1%) |
| Protein Ubiquitination Pathway | 1.33 × 10−15 | 72/245 (29.4%) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Narra, H.P.; Sahni, A.; Khanipov, K.; Fofanov, Y.; Sahni, S.K. Global Transcriptomic Profiling of Pulmonary Gene Expression in an Experimental Murine Model of Rickettsia conorii Infection. Genes 2019, 10, 204. https://doi.org/10.3390/genes10030204
Narra HP, Sahni A, Khanipov K, Fofanov Y, Sahni SK. Global Transcriptomic Profiling of Pulmonary Gene Expression in an Experimental Murine Model of Rickettsia conorii Infection. Genes. 2019; 10(3):204. https://doi.org/10.3390/genes10030204
Chicago/Turabian StyleNarra, Hema P., Abha Sahni, Kamil Khanipov, Yuriy Fofanov, and Sanjeev K. Sahni. 2019. "Global Transcriptomic Profiling of Pulmonary Gene Expression in an Experimental Murine Model of Rickettsia conorii Infection" Genes 10, no. 3: 204. https://doi.org/10.3390/genes10030204
APA StyleNarra, H. P., Sahni, A., Khanipov, K., Fofanov, Y., & Sahni, S. K. (2019). Global Transcriptomic Profiling of Pulmonary Gene Expression in an Experimental Murine Model of Rickettsia conorii Infection. Genes, 10(3), 204. https://doi.org/10.3390/genes10030204






