Identification of Reference Genes for Quantitative Gene Expression Studies in Three Tissues of Japanese Quail
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Sample Collection
2.2. RNA Extraction
2.3. Reverse Transcription and Quantitative Real-Time PCR
2.4. Gene Stability Analyses
2.5. Statistical Analyses
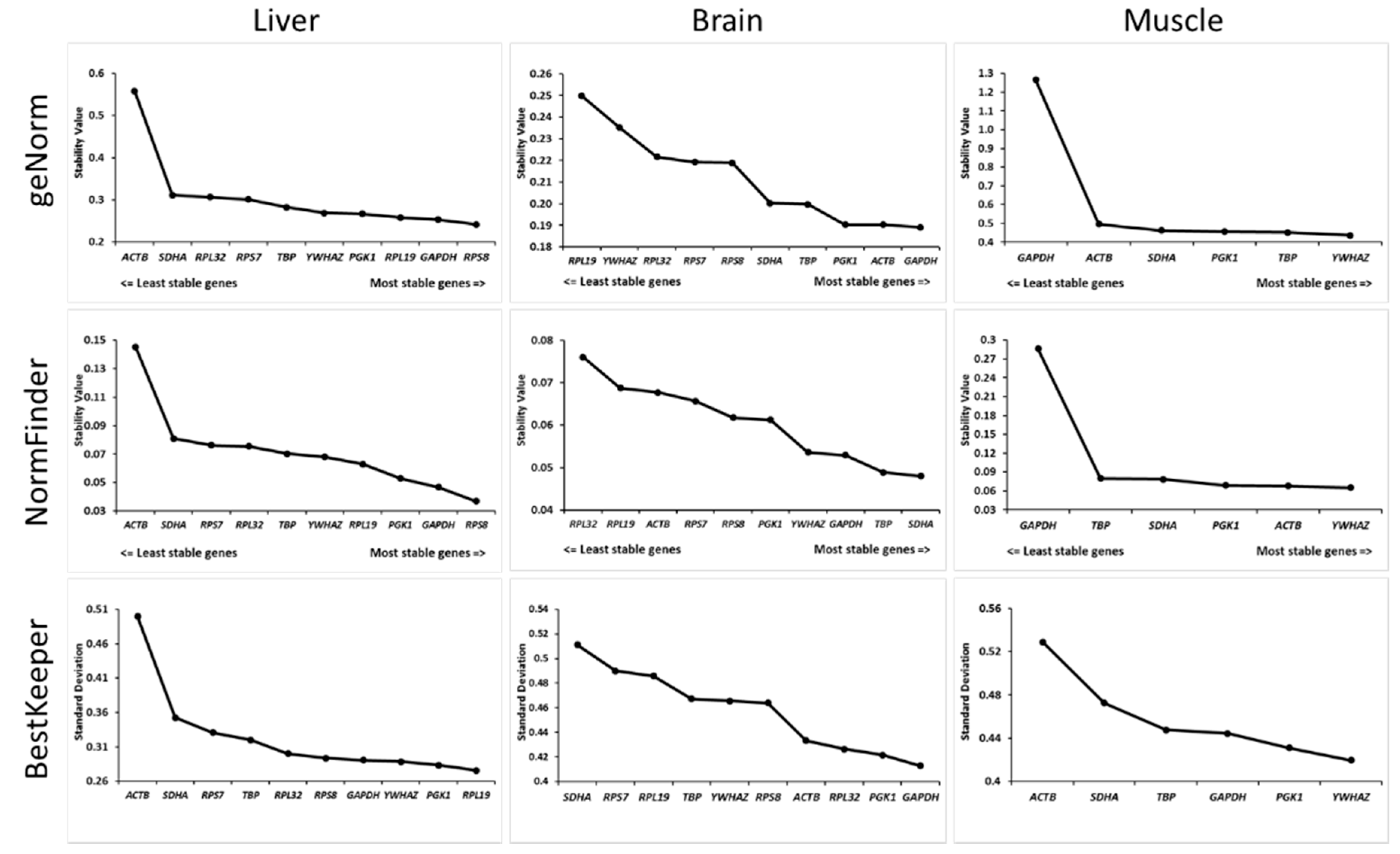
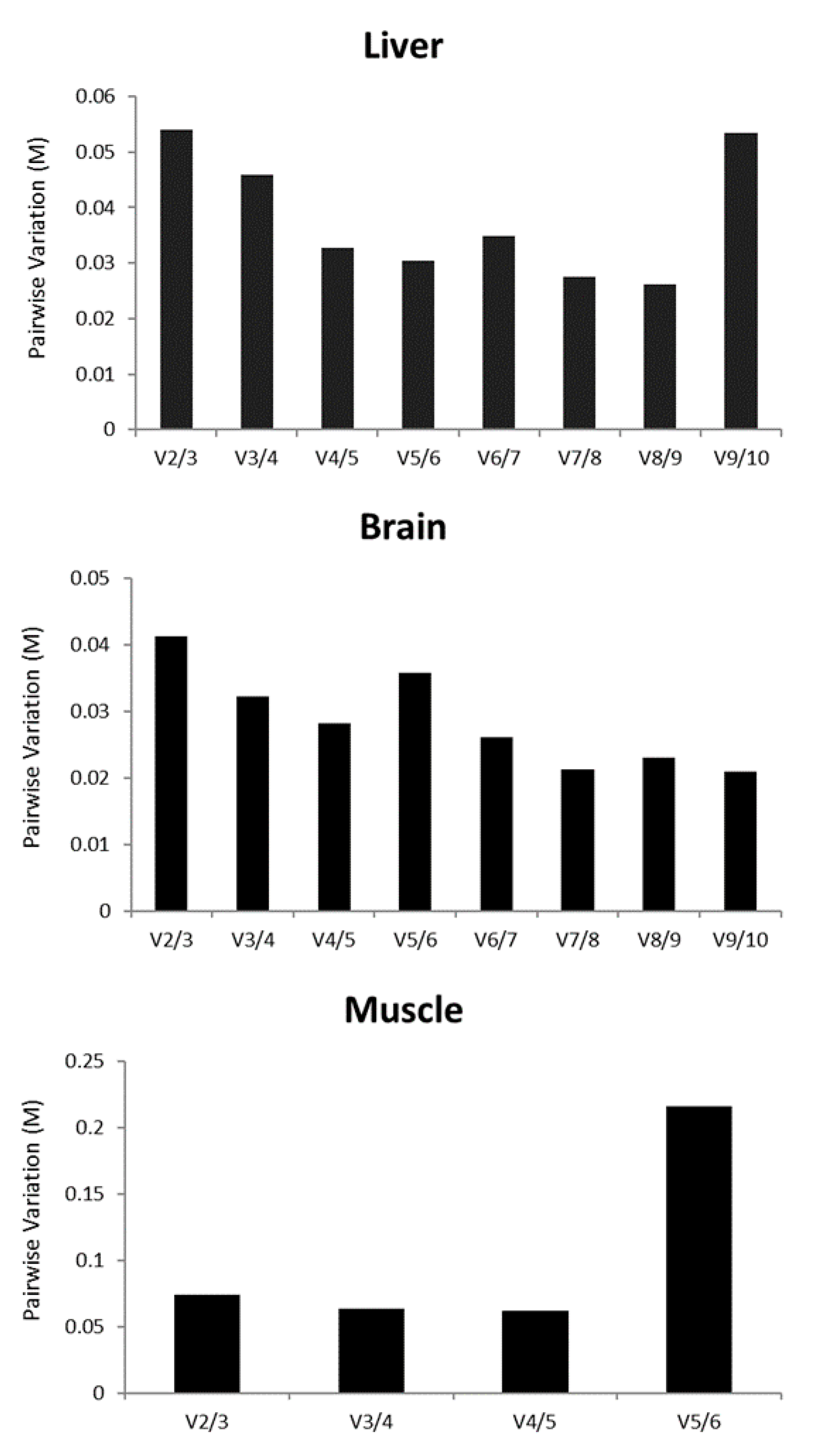
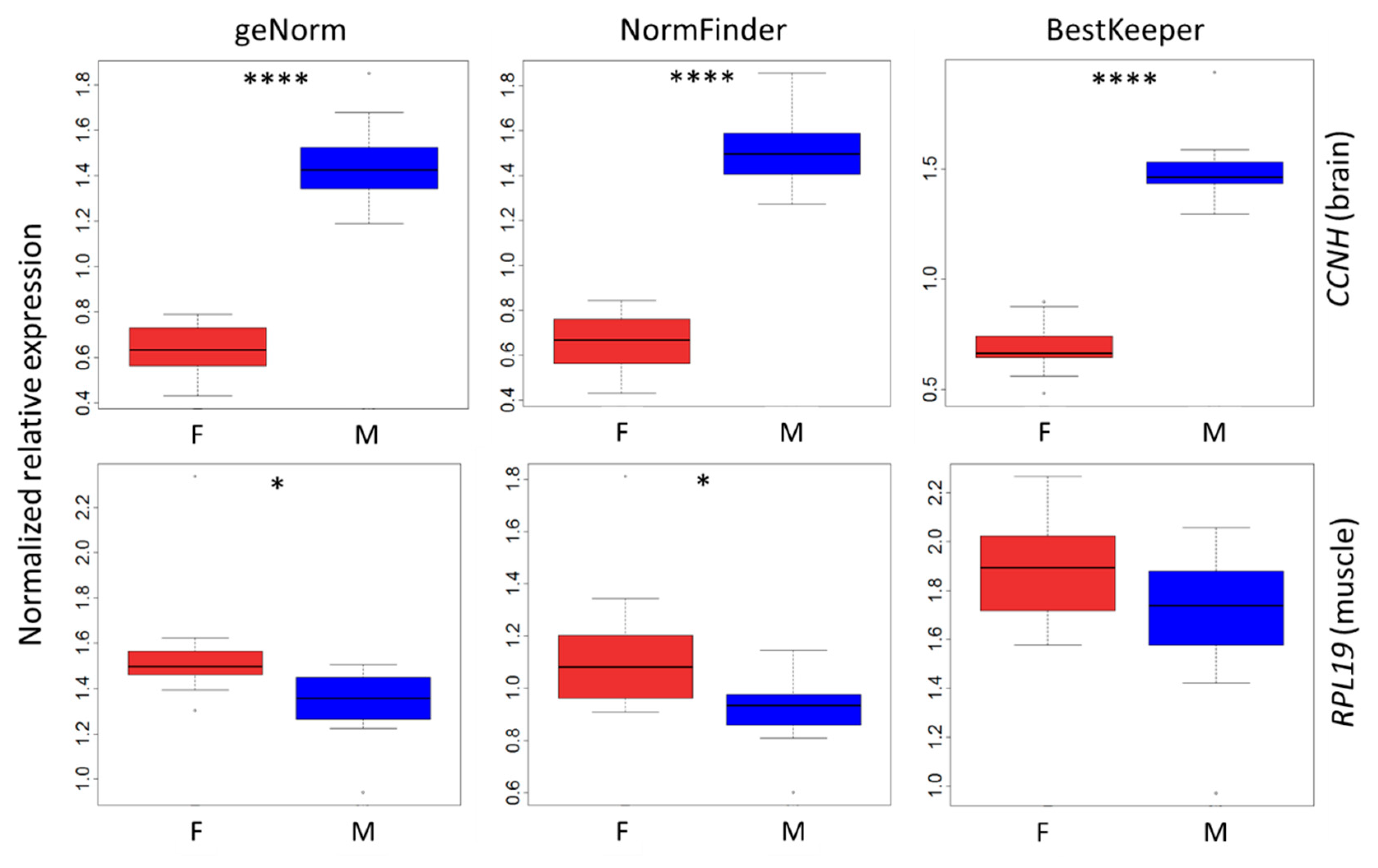
3. Results
3.1. Primer Design, Real-Time qPCR Experiment and PCR Efficiency
3.2. Impact of Sex on the Expression of Putative Reference Genes
3.3. Definition of the Most Stable Gene
3.4. Identification of the Combination of the Most Stable Genes
3.5. Validation of Reference Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Abd El-Gawad, A.E.A.; El-Wardany, I.; El-Daly, E.F.; Abd El-Azeem, N.A.; Hemid, A.H. Alleviating the Effect of Some Environmental Stress Factors on Productive Performance in Japanese Quail 1. Growth Performance. World J. Agric. Sci. 2008, 4, 605–611. [Google Scholar]
- Caetano-Anolles, K.; Seo, M.; Rodriguez-zas, S.; Oh, J.; Han, J.Y.; Lee, K.; Park, T.S.; Shin, S.; Jiao, Z.J.; Ghosh, M.; et al. Comprehensive Identification of Sexual Dimorphism-Associated Differentially Expressed Genes in Two-Way Factorial Designed RNA-Seq Data on Japanese Quail (Coturnix coturnix japonica). PLoS ONE 2015, 10, e0139324. [Google Scholar]
- Le Douarin, N. A biological cell labeling technique and its use in experimental embryology. Dev. Biol. 1973, 30, 217–222. [Google Scholar] [CrossRef]
- Huss, D.; Poynter, G.; Lansford, R. Japanese quail (Coturnix japonica) as a laboratory animal model. Lab Anim. 2008, 37, 513–519. [Google Scholar] [CrossRef] [PubMed]
