Abstract
Serotonin is a neurotransmitter involved in various physiological processes in the central and peripheral nervous systems. Serotonin is also a precursor for melatonin biosynthesis, which mainly occurs in the pineal gland of vertebrates. Tryptophan hydroxylase (TPH) acts as the rate-limiting enzyme in serotonin biosynthesis and is the initial enzyme involved in the synthesis of melatonin. Recently, two enzymes—TPH1 and TPH2—were reported to form the TPH family in vertebrates and to play divergent roles in serotonergic systems. Here, we examined the evolution of the TPH family from 70 vertebrate genomes. Based on the sequence similarity, we extracted 184 predicted tph homologs in the examined vertebrates. A phylogenetic tree, constructed on the basis of these protein sequences, indicated that tph genes could be divided into two main clades (tph1 and tph2), and that the two clades were further split into two subgroups of tetrapods and Actinopterygii. In tetrapods, and some basal non-teleost ray-finned fishes, only two tph isotypes exist. Notably, tph1 in most teleosts that had undergone the teleost-specific genome duplication could be further divided into tph1a and tph1b. Moreover, protein sequence comparisons indicated that TPH protein changes among vertebrates were concentrated at the NH2-terminal. The tertiary structures of TPH1 and TPH2 revealed obvious differences in the structural elements. Five positively selected sites were characterized in TPH2 compared with TPH1; these sites may reflect the functional divergence in enzyme activity and substrate specificity. In summary, our current work provides novel insights into the evolution of tph genes in vertebrates from a comprehensive genomic perspective.
1. Introduction
As one of the major monoaminergic neurotransmitters in the central nervous system, serotonin is involved in various physiological processes, such as the regulation of appetite, aggressiveness, body temperature, fear, mood, pain, reproduction, sleep, and vascular functions [1,2]. Before it becomes a neurotransmitter, serotonin also influences the development and maturation of the mammalian brain [3]. In addition, serotonin acts as a precursor for the synthesis of melatonin, which is mainly produced in the pineal gland of vertebrates. Tryptophan hydroxylase (TPH, EC 1.14.16.4) is the rate-limiting enzyme for serotonin biosynthesis and is the initial activated enzyme in melatonin synthesis. TPH transforms tryptophan (Trp) into 5-hydroxytryptophan (5-HTP) [4], and subsequently, 5-HTP is catalyzed to serotonin (5-hydroxytryptamine, 5-HT) by l-aromatic amino acid decarboxylase (AAAD, EC 4.1.1.28) [5]. Serotonin is further catalyzed by aralkylamine N-acetyltransferase (AANAT, EC 2.3.1.87) [6] and acetylserotonin-O-methyltransferase (ASMT; EC 2.1.1.4) [7] to finally generate melatonin (MT; see Figure 1).
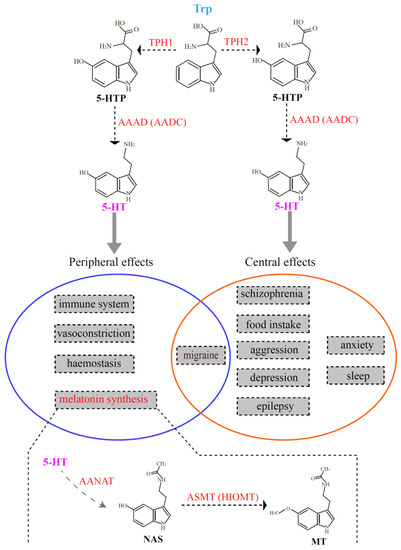
Figure 1.
Schematic representation of the two serotonin systems in vertebrates. The top part denotes the process of serotonin biosynthesis, which is also the first two steps in melatonin synthesis. The middle part (in the circles) summarizes the functions regulated by the two serotonin systems. The bottom part interprets the last two steps for melatonin synthesis. 5-HT, 5-hydroxytryptamine; AAAD (AADC), l-aromatic amino acid decarboxylase; AANAT, aralkylamine N-acetyltransferase; ASMT, acetylserotonin-O-methyltransferase; HIOMT, hydroxyindole-O-mehyltransferase; MT, melatonin; NAS, N-acetylserotonin; TPH, tryptophan hydroxylase.
Tryptophan hydroxylase belongs to the superfamily of aromatic amino acid hydroxylase that includes tyrosine hydroxylase (TH) and phenylalanine hydroxylase (PAH) [8]. Compared with the increasing number of reports concerning the families of TH and PAH [9,10,11,12], studies of TPH appear sparse [13,14], and a comprehensive investigation in vertebrates is unavailable at present. For more than a decade, the TPH family was initially thought to have only one member. However, in 2003, Walther et al. knocked out the tph gene (now called tph1) in mice. Surprisingly, they discovered that the tph1-deficient mice lacked serotonin in the pineal gland and certain peripheral tissues (such as gut and blood). They also observed that the serotonin content in the brainstem of the tph1-deficient mice was slightly reduced in comparison to the wild type, suggesting the existence of another tph member contributing to the stable concentration of serotonin in the brainstem [13]. Subsequently, they discovered and verified a novel member of the TPH family that they named tph2, and thus the classical tph gene was renamed tph1. It has been shown that tph1 is mainly distributed in the pineal gland and peripheral gut, spleen, and thymus; tph2 is predominantly expressed in structures of the central nervous system, such as the brainstem [15]. As summarized in Figure 1, there are two independent serotonin systems in vertebrates that are regulated by two different TPH enzymes. Therefore, these serotonin systems regulated by tph1 and tph2 have distinct functions. tph1 usually plays important roles in peripheral effects such as hemostasis, the immune system, melatonin synthesis, migraine, and vasoconstriction; tph2 is involved in effects of the central nervous system, such as aggression, anxiety, depression, epilepsy, food intake, migrate and sleep. Only the pathogenesis of migraine may involve both serotonin systems (the overlapped middle part in Figure 1).
Fish are the most diverse vertebrate taxa, and their serotonergic neurons can be identified based on corresponding 5-HT levels. A previous review [16] described tph as a specific marker for 5-HT generation. Teleosts experienced an episode of whole genome duplication (WGD) approximately 320 million years ago (Mya) [17,18], resulting in the duplication of certain genes related to 5-HT. After the WGD event, however, these duplicated genes occasionally were lost or became pseudogenes [19]. Hence, more tph copies (tph1a, tph1b, and tph2) have been reported in teleost species such as zebrafish, sticklebacks, and medaka [16,20].
Two members (tph1 and tph2) of the TPH family with distinct catalytic activity or substrate specificity had been reported previously [13]. Certain studies already indicated that distinct functions of different family members may have originated from different selection pressures during evolution [21,22]. Investigations of codon substitutions within vertebrate groups also suggested that certain protein families had experienced positive selection, which was considered as an important driving force in protein evolution both from structural and functional views [23,24,25]. To date, although additional copies of tph have been reported in fishes, a detailed phylogenetic analysis of these tph members among teleosts and tetrapods is lacking, and functional changes related to possible positive selection on different tph copies is poorly understood.
In the present study, we focused on 70 vertebrate species with high-quality genome assemblies for extraction of their tph family members. Meanwhile, we examined a number of special teleosts that possess some unique living habitats: cavefish (Sinocyclocheilus anshuiensis, Sa), amphibious mudskippers (blue-spotted Boleophthalmus pectinirostris (BP) and giant-fin Periophthalmus magnuspinnatus (PM)), migratory fish (Atlantic salmon (Salmo salar), and rainbow trout (Oncorhynchus mykiss)). We constructed a comprehensive phylogenetic tree from encoding sequences and implemented synteny analysis among the examined species. Sequence and structural divergences between different TPH members were also compared. Moreover, the biochemical and structural properties of possible positively selected sites in the TPH family were summarized. We attempted to answer the following questions. (1) What are the main structural differences in different members of the TPH family among various taxa of vertebrates? (2) Have some species experienced loss of certain tph member(s) during evolution? (3) Did positive selection affect functional changes of the TPH members during evolution?