- Pusch, E.A.; Bentz, A.B.; Becker, D.J.; Navara, K.J. Behavioral phenotype predicts physiological responses to chronic stress in proactive and reactive birds. Gen. Comp. Endocrinol. 2018, 255, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Recoquillay, J.; Pitel, F.; Arnould, C.; Leroux, S.; Dehais, P.; Moreno, C.; Calandreau, L.; Bertin, A.; Gourichon, D.; Bouchez, O.; et al. A medium density genetic map and QTL for behavioral and production traits in Japanese quail. BMC Genom. 2015, 16, 10. [Google Scholar] [CrossRef] [PubMed]
- Kawahara-Miki, R.; Sano, S.; Nunome, M.; Shimmura, T.; Kuwayama, T.; Takahashi, S.; Kawashima, T.; Matsuda, Y.; Yoshimura, T.; Kono, T. Next-generation sequencing reveals genomic features in the Japanese quail. Genomics 2013, 101, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Alkan, S.; Karsli, T.; Karabag, K.; Galic, A.; Balcioglu, M.S. The effects of thermal manipulation during early and late embryogenesis on hatchability, hatching weight and body weight in Japanese quails (Coturnix coturnix japonica). Arch. Tierzucht 2013, 56, 789–796. [Google Scholar] [CrossRef]
- Ozcelik, M.; Ozbey, O. The effect of the high environmental temperature on some blood parameters and the laying performance of Japanese quails with different body weights* (short communication). Arch. Anim. Breed. 2004, 47, 93–98. [Google Scholar] [CrossRef]
- Higuchi, R.; Dollinger, G.; Sean Walsh, P.; Griffith, R. Simultaneous amplification and detection of specific DNA sequences. Bio/Technology 1992, 10, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Wittwer, C.T.; Herrmann, M.G.; Moss, A.A.; Rasmussen, R.P. Continuous Fluorescence Monitoring of Rapid Cycle DNA Amplification. Biotechniques 2013, 54, 314–320. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A. Absolute quantification of mrna using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000, 25, 169–193. [Google Scholar] [CrossRef] [PubMed]