2. Materials and Methods
2.1. Sequence Collection
In this study, a total of 70 vertebrate genomes were chosen to extract tph genes. Genome sequences of 68 species, including 11 mammals, 18 aves, 9 reptiles, 3 amphibians, and 27 Actinopterygii, with high-quality assemblies, were downloaded from the National Center for Biotechnology Information (NCBI), with the exception of two fish species (the American paddlefish and the Chinese sturgeon), which were assembled based on Illumina and PacBio sequencing using Platanus (version 1.2.4 [26], parameters: −k 31 −s 10 −a 10 −u 0.1 −d 0.4) and DBG2OLC (default version, [27], parameters: k 17 KmerCovTh 6 MinOverlap 80 AdaptiveTh 0.012 RemoveChimera 1) by our lab (Table S1). To further collect the entire length of tph1 and tph2 coding sequences in each genome, those reported and verified tph genes in human (Homo sapiens), chicken (Gallus gallus), painted turtle (Chrysemys picta bellii), African clawed frog (Xenopus laevis), and zebrafish (Danio rerio) were downloaded from NCBI as the queries. In general, we selected the sequences from organisms close to the target species as the queries; for most sequences of tph2 in the amphibians we used human tph2 (NP_775489.2) as the query. Detailed information concerning the queries is provided in Table S2.
We constructed a local database for each genome and subsequently aligned the sequences through BLAST (version 2.2.28) [28] using the protein-nucleotide aligned strategy with an E-value of 10−5. The alignment results were further processed by a Perl script to obtain the best hit of each alignment. Finally, EXONERATE (version 2.2.0) [29] was employed to predict the full length of each tph gene.
2.2. Sequence Alignment and Phylogenetic Reconstruction
The predicted nucleotide sequences of tph genes were initially used for multiple codon-based alignments using the Muscle module in MEGA (version 7.0) [30] and, subsequently, were translated to protein sequences for phylogenetic analyses. ProtTest (version 3.4.2) [31] was used to predict amino acid (aa) substitutions under the Akaike information criterion (AIC). The parameters within the best amino acid substitution of JTT+I+G were input to PhyML (version 3.1) [32] to construct phylogenetic topologies using maximum likelihood (ML) with 1000 bootstrap replicates for the evaluation of their branch supports.
To further confirm the topology yielded by ML, our alignment sequences were also analyzed using Bayesian inference (BI) in MrBayes (version 3.2.6) [33]. We conducted two independent Bayesian searches for 1 × 106 generations with one cold chain and three heated chains per run, and every 100 generations was sampled for each run. The initial 25% of the runs were discarded as a burn-in. A consensus tree was generated with the remaining runs, and Bayesian posterior probabilities were calculated to evaluate the branch supports. The maximum clade credibility tree was identified from the remaining samples using TreeAnnotator (version 1.7.5) [34].
2.3. Identification of Conserved Synteny
To examine the conservation of tph genes in vertebrates, we searched several genes located upstream and downstream of each tph paralog within tetrapods and teleosts through Ensemble datasets. Those genes located upstream and downstream of each tph paralog in humans and zebrafish were employed as the reference sequences for searching syntenic locations in the tetrapod and teleost sequences, respectively. Subsequently, we employed the protein aligned to nucleotides strategy to examine these extracted synteny genes in tetrapods and teleosts. Genome assemblies of different species were searched using BLAST with an E-value cutoff at 10−5. The best hit was used to identify an occurrence of a collinear gene among the examined species.
2.4. Detection of Differences between TPH Protein Sequences
We first selected the two human TPH isoforms to perform the alignment, and then analyzed the differences between TPH1 and TPH2. To further analyze secondary structures of these TPH proteins, we also downloaded the common human TPH1 (5L01) and TPH2 (4V06) protein templates from the Protein Data Bank (PDB) for comparing the differences between TPH1 and TPH2 in vertebrates. One or two representative species from each class were chosen to align with the human TPH1 and TPH2 templates. The examined species included cattle (Bos taurus), chicken, zebra finch (Taeniopygia guttata), American alligator (Alligator mississippiensis), Chinese alligator (A. sinensis), tropical clawed frog (Xenopus tropicalis), African clawed frog, zebrafish, amphibious teleosts (mudskippers), a Sinocyclocheilus cavefish (Sa), and tetraploid Sinocyclocheilus fish (Sg and Sr). Previous studies had reported that teleosts such as zebrafish possessed two tph1 genes (tph1a and tph1b) [20]; thus, we further chose some representative species from the teleosts to identify the differences between TPH1a and TPH1b. All the alignment results were colorized by TEXshade (version 1.24) [35].
2.5. Prediction of Tertiary Structures of TPH Proteins
To understand whether these possible variations could shape the tertiary structures, we predicted the tertiary structure and function of each TPH protein in zebrafish, chicken, and one mudskipper (BP) using SWISS-MODEL [36], an online modeling tool for automated comparison of protein homology. We first uploaded our target sequences and searched for a template that best matched our target sequences in terms of coverage and identity. The best templates of the TPH protein model in human (TPH1: 5L01; TPH2: 4V06) from PDB were downloaded to compare the tertiary structure differences among TPH1a, TPH1b, and TPH2. We subsequently conducted model–template alignment for structural comparisons. Finally, PyMOL (version 2.2) [37] was applied to visualize the predicted TPH protein tertiary structures, and these were adjusted in a similar pattern to identify variations.
2.6. Identification of Putative Positively Selected Sites
To evaluate variation in selection pressure between lineages of TPH1 and TPH2, we selected a subset of five species from those shown in Figure S1 to represent the five groups of vertebrates, and we constructed a BI tree for selection pressure analyses. We estimated the ratio of dN/dS (ratio of nonsynonymous substitutions (dN) to that of synonymous substitutions (dS)) under two prior assumptions using branch models in the CODEML modules of PAML (version 4) [38]. We first ran a one-ratio model for which one ω value was assumed for all clades. We then ran a two-ratio model for which ω values could change between the TPH1 clade and the TPH2 clade (setting the TPH2 clade as an independently varying target ω2; setting the TPH1 clade as the constant ω1). Likelihood ratio (LR) tests and Chi-square (χ2) tests were conducted by comparing the probabilities of the one-ratio and two-ratio models to verify which model best fit the data [39]. Finally, a branch-site model, allowing ω to have more than one change in the target branches, was used to identify which codon(s) had likely undergone positive selection. Those sites with significance (p-value less than 0.05) based on the Empirical Bayes method [40] were regarded as positively selected.
3. Results
3.1. Variation of tph Copy Number in Vertebrates
A total of 184 tph nucleotide sequences, including 105 tph1 (37 tph1a and 26 tph1b in teleosts, and 42 tph1 in tetrapods and non-teleosts) and 79 tph2 were successfully derived from 70 vertebrate species (see Table 1). These sequences and their encoding protein sequences were pooled for further analysis.

Table 1.
Distribution of the tryptophan hydroxylase (TPH) family in the selected vertebrate genomes.
In this study, we determined that mammals, birds, reptiles, amphibians, and some basal non-teleost ray-finned fishes, such as the spotted gar, American paddlefish, and Chinese sturgeon, had two tph isotypes (tph1 and tph2). For most tetrapods, we observed only one copy of tph1 and one copy of tph2, the one exception being the African clawed frog that had two copies of both tph1 and tph2 (see Figure 2 and Table S3). Two copies of tph1 and tph2 in the American paddlefish and eight copies of tph1 and five copies of tph2 in the Chinese sturgeon were also identified.
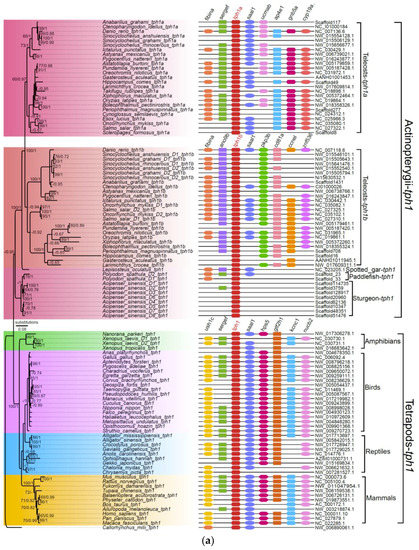
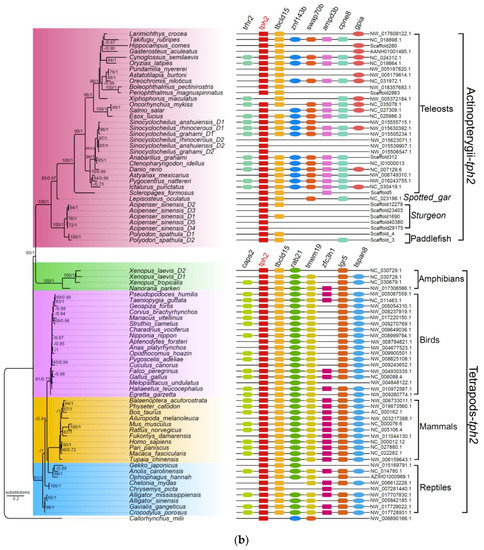
Figure 2.
Phylogenetic trees and genome synteny of tph1 and tph2 in vertebrates. (a) The phylogenetic tree was generated from 105 TPH1 protein sequences (the left part), and the synteny data of tph1 (the right part) are presented for confirmation. (b) The phylogenetic tree was generated from 79 TPH2 protein sequences (the left part), and the synteny data of tph2 (the right part) are presented for validation. Numbers on the branches from left to right are bootstrap values generated in the PhyML reconstruction and the Bayesian posterior probabilities obtained in the Bayesian inference, respectively. The bootstrap values under 60% and posterior probabilities less than 0.65 are not shown.
Previous studies reported the existence of three tph members (defined as tph1a, tph1b, and tph2) in teleosts such as zebrafish, medaka, and sticklebacks [16,20]. In our current work, we further strengthened the finding that most teleosts with the teleost-specific genome duplication (TSGD) presented three tph isotypes (tph1a, tph1b, and tph2) with the exception of the Asian arowana (Scleropages formosus), fugu (Takifugu rubripes), Northern pike (Esox lucius), and tongue sole (Cynoglossus semilaevis), for which we failed to find tph1b. In addition, certain tetraploid fishes that had undergone more than one WGD event, such as the Chinese golden-line fish (Sa, Sr, and Sg), had five tph copies (not the expected six); no duplicates of tph1a were identified. Other typical tetraploid teleosts such as the rainbow trout and Atlantic salmon had four copies of the tph genes, but the duplicated tph1a and tph2 loci were not found (Figure 2 and Table S3).
3.2. Phylogenetic Relationships among the tph Genes in Vertebrates
To understand the relationships among these tph genes in vertebrates, we constructed a robust phylogenentic tree based on the Bayesian inference (BI) method from the deduced 184 TPH protein sequences. The TPH2 protein sequence from the elephant shark (Callorhynchus milii) was employed as the outgroup. According to the well-supported phylogenetic topology (Figure S1), all tph genes could be divided into two main groups (tph1 and tph2), and both groups were further divided into two subgroups of tetrapods and Actinopterygii (Figure S1). The tetrapod subgroup was further divided into four main clades (amphibians, mammals, reptiles, and birds).
To avoid overestimation based on the BI method, we also performed a phylogenetic analysis (the left areas of Figure 2a,b) using the maximum likelihood (ML) method on 105 TPH1 and 79 TPH2 protein sequences. We observed that the topologies generated from both ML and BI were similar, indicating a relative stability of the phylogenetic trees. Finally, we employed the ML tree for presentation, and the Bayesian posterior probabilities from the BI analysis were labeled onto each node to indicate the degree of support (Figure 2).
Based on these results, we determined that both tph1 and tph2 were divided into two main clades, one for tetrapods and another for Actinopterygii. We detected in tetrapods and some basal non-teleost ray-finned fishes, namely, the spotted gar, American paddlefish, and Chinese sturgeon, only one isform of tph1. Notably, tph1, in the majority of teleosts that had undergone TSGD, could be further divided into tph1a and tph1b (see more details in the left part of Figure 2a).
3.3. Synteny Data
Our synteny data demonstrated that tph1a, tph1b, and tph2 share a conserved suite of genes bounding the sequences in both sides, although several genes were not identified in certain species. Genes located upstream and downstream of tph between tetrapods and Actinopterygii were different. In general, seven genes (gtf2h1, hps5, kcnc1, nucb2, saal1, sergef, and ush1c) were in the neighborhood of the tph1 gene in tetrapods. Among these seven genes, only two, saal1 and sergef, were identified near the tph1 gene in Actinopterygii. Similarly, seven genes (ap4e1, cyp19a1, fibina, gnb5a, saal1, sergef, and ucmab) were identified near tph1a, and seven genes (ano9b, ccne1, cd81a, fibinb, pkp3b, saal1, and znf536) were identified near tph1b in Actinopterygii. However, only fibina and saal1 were common to the loci neighboring tph1a and tph1b (see more details in the right part of Figure 2a). The genes located near tph2 differed between tetrapods and Actinopterygii. Seven genes (caps2, lgr5, rab21, tbc1d15, tmem19, tspan8, and zfc3h1) and seven genes (ampd3b, cpne8, gpia, swap70b, tbc1d15, trhr2, and znf143b) were located near tph2 in tetrapods and Actinopterygii, respectively. Only tbc1d15 was shared between tetrapods and Actinopterygii (see the right part of Figure 2b). In non-teleost ray-finned fishes such as sturgeons and paddlefishes, many genes near to tph were not identified in teleosts. However, the genes adjacent to tph throughout the tetrapods were identified to a greater degree than in ray-finned fishes based on the results of synteny analyses, indicating that the adjacent regions in tetrapods were more conservative than those in ray-finned fishes (see more details in the right areas of Figure 2).
3.4. Sequence Variations and Secondary Structures of the tph Genes
We first aligned the protein sequences of human tph1 and tph2, which were located on chromosomes 11 and 12, respectively. Depending on the alignment results, we observed that TPH1 and TPH2 were highly conserved in humans, while TPH2 has an extra length of 41 aa at the NH2-terminal (Figure 3). Ser 19 in TPH2 and Ser 58 in TPH1 have been reported as two of the most important phosphorylation sites [13,41]. The TPH enzymes are activated through phosphorylation, and then the phosphorylated TPHs bind to 14-3-3 proteins (a family of acidic, highly homogenous proteins) to increase the stability of these enzymes against degradation [42]. Thus, the absent region in TPH1 in comparison to TPH2 may imply functional divergence despite the existence of other conserved sequence features. The phosphorylation site of Ser 58 in TPH1 was also conserved in TPH2, while TPH2 had an additional phosphorylation site at Ser 19, which may suggest that TPH2 would be more stable and have stronger inhibition of dephosphorylation compared with TPH1. Both TPH1 and TPH2 have a conserved pentapeptide (Val–Pro–Trp–Phe–Pro) catalytic domain that begins at the residue 151 of human TPH2 (see Figure 3).
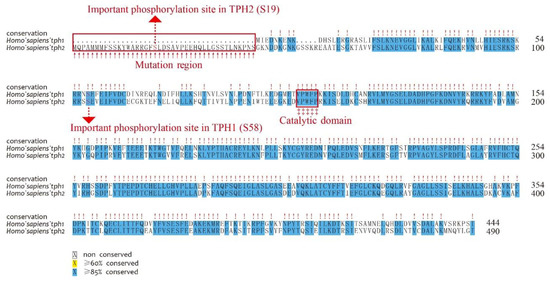
Figure 3.
Alignment of human TPH1 and TPH2 protein sequences. Important residues include the mutation region (in the NH2-terminal red box), phosphorylation sites (dashed lines with red arrows), and catalytic domains (‡ with the red box).