- Kubista, M.; Andrade, J.M.; Bengtsson, M.; Forootan, A.; Jonák, J.; Lind, K.; Sindelka, R.; Sjöback, R.; Sjögreen, B.; Strömbom, L.; et al. The real-time polymerase chain reaction. Mol. Aspects Med. 2006, 27, 95–125. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, R.; Fockler, C.; Dollinger, G.; Watson, R. Kinetic PCR analysis: Real-time monitoring of DNA amplification reactions. Nat. Biotechnol. 1993, 11, 1026–1030. [Google Scholar] [CrossRef]
- David, S.-A.; Mersch, M.; Foissac, S.; Collin, A.; Pitel, F.; Coustham, V. Genome-Wide Epigenetic Studies in Chicken: A Review. Epigenomes 2017, 1, 20. [Google Scholar] [CrossRef]
- Git, A.; Dvinge, H.; Salmon-Divon, M.; Osborne, M.; Kutter, C.; Hadfield, J.; Bertone, P.; Caldas, C. Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA 2010, 16, 991–1006. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The {MIQE} Guidelines: Minimum Information for Publication of Quantitative {Real-Time} {PCR} Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Perkins, J.R.; Dawes, J.M.; McMahon, S.B.; Bennett, D.L.H.; Orengo, C.; Kohl, M. ReadqPCR and NormqPCR: R packages for the reading, quality checking and normalisation of RT-qPCR quantification cycle (Cq) data. BMC Genom. 2012, 13, 296. [Google Scholar] [CrossRef] [PubMed]
- De Spiegelaere, W.; Dern-Wieloch, J.; Weigel, R.; Schumacher, V.; Schorle, H.; Nettersheim, D.; Bergmann, M.; Brehm, R.; Kliesch, S.; Vandekerckhove, L.; et al. Reference gene validation for RT-qPCR, a note on different available software packages. PLoS ONE 2015, 10, e0122515. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; Van Roy, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3. [Google Scholar] [CrossRef]
- McBryan, J.; Hamill, R.M.; Davey, G.; Lawlor, P.; Mullen, A.M. Identification of suitable reference genes for gene expression analysis of pork meat quality and analysis of candidate genes associated with the trait drip loss. Meat Sci. 2010, 86, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Robinson, T.L.; Sutherland, I.A.; Sutherland, J. Validation of candidate bovine reference genes for use with real-time PCR. Vet. Immunol. Immunopathol. 2007, 115, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, M.; Bernard, L.; Bes, S.; Leroux, C. Selection of reference genes for quantitative real-time PCR normalisation in adipose tissue, muscle, liver and mammary gland from ruminants. Animal 2013, 7, 1344–1353. [Google Scholar] [CrossRef] [PubMed]
- Walker, C.G.; Meier, S.; Mitchell, M.D.; Roche, J.R.; Littlejohn, M. Evaluation of real-time PCR endogenous control genes for analysis of gene expression in bovine endometrium. BMC Mol. Biol. 2009, 10, 100. [Google Scholar] [CrossRef] [PubMed]
- Brinkhof, B.; Spee, B.; Rothuizen, J.; Penning, L.C. Development and evaluation of canine reference genes for accurate quantification of gene expression. Anal. Biochem. 2006, 356, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Olias, P.; Adam, I.; Meyer, A.; Scharff, C.; Gruber, A.D. Reference genes for quantitative gene expression studies in multiple avian species. PLoS ONE 2014, 9, e99678. [Google Scholar] [CrossRef] [PubMed]
- Bages, S.; Estany, J.; Tor, M.; Pena, R.N. Investigating reference genes for quantitative real-time PCR analysis across four chicken tissues. Gene 2015, 561, 82–87. [Google Scholar] [CrossRef] [PubMed]