To provide estimates for the divergence of TPH1 and TPH2 in vertebrates, we aligned the human structural templates of 5L01 and 4V06 with TPH1 and TPH2 from different species. TPH2 sequences from six representative species in vertebrates were selected to perform the alignment using human TPH2 as the template. We found that variations of TPH2 in the examined vertebrates were mainly present in the NH2-terminal and in residues around the first α-helix. The mutation regions of TPH2 among vertebrates are marked with the rose box in Figure 4a. Seven phosphorylation sites (S19, K79, S104, Y249, Y252, S382, and S392) in TPH2, according to the human template, are marked with red boxes in Figure 4a. Among these phosphorylation sites, K79 in humans was changed to Q79 in the amphibians and to R79 in the amphibious mudskippers (BP and PM); S104 in humans was changed to N104 in teleosts; and Y249 in humans was also changed to H249 in teleosts. In general, these TPH2 protein sequences contained one Biopterin_H- and one Biopterin-dependent aromatic amino acid hydroxylase (BDAAAH) domain, and their secondary structures consisted of 17 α-helices and 10 β-strands (Figure 4a). Alignment and secondary structures of TPH2 in additional vertebrate species are provided in Figure S2.
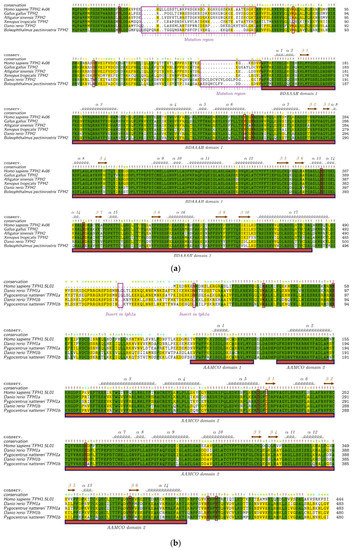
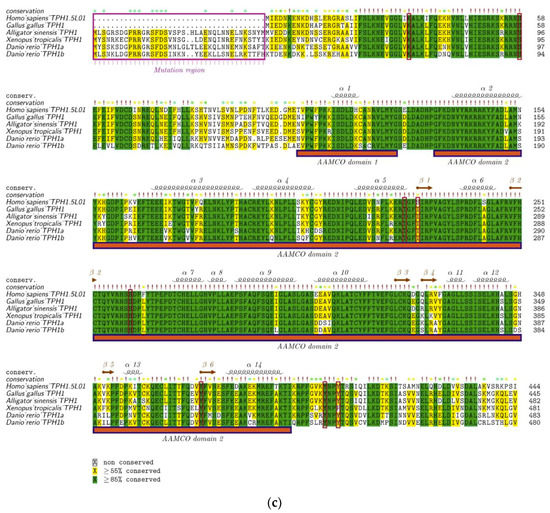
Figure 4.
Alignment and secondary structures of TPH protein sequences in vertebrates. (a) TPH2 protein sequences from representative vertebrate species were aligned with the human TPH2 and its secondary structure template 4V06. (b) Sequence alignment between TPH1a and TPH1b in some representative teleosts is provided for comparison. (c) TPH1 protein sequences from representative vertebrates were aligned with the human TPH1 and its secondary structure template 5L01. The mutation regions are marked with rose boxes and upper arrows in each figure. The red boxes denote the phosphorylation sites based on the human templates. Related alignment and secondary structures in more vertebrate species are provided in Figure S2.
As mentioned above, most teleosts that had undergone TSGD had two isotypes of tph1 named tph1a and tph1b. To further detect the differences between tph1a and tph1b, we selected two representative species from the teleosts to perform the alignment with the human TPH1 (protein ID: 5L01) as the template. TPH1a contains 483 aa, while TPH1b contains 480 aa. TPH1a has two aa (LG or IG) inserted at sites 20 and 21 and one aa of N inserted at site 47 compared to TPH1b. These three insertion sites in TPH1a are marked with rose boxes in Figure 4b. Many variable sites between TPH1a and TPH1b were also identified near the first α-helix. However, the phosphorylation sites (marked within red boxes) are almost identical between TPH1a and TPH1b. In general, the domain regions were highly conserved between TPH1a and TPH1b in teleosts (Figure 4b). Alignment and secondary structures of TPH1a and TPH1b in additional teleost species are presented in Figure S2.
Similarly, alignment of TPH1 proteins in five representative species of vertebrates is shown in Figure 4c. The alignment indicated that fishes, amphibians, and reptiles had about 39 more aa compared with mammals and birds at the NH2-terminal. Eight phosphorylation sites (K33, S58, T205, S228, S260, Y373, Y401, and Y404 in TPH1) are marked with red boxes in Figure 4c. Among these phosphorylation sites, only S288 in humans was changed to T228 in other tetrapods and fishes, and Y404 in human was changed to F404 in amphibians. Generally, TPH1 contains two aromatic amino acid monoxygenases, catalytic and oligomerization (AAMCO) domains, and the secondary structures of TPH1 consist of 14 α-helices and six β-strands (see Figure 4c). Alignment and secondary structures of TPH1 in additional teleost species are shown in Figure S2.
3.5. Predicted Three-Dimensional (3D) Structures of the TPH Proteins
In comparing the three-dimensional (3D) structure of TPH1 between zebrafish and chickens, we observed that both shared the same conformation of the structural elements (α-helices and β-strands), while some slight structural modifications were present in the loops (Figure 5a,c). The 3D structures of TPH1a and TPH1b in zebrafish were also strikingly similar, except for minor differences in the loop regions (Figure 5a,b).
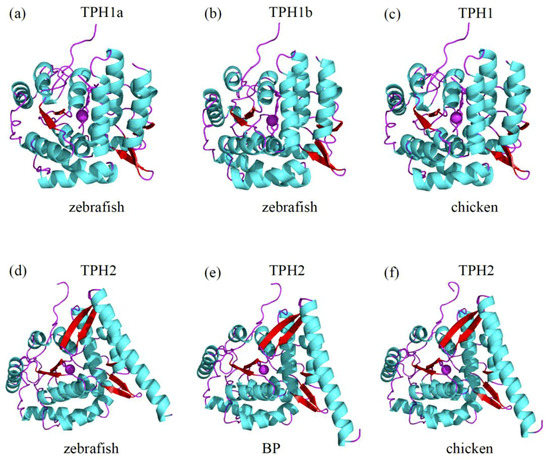
Figure 5.
Predicted 3D structures of representative TPH proteins. Comparisons of the 3D structures of zebrafish TPH1a (a), zebrafish TPH1b (b), chicken TPH1 (c), zebrafish TPH2 (d), BP TPH2 (e), and chicken TPH2 (f) are illuminated in Section 3.5. Helices of the catalytic domains and β-strands are colored sky blue and red, respectively. Loop regions are marked in purple.
Similarly, we compared the 3D structures of TPH2 among zebrafish, amphibious mudskippers (BP), and chickens. The structural elements of α-helices and β-strands also shared the same conformation, except for some slight variation in the loops (Figure 5d–f).
It seems that the loop regions in these TPH proteins were relatively flexible. However, the 3D structures of TPH1 and TPH2 showed obvious differences, and the divergences were located not only in the structural elements of α-helices and β-strands but also in the loops (see Figure 5), which was consistent with the functional differences between tph1 and tph2.
3.6. Detection of Putative Positively Selected Sites in the TPH Family
Researchers had reported that tph1 from peripheral tissues and tph2 from the brainstem demonstrated biochemical and functional differences [13,41]. In our present study, we also verified that TPH1 and TPH2 had structural differences based on our aforementioned prediction of 3D structures. However, positive selection has been identified as an important driving force in protein evolution of both structure and function based on phylogenetic analyses [43]. We therefore characterized the selection pressures of different members in the TPH family.
A phylogenetic tree for selection pressure analysis was constructed based on a subset of five TPH1 and five TPH2 protein sequences that were selected from those shown in Figure S1 to represent the five major groups of vertebrates (Figure 6). We employed a one-ratio or two-ratio branch model in PAML to check for significant differences in selection pressure between the clades of TPH1 and TPH2 (Figure 6). The log-likelihood values under one-ratio and two-ratio models were ln L = −8188.872713 and ln L = −8182.688839, respectively. Our likelihood ratio tests suggested rejection of the one-ratio model (p < 0.001); the branch model test suggested that selection pressure significantly differed between the clades of TPH1 and TPH2. We therefore concluded that selection pressure varied between the clades of TPH1 and TPH2.