- De Boever, S.; Vangestel, C.; De Backer, P.; Croubels, S.; Sys, S.U. Identification and validation of housekeeping genes as internal control for gene expression in an intravenous LPS inflammation model in chickens. Vet. Immunol. Immunopathol. 2008, 122, 312–317. [Google Scholar] [CrossRef] [PubMed]
- Yue, H.; Lei, X.W.; Yang, F.L.; Li, M.Y.; Tang, C. Reference gene selection for normalization of PCR analysis in chicken embryo fibroblast infected with H5N1 AIV. Virol. Sin. 2010, 25, 425–431. [Google Scholar] [CrossRef] [PubMed]
- De Winter, P.; Sugden, D.; Baggott, G.K. Effect of egg turning and incubation time on carbonic anhydrase gene expression in the blastoderm of the Japanese quail (Coturnix c. japonica). Br. Poult. Sci. 2008, 49, 566–573. [Google Scholar] [CrossRef] [PubMed]
- RStudio. RStudio: Integrated development environment for R (Version 0.97.311). J. Wildl. Manag. 2011, 75, 1753–1766. [Google Scholar]
- Liu, L.L.; Zhao, H.; Ma, T.F.; Ge, F.; Chen, C.S.; Zhang, Y.P. Identification of valid reference genes for the normalization of RT-qPCR expression studies in human breast cancer cell lines treated with and without transient transfection. PLoS ONE 2015, 10, e0117058. [Google Scholar] [CrossRef] [PubMed]

| Gene Symbol | Gene Name | Primer (5′–3′) | Accession Number | Amplicon Size (bp) |
|---|---|---|---|---|
| ACTB | Actin β | F: TGACCGCGGTACAAACACAG | XM_015876619.1 | 167 |
| R: CATACCAACCATCACACCCTGA | ||||
| CCNH | Cyclin H | F: GTCTGTAGTGGGAACGGCTT | XM_015849748.1 | 177 |
| R: TGTCCAACAGGGCTTTCTCG | ||||
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | F: TCTCTGTTGTTGACCTGACCTG | XM_015873412.1 | 154 |
| R: ATGGCTGTCACCATTGAAGTC | ||||
| PGK1 | Phosphoglycerate kinase 1 | F: CAAGCTCACCCTGGACAAGT | XM_015860450.1 | 119 |
| R: GGACGGCTGCCTTGATTCTT | ||||
| RPL19 | Ribosomal protein L19 | F: GCATCGGTAAGAGGAAGGGT | XM_015885843.1 | 163 |