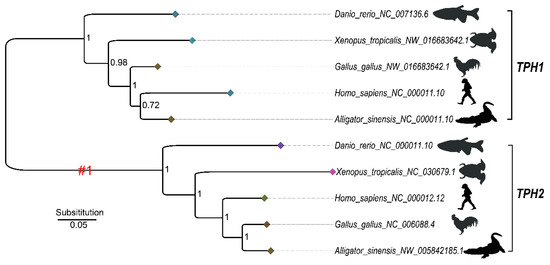
Figure 6.
The Bayesian inference (BI) tree based on five TPH1 and TPH2 proteins selected from those shown in Figure S1 to represent the five major groups of vertebrates for selection pressure analysis. #1 denotes the foreground of the TPH2 clade. Numbers in the topology indicate the Bayesian posterior probabilities. The diamonds at the tips of each branch are colored based on the branch length, with a darker color representing a shorter evolutionary branch.
The mean ω (dN/dS) values for the TPH1 and TPH2 clades were 0.047 and 4.592, respectively, indicating that the TPH2 clade may have experienced positive selection. Furthermore, we estimated that some codons had undergone positive selection. Based on Empirical Bayes analysis, we further identified five positively selected candidate sites (D159, S285, I358, H426, and K441; see Figure 7), and the posterior probabilities of these sites were greater than 0.99. The positively selected site D159 was near to the pentapeptide (Val–Pro–Trp–Phe–Pro) catalytic active domain, while other positive sites were next to the phosphorylation site region or α-helix and loops of the TPH family (see Figure 7).
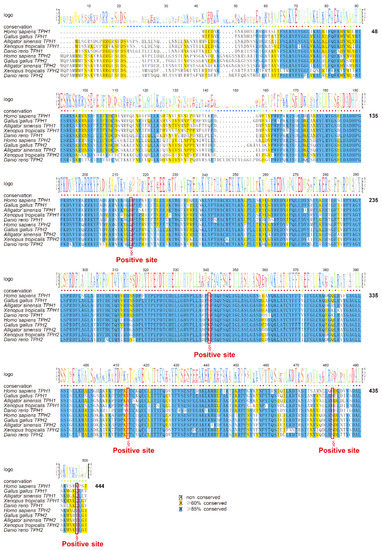
Figure 7.
Alignment of the datasets for selection pressure analysis. The five positively selected sites predicted by the branch-site model test are marked with red boxes and emphasized with section marks (§).
We also added ten additional vertebrate sequences, with five belonging to TPH1 and five belonging to TPH2 (see Figure S3) to form a more comprehensive dataset to confirm the analysis of positive selection. The results also supported the hypothesis that the branch of TPH2 possibly experienced positive selection in comparison to TPH1 (the log-likelihood values under one-ratio and two-ratio models were ln L = −11719.144671 and ln L = −11713.763977, respectively, and the p-value from the χ2 test was less than 0.001). We performed the analysis again under branch site models and determined that the list of inferred positively selected sites was the same as in the previous version.
4. Discussion
4.1. Possible Reasons for Variation of tph Copy Number in Vertebrates
Whole genome duplication has been proposed to provide additional genetic material for the appearance of new genes, allowing organisms to acquire novel phenotypes to survive natural challenges [44]. In vertebrates, two rounds of WGD occurred in their common ancestor [45,46]. Specifically, the first round happened before the split between Gnathostomes and Agnatha (jawless vertebrates), and the second round occurred before the split between Chondrichthyes and Osteichthyes [47,48]. Furthermore, the teleost lineage had undergone an additional WGD known as the TSGD approximately 320 Mya [17,18]. Besides the TSGD at the base of the teleost lineage, recent genome duplication events had occurred within some teleost lineages, including the salmonid-specific genome duplication 80 Mya [49,50], and the common carp lineage appeared approximately 8 Mya [51]. In addition to teleosts, some basal ray-finned fish lineages, such as sturgeons and paddlefishes in Acipenserifomes, also had experienced tetraploidization events around 184 Mya [52].
According to our results, most tetrapods examined in this study and some basal non-teleost ray-finned fishes (e.g., the spotted gar) had single copies of tph1 and tph2, with the exception of Xenopus laevis, which had two copies of both tph1 and tph2. It has been reported that X. laevis had undergone recent tetraploidization approximately 40 Mya [53], and thus the doubled copies of tph1 and tph2 were generated in this frog species. This WGD event in X. laevis might have been propitious for adapting to various environmental factors such as salt, drought, cold, and disease compared to the closely-related diploid species such as X. tropicalis. Spotted gar diverged from the teleost lineage before the TSGD and thus was not affected by the TSGD event. Furthermore, Acipenserifomes experienced their own lineage-specific polyploidization event; thus we detected two copies of tph1 and tph2 in the American paddlefish and eight copies of tph1 and five copies of tph2 in the Chinese sturgeon.
Researchers had reported that zebrafish, medaka, and sticklebacks had three tph isotypes named tph1a, tph1b, and tph2 [16,20]. Interestingly, based on a comparative genome-wide survey, we found that most teleosts with the TSGD had three tph isotypes, except for Asian arowana, fugu, Northern pike, and tongue sole, which had lost the tph1b gene. For the common teleosts, all three tph isotypes were present in single copies. However, tetraploid fishes such as the golden-line fishes (Sa, Sr, and Sg) had five copies (not the expected six) of the three tph isotypes, including three copies of tph1 and two copies of tph2 (with the possible loss of one tph1a). Other typical tetraploid teleosts such as the rainbow trout and Atlantic salmon, which had undergone the salmonid-specific genome duplication, had four copies of tph genes, but copies of duplicated tph1a and tph2 were not found. The most frequent fate of duplicated genes after WGD is nonfunctionalization; therefore, these duplicated genes often experienced gene loss [54]. We propose that the main cause of copy number variation of tph in vertebrates was a combination of WGD and gene loss, as in the cases of aaad [55], aanat [56], and asmt [57], which encode the remaining three enzymes for melatonin biosynthesis (Figure 1). Moreover, incomplete annotation of some genomes may be one of the reasons for the lack of identification of certain genes in some species. Copy numbers between diploid and tetraploid species do not always correspond to a 1:2 ration because of selective loss of certain copies. In fact, this phenomenon seems to be particularly common in teleosts.
Moreover, our synteny analysis revealed that almost all tph1 and tph2 genes were localized on different scaffolds in the same species, except for fugu, Northern pike, and tongue sole. Interestingly, we found that the synteny genes were not conserved between tetrapods and Actinopterygii. Only two genes, saal1 and sergef, near the tph1 gene, were shared in Actinopterygii and tetrapods. Furthermore, synteny genes located near tph1a and tph1b in the teleosts were not completely consistent. Only tbc1d15 was shared by tetrapods and Actinopterygii among genes localized near the downstream and upstream areas of tph2. It seems that the tph gene family experienced rearrangement during the evolution from teleosts to tetrapods. Meanwhile, we also observed that the synteny of tph in tetrapods was generally greater than that of teleosts. This phenomenon may be due to the fact that teleost lineages are more prone to interchromosomal rearrangements than tetrapods, resulting in shorter conserved syntenic blocks in teleosts compared with tetrapods [58,59]. However, the TSGD events had also led to shorter syntenic blocks through differential gene loss without rearrangements. Differential loss of blocks of duplicated genes can cause synteny disruption, contributing to shorter syntenic blocks in teleosts than in non-teleost vertebrates [54]. The disrupted syntenic blocks are very common in teleost genomes. For example, Ravi et al. (2013) reported that the syntenic block of the Pax6 locus contained six genes, and this block was completely conserved in tetrapods. However, in zebrafish, Pax6 was duplicated to pax6a and pax6b, owing to the TSGD, and the blocks of pax6a and pax6b retained three and four genes, respectively [60].
4.2. Adaptive Evolution of TPHs in Vertebrates and Impact on Human Health
Elucidating the mechanisms of protein functional diversity and understanding how enzyme families evolve are core issues in evolutionary biology. Catalytic activity toward various substrates and substrate specificity are the key functional properties of an enzymatic protein. Enzyme families with divergent functions usually acquire specific activities relating to structural and functional variations caused by amino acid substitutions during evolution [61]. Numerous phylogenetic analyses based on amino acid substitutions have suggested that many enzymatic protein families have undergone positive selection [24,62], which has been considered as an important driving force in protein evolution.
The TPH family has tissue-specific expression patterns: tph1 is distributed in the pineal gland and peripheral tissues (gut, spleen, and thymus), while tph2 is predominantly expressed in the central tissue of the brainstem [15]. Thus, there are two serotonin systems in vertebrates independently regulating distinct functions (Figure 1). However, the lack of comprehensive structural and functional analyses has generally hindered attempts to elaborate this pattern. Structural and functional characteristics of proteins are defined into rigid and flexible regions. Rigidity plays an important role in the integrity of protein folding, while flexibility is important for binding and catalyzing different substrates [63]. In the present study, we compared the 3D structures of TPHs in representative species of vertebrates. The 3D structures of TPH1 and TPH2 indicated that the structural elements of α-helices and β-strands generally shared the same conformation, but some slight structural modifications were present in the loops. However, when we compared the 3D structures of TPH1 and TPH2, we observed obvious differences. The divergence was not only in the loops but also in the structural elements of α-helices and β-strands. These divergences are consistent with the differences in function between tph1 and tph2.
Positive selection acts as an important driving force in protein evolution in both structure and function. Estimating the ratio of dN to that of dS between homologous protein coding genes is a significant indictor of positive selection at the molecular level. Ratios of dN/dS (ω) in lineages that have experienced selection may be different from those of other lineages. Previous studies reported evolutionary divergence from both structural and functional views in the TPH family [8,21]. Thus, we further analyzed the selective aspects of the functional and structural divergence of the TPH family and attempted to elucidate patterns of positively selected sites. We detected five positively selected sites in TPH2. Conventional theory emphasizes that replacements in active sites of amino acids will lead to changes in a protein’s function. TPH1 and TPH2 have a conserved pentapeptide (Val–Pro–Trp–Phe–Pro) catalytic active domain in humans (see Section 2.4). The five positively selected sites detected in our present study, however, were not in the catalytic domain. As we reported, the positively selected site of D159 was near the pentapeptide catalytic domain, while other positively selected sites were near the phosphorylation sites or the structural elements (α-helix and loops) of TPHs. Our present study suggests that mutations outside the active regions may have effects on the proteins, despite that mutations close to the active sites were apparently more effective than those more distant. This phenomenon resembles the pine glutathione S-transferase (GST) enzyme family investigated by Lan et al. (2013), in which three of the five positively selected sites detected in their study were located in the nonactive sites [43]. Similarly, positively selected sites were also detected outside the active region of the JGW family in Drosophila, and this generated a new dehydrogenase with varied substrate specificity compared to the ancestral protein [64]. The positive selection acting on residues adjacent to the active sites, rather than direct action on the active sites, may be a general mechanism for the functional diversification of the enzyme families [43,64]. Meanwhile, protein loop regions and α-helix regions are relatively flexible compared with β-strands. In our present study, all five positively selected sites in the TPH family were in the loops and α-helix, suggesting that the TPH proteins could tolerate structural modification. In summary, the positively selected sites detected in the TPH family may affect TPH activity and substrate specificity.
In addition, recent advances have implicated serotonin as a regulator of inflammation [65], proliferation [66], regeneration [67], and repair [68]. Interestingly, a recent population-based study showed that a high intake of selective serotonin reuptake inhibitors was correlated with a reduced incidence of colorectal cancer, suggesting a biological role of serotonin in colorectal cancer growth in vivo [69]. Furthermore, the role of serotonin in tumor biology in vivo has been elucidated using a genetic model of serotonin deficiency (tph1-/-) in mice, and researchers concluded that serotonin regulated angiogenesis in colorectal cancer allografts by influencing matrix metalloproteinase 12 expression in tumor-infiltrating macrophages, thereby affecting the production of circulating angiostatin [70]. Therefore, serotonin-related pathways may represent a new therapeutic target for cancer treatment. Our current study provides basic information necessary for further study of serotonin’s role in human health.
5. Conclusions
Our present study is the first comprehensive report concerning tph evolution in vertebrates. We investigated many features of the tph family to provide novel insights into the evolution of tph genes from a genomic view. Through genome-wide alignment, we found that tetrapods and non-teleosts had two tph isotypes (tph1 and tph2); however, in the teleost lineage, tph1 had further diverged into tph1a and tph1b. Therefore, most of the teleosts that had undergone the TSGD event had three tph isotypes: tph1a, tph1b, and tph2. Copy numbers between diploid and tetraploid species did not always correspond to a 1:2 ratio, and we estimated that the WGD and gene loss generated the corresponding variation in tph gene numbers in vertebrates. Several important sites of TPH proteins were analyzed for structural comparison, and differences of TPHs in vertebrates were mainly focused at the NH2-terminal. Tertiary structures of TPH1 and TPH2 showed obvious differences in the structural elements (α-helix and β-strands) and loops. Five positively selected sites were characterized in the TPH family, and these sites may significantly affect the enzyme activity and substrate specificity. This result was consistent with tissue-specific transcription patterns of tph1 and tph2 and differential roles in the serotonin systems of vertebrates. In summary, through these analyses and comparisons, we provided novel insights into the molecular evolution of the TPH family in vertebrates.
Supplementary Materials
Supplementary materials can be found online at https://www.mdpi.com/2073-4425/10/3/203/s1. Table S1. GenBank ID of the selected 70 vertebrate genomes. Table S2. Queries used for extraction of tph genes from the examined 70 vertebrate species. Table S3. Copy number variation in tph genes in the selected vertebrate genomes. Figure S1. BI tree deduced from the 184 vertebrate TPH protein sequences. Figure S2. Alignment and secondary structures of TPH protein sequences in additional vertebrate species. Figure S3. The BI tree based on TPH1 and TPH2 proteins in additional vertebrate species for selection pressure analyses.
Author Contributions
Q.S., J.X., Y.L. (Yanping Li) and H.T. conceived and designed the study. Y.L. (Yanping Li) and Y.L. (Yunyun Lv) performed the data analysis and figure preparation. C.B. and X.Y. participated in data analysis and discussions. Y.L. (Yanping Li) and J.X. prepared the manuscript. Q.S., Y.L. (Yunyun Lv) H.T., and D.E. revised the manuscript.
Funding
This research was funded by the Office of Fisheries Supervision and Management for the Yangtze River Basin, MARA, PRC (grant number: 171821301354051046) and Yunnan Innovation and Enhancement Program of Provincial Science and Technology Department (grant number: 2016AB024).
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
ampd3b: Adenosine Monophosphate Deaminase 3; ano9b, anoctamin-9-like; ap4e1, Adaptor Related Protein Complex 4 Subunit Epsilon 1; caps2, Calcyphosine 2; ccne1, Cyclin E1; cd81a, CD81 antigen; cpne8, Copine 8; cyp19a1, Cytochrome P450 Family 19 Subfamily A Member 1; dS/dN, ratio of nonsynonymous substitutions (dN) to that of synonymous substitutions (dS); fibina, Fin Bud Initiation Factor Homolog; gnb5a, G Protein Subunit β 5 A; gpia, Platelet membrane glycoprotein Ia; gtf2h1, General Transcription Factor IIH Subunit 1; hps5, Biogenesis of Lysosomal Organelles Complex 2 Subunit 2; kcnc1, Potassium Voltage-Gated Channel Subfamily C Member 1; lgr5, Leucine Rich Repeat Containing G Protein-Coupled Receptor 5; nucb2, Nucleobindin 2; pkp3b, Plakophilin 3; rab21, RAB21, Member RAS Oncogene Family; saal1, Serum Amyloid A Like 1; sergef, Secretion Regulating Guanine Nucleotide Exchange Factor; swap70b, Switching B Cell Complex Subunit SWAP70; tbc1d15, TBC1 Domain Family Member 15; tmem19, Transmembrane Protein 19; trhr2, Thyrotropin Releasing Hormone Receptor 2; TSGD, teleost-specific genome duplication; tspan8, Tetraspanin 8; ucmab, Upper Zone of Growth Plate and Cartilage Matrix Associated B; ush1c, USH1 Protein Network Component Harmonin; WGD, whole genome duplication; zfc3h1, Zinc Finger C3H1-Type Containing; znf143b, Zinc Finger Protein 143b; znf536, Zinc Finger Protein 536.
References
- Lucki, I. The spectrum of behaviors influenced by serotonin. Biol. Psychiatry 1998, 44, 151–162. [Google Scholar] [CrossRef]
- Jacobs, B.L.; Azmitia, E.C. Structure and function of the brain serotonin system. Physiol. Rev. 1992, 72, 165–229. [Google Scholar] [CrossRef] [PubMed]
- Côté, F.; Fligny, C.; Bayard, E.; Launay, J.-M.; Gershon, M.D.; Mallet, J.; Vodjdani, G. Maternal serotonin is crucial for murine embryonic development. Proc. Natl. Acad. Sci. USA 2007, 104, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Chong, N.W.; Bernard, M.; Klein, D.C. Characterization of the chicken serotonin N-acetyltransferase gene. J. Biol. Chem. 2000, 275, 32991–32998. [Google Scholar] [CrossRef] [PubMed]
- Lovenberg, W.; Weissbach, H.; Udenfriend, S. Aromatic l-amino acid decarboxylase. J. Biol. Chem. 1962, 237, 89. [Google Scholar] [PubMed]
- Bernard, M.; Iuvone, P.M.; Cassone, V.M.; Roseboom, P.H.; Coon, S.L.; Klein, D.C. Avian melatonin synthesis: Photic and circadian regulation of serotonin N-acetyltransferase mRNA in the chicken pineal gland and retina. J. Neurochem. 1997, 68, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Ishida, I.; Obinata, M.; Deguchi, T. Molecular cloning and nucleotide sequence of cDNA encoding hydroxyindole o-methyltransferase of bovine pineal glands. J. Biol. Chem. 1987, 262, 2895–2899. [Google Scholar]
- McKinney, J.; Teigen, K.; Frøystein, N.Å.; Salaün, C.; Knappskog, P.M.; Haavik, J.; Martínez, A. Conformation of the substrate and pterin cofactor bound to human tryptophan hydroxylase. Important role of Phe313 in substrate specificity. Biochemistry 2001, 40, 15591–15601. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, P.F. Tetrahydropterin-dependent amino acid hydroxylases. Annu. Rev. Biochem. 1999, 68, 355. [Google Scholar] [CrossRef]
- Erlandsen, H.; Bjørgo, E.; Flatmark, T.; Stevens, R.C. Crystal structure and site-specific mutagenesis of pterin-bound human phenylalanine hydroxylase. Biochemistry 2000, 39, 2208–2217. [Google Scholar] [CrossRef] [PubMed]
- Daubner, S.C.; Le, T.; Wang, S. Tyrosine hydroxylase and regulation of dopamine synthesis. Arch. Biochem. Biophys. 2011, 508, 1–12. [Google Scholar] [CrossRef]
- Andersen, O.A.; Flatmark, T.; Hough, E. Crystal structure of the ternary complex of the catalytic domain of human phenylalanine hydroxylase with tetrahydrobiopterin and 3-(2-thienyl)-l-alanine, and its implications for the mechanism of catalysis and substrate activation. J. Mol. Biol. 2002, 320, 1095–1108. [Google Scholar] [CrossRef]
- Walther, D.J.; Peter, J.U.; Bashammakh, S.; Hörtnagl, H.; Voits, M.; Fink, H.; Bader, M. Synthesis of serotonin by a second tryptophan hydroxylase isoform. Science 2003, 299, 76. [Google Scholar] [CrossRef]
- Mockus, S.M.; Vrana, K.E. Advances in the molecular characterization of tryptophan hydroxylase. J. Mol. Neurosci. 1998, 10, 163–179. [Google Scholar] [CrossRef] [PubMed]
- Cornide-Petronio, M.E.; Anadón, R.; Rodicio, M.C.; Barreiroiglesias, A. The sea lamprey tryptophan hydroxylase: New insight into the evolution of the serotonergic system of vertebrates. Brain Struct. Funct. 2013, 218, 587–593. [Google Scholar] [CrossRef]
- Lillesaar, C. The serotonergic system in fish. J. Chem. Neuroanat. 2011, 41, 294–308. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.S.; Braasch, I.; Frickey, T.; Meyer, A.; Peer, Y.V.D. Genome duplication, a trait shared by 22,000 species of ray-finned fish. Genome Res. 2003, 13, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Jaillon, O.; Aury, J.M.; Brunet, F.; Petit, J.L.; Stangethomann, N.; Mauceli, E.; Bouneau, L.; Fischer, C.; Ozoufcostaz, C.; Bernot, A. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 2004, 431, 946. [Google Scholar] [CrossRef] [PubMed]
- Force, A.; Lynch, M.; Pickett, F.B.; Amores, A.; Yan, Y.L.; Postlethwait, J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 1999, 151, 1531–1545. [Google Scholar] [PubMed]
- Bellipanni, G.; Rink, E.; Ballycuif, L. Cloning of two tryptophan hydroxylase genes expressed in the diencephalon of the developing zebrafish brain. Gene Expr. Patterns 2002, 119, S215–S220. [Google Scholar] [CrossRef]
- Varadarajan, N.; Gam, J.; Olsen, M.J.; Georgiou, G.; Iverson, B.L. Engineering of protease variants exhibiting high catalytic activity and exquisite substrate selectivity. Proc. Natl. Acad. Sci. USA 2005, 102, 6855–6860. [Google Scholar] [CrossRef] [PubMed]
- Barkman, T.J.; Martins, T.R.; Sutton, E.; Stout, J.T. Positive selection for single amino acid change promotes substrate discrimination of a plant volatile-producing enzyme. Mol. Biol. Evol. 2007, 24, 1320–1329. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Bielawski, J.P. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 2000, 15, 496–503. [Google Scholar] [CrossRef]
- Smith, N.G.C.; Eyre-Walker, A. Adaptive protein evolution in Drosophila. Nature 2002, 415, 1022–1024. [Google Scholar] [CrossRef] [PubMed]
- Bishop, J.G.; Dean, A.M.; Mitchellolds, T. Rapid evolution in plant chitinases: Molecular targets of selection in plant-pathogen coevolution. Proc. Natl. Acad. Sci. USA 2000, 97, 5322–5327. [Google Scholar] [CrossRef]
- Kajitani, R.; Toshimoto, K.; Noguchi, H.; Toyoda, A.; Ogura, Y.; Okuno, M.; Yabana, M.; Harada, M.; Nagayasu, E.; Maruyama, H. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 2014, 24, 1384–1395. [Google Scholar] [CrossRef]
- Ye, C.; Hill, C.M.; Wu, S.; Ruan, J.; Ma, Z.S. Dbg2olc: Efficient assembly of large genomes using long erroneous reads of the third generation sequencing technologies. Sci. Rep. 2016, 6, 31900. [Google Scholar] [CrossRef]
- Mount, D.W. Using the basic local alignment search tool (blast). CSH Protoc. 2007, 2007. [Google Scholar] [CrossRef] [PubMed]
- Slater, G.S.C.; Birney, E. Automated generation of heuristics for biological sequence comparison. BMC Bioinform. 2005, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. Prottest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. Mrbayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Drummond, A.J.; Rambaut, A. Beast: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef] [PubMed]
- Beitz, E. Texshade: Shading and labeling of multiple sequence alignments using latex2e. Bioinformatics 2000, 16, 135–139. [Google Scholar] [CrossRef]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: an automated protein homology-modeling server. Nucleic. Acids. Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef] [PubMed]
- Delano, W.L. Pymol: An open-source molecular graphics tool. CCP4 News. On Prot. Crys. 2002, 40, 82–92. [Google Scholar]
- Yang, Z. Paml 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Nielsen, R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 2000, 17, 32–43. [Google Scholar] [CrossRef]
- Yang, Z.; Wong, W.S.; Nielsen, R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 2005, 22, 1107–1118. [Google Scholar] [CrossRef]
- Carkaci-Salli, N.; Flanagan, J.M.; Martz, M.K.; Salli, U.; Walther, D.J.; Bader, M.; Vrana, K.E. Functional domains of human tryptophan hydroxylase 2 (htph2). J. Biol. Chem. 2006, 281, 28105–28112. [Google Scholar] [CrossRef] [PubMed]
- Winge, I.; McKinney, J.A.; Ying, M.; D’Santos, C.S.; Kleppe, R.; Knappskog, P.M.; Haavik, J. Activation and stabilization of human tryptophan hydroxylase 2 by phosphorylation and 14-3-3 binding. Biochem. J. 2008, 410, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Lan, T.; Wang, X.R.; Zeng, Q.Y. Structural and functional evolution of positively selected sites in pine glutathione s-transferase enzyme family. J. Biol. Chem. 2013, 288, 24441–24451. [Google Scholar] [CrossRef] [PubMed]
- Kaessmann, H. Origins, evolution, and phenotypic impact of new genes. Genome Res. 2010, 20, 1313–1326. [Google Scholar] [CrossRef] [PubMed]
- Guyomard, R.; Boussaha, M.; Krieg, F.; Hervet, C.; Quillet, E. A synthetic rainbow trout linkage map provides new insights into the salmonid whole genome duplication and the conservation of synteny among teleosts. BMC Genet. 2012, 13, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Glasauer, S.M.K.; Neuhauss, S.C.F. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol. Genet. Genom. 2014, 289, 1045. [Google Scholar] [CrossRef]
- Meyer, A.; Van de Peer, Y. From 2r to 3r: Evidence for a fish-specific genome duplication (fsgd). Bioessays 2005, 27, 937–945. [Google Scholar] [CrossRef]
- Kasahara, M.; Naruse, K.; Sasaki, S.; Nakatani, Y.; Qu, W.; Ahsan, B.; Yamada, T.; Nagayasu, Y.; Doi, K.; Kasai, Y.; et al. The medaka draft genome and insights into vertebrate genome evolution. Nature 2007, 447, 714–719. [Google Scholar] [CrossRef]
- Macqueen, D.J.; Johnston, I.A. A well-constrained estimate for the timing of the salmonid whole genome duplication reveals major decoupling from species diversification. Proc. R. Soc. B Biol. Sci. 2014, 281, 20132881. [Google Scholar] [CrossRef]
- Lien, S.; Koop, B.F.; Sandve, S.R.; Miller, J.R.; Kent, M.P.; Nome, T.; Hvidsten, T.R.; Leong, J.S.; Minkley, D.R.; Zimin, A. The Atlantic salmon genome provides insights into rediploidization. Nature 2016, 533, 200–205. [Google Scholar] [CrossRef]
- Xu, P.; Zhang, X.; Wang, X.; Li, J.; Liu, G.; Kuang, Y.; Xu, J.; Zheng, X.; Ren, L.; Wang, G. Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nat. Genet. 2014, 46, 1212–1219. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Ludwig, A.; Wang, D.; Diogo, R.; Wei, Q.; He, S. Age and biogeography of major clades in sturgeons and paddlefishes (pisces: Acipenseriformes). Mol. Phylogenet. Evol. 2007, 42, 854–862. [Google Scholar] [CrossRef] [PubMed]
- Hellsten, U.; Khokha, M.K.; Grammer, T.C.; Harland, R.M.; Richardson, P.; Rokhsar, D.S. Accelerated gene evolution and subfunctionalization in the pseudotetraploid frog Xenopus laevis. BMC Biol. 2007, 5, 31. [Google Scholar] [CrossRef] [PubMed]
- Ravi, V.; Venkatesh, B. The divergent genomes of teleosts. Annu. Rev. Anim. Biosci. 2018, 6, 47–68. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lv, Y.; Bian, C.; You, X.; Deng, L.; Shi, Q. A comparative genomic survey provides novel insights into molecular evolution of l-aromatic amino acid decarboxylase in vertebrates. Molecules 2018, 23, 917. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; You, X.; Bian, C.; Yu, H.; Coon, S.L.; Shi, Q. Molecular evolution of aralkylamine N-acetyltransferase in fish: A genomic survey. Int. J. Mol. Sci. 2015, 17, 51. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Ruan, Z.; Li, J.; Bian, C.; You, X.; Coon, S.L.; Shi, Q. A comparative genomic and transcriptomic survey provides novel insights into N-acetylserotonin methyltransferase (asmt) in fish. Molecules 2017, 22, 1653. [Google Scholar] [CrossRef]
- Braasch, I.; Gehrke, A.R.; Smith, J.J.; Kawasaki, K.; Manousaki, T.; Pasquier, J.; Amores, A.; Desvignes, T.; Batzel, P.; Catchen, J.; et al. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat. Genet. 2016, 48, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Amores, A.; Catchen, J.; Ferrara, A.; Fontenot, Q.; Postlethwait, J.H. Genome evolution and meiotic maps by massively parallel DNA sequencing: Spotted gar, an outgroup for the teleost genome duplication. Genetics 2011, 188, 799–808. [Google Scholar] [CrossRef]
- Ravi, V.; Bhatia, S.; Gautier, P.; Loosli, F.; Tay, B.H.; Tay, A.; Murdoch, E.; Coutinho, P.; van Heyningen, V.; Brenner, S.; et al. Sequencing of pax6 loci from the elephant shark reveals a family of pax6 genes in vertebrate genomes, forged by ancient duplications and divergences. PLoS Genet. 2013, 9, e1003177. [Google Scholar] [CrossRef]
- Yoshikuni, Y.; Ferrin, T.E.; Keasling, J.D. Designed divergent evolution of enzyme function. Nature 2006, 440, 1078–1082. [Google Scholar] [CrossRef] [PubMed]
- Weinberger, H.; Moran, Y.; Gordon, D.; Turkov, M.; Kahn, R.; Gurevitz, M. Positions under positive selection--key for selectivity and potency of scorpion α-toxins. Mol. Biol. Evol. 2009, 27, 1025. [Google Scholar] [CrossRef] [PubMed]
- Huber, R. Flexibility and rigidity of proteins and protein–pigment complexes. Angew. Chem. Int. Ed. 2010, 27, 79–88. [Google Scholar] [CrossRef]
- Zhang, J.; Dean, A.M.; Brunet, F.; Long, M. Evolving protein functional diversity in new genes of drosophila. Proc. Natl. Acad. Sci. USA 2004, 101, 16246–16250. [Google Scholar] [CrossRef] [PubMed]
- Nocito, A.; Dahm, F.; Jochum, W.; Jang, J.H.; Georgiev, P.; Bader, M.; Renner, E.L.; Clavien, P.A. Serotonin mediates oxidative stress and mitochondrial toxicity in a murine model of nonalcoholic steatohepatitis. Gastroenterology 2007, 133, 608–618. [Google Scholar] [CrossRef] [PubMed]
- Marzioni, M.; Glaser, S.; Francis, H.; Marucci, L.; Benedetti, A.; Alvaro, D.; Taffetani, S.; Ueno, Y.; Roskams, T.; Phinizy, J.L. Autocrine/paracrine regulation of the growth of the biliary tree by the neuroendocrine hormone serotonin. Gastroenterology 2005, 128, 121–137. [Google Scholar] [CrossRef] [PubMed]
- Lesurtel, M.; Graf, R.; Aleil, B.; Walther, D.J.; Tian, Y.; Jochum, W.; Gachet, C.; Bader, M.; Clavien, P.-A. Platelet-derived serotonin mediates liver regeneration. Science 2006, 312, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Nocito, A.; Georgiev, P.; Dahm, F.; Jochum, W.; Bader, M.; Graf, R.; Clavien, P.A. Platelets and platelet-derived serotonin promote tissue repair after normothermic hepatic ischemia in mice. Hepatology 2007, 45, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Tamim, H.; Shapiro, S.; Stang, M.R.; Collet, J.-P. Use of antidepressants and risk of colorectal cancer: A nested case-control study. Lancet Oncol. 2006, 7, 301–308. [Google Scholar] [CrossRef]
- Nocito, A.; Dahm, F.; Jochum, W.; Jang, J.H.; Georgiev, P.; Bader, M.; Graf, R.; Clavien, P.-A. Serotonin regulates macrophage-mediated angiogenesis in a mouse model of colon cancer allografts. Cancer Res. 2008, 68, 5152–5158. [Google Scholar] [CrossRef]
Sample Availability: Not available. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).