| R: ACGTTGCCCTTGACCTTCAG | ||||
| RPL32 | Ribosomal protein L32 | F: ATGGGAGCAACAAGAAGACA | XM_015875135.1 | 139 |
| R: TTGGAAGACACGTTGTGAGC | ||||
| RPS7 | Ribosomal protein S7 | F: TGTGGTGTTCATTGCTCAGAGA | XM_015859359.1 | 179 |
| R: TGCCATCCAGTTTTACGCGG | ||||
| RPS8 | Ribosomal protein S8 | F: GCTGACACCTGAGGAAGAAGA | XM_015870342.1 | 196 |
| R: CTTGCCTTCCAACACGTAGC | ||||
| SDHA | Succinate dehydrogenase complex, subunit A | F: TACGGGAAGGAAGGGGTTGT | XM_015854268.1 | 167 |
| R: CACAGTAGGCAGAACGGGAA | ||||
| TBP | TATA box binding protein | F: CCGGAATCATGGATCAGAAC | XM_015857924.1 | 85 |
| R: GGAATTCCAGGAGTCATTGC | ||||
| YWHAZ | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta | F: CGAACAAAAGACGGAAGGCG | XM_015856086.1 | 154 |
| R: AACTTTGCTTTCTGCTTGCGA |
| Tissue | Liver | Brain | Muscle | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | LDR | PCR eff. (%) | R² | LDR | PCR eff. (%) | R² | LDR | PCR eff. (%) | R² |
| ACTB | 18–27 | 93 | 0.98 | 17–28 | 98 | 0.99 | 19–28 | 98 | 0.96 |
| GAPDH | 17–26 | 99 | 0.99 | 16–26 | 103 | 0.99 | 14–22 | 95 | 0.98 |
| PGK1 | 20–29 | 99 | 0.98 | 20–29 | 99 | 0.99 | 17–26 | 100 | 0.98 |
| RPL19 | 19–28 | 101 | 0.95 | 20–30 | 106 | 0.99 | 20–29 | 97 | 0.96 |
| RPL32 | 21–30 | 104 | 0.98 | 21–31 | 98 | 0.99 | 23–31 | 102 | 0.98 |
| RPS7 | 20–29 | 103 | 0.97 | 20–29 | 105 | 0.99 | 22–30 | 99 | 0.97 |
| RPS8 | 20–29 | 105 | 0.98 | 20–29 | 113 | 0.99 | 21–29 | 100 | 0.99 |
| SDHA | 22–31 | 98 | 0.98 | 20–29 | 106 | 0.99 | 21–30 | 101 | 0.98 |
| TBP | 19–34 | 102 | 0.97 | 20–29 | 93 | 0.99 | 25–34 | 99 | 0.94 |
| YWHAZ | 23–32 | 99 | 0.98 | 19–28 | 99 | 0.99 | 23–32 | 100 | 0.95 |
| Gene | Liver | Brain | Muscle |
|---|---|---|---|
| ACTB | 0.099 | 0.213 | 0.751 |
| GAPDH | 0.363 | 0.254 | 0.800 |
| PGK1 | 0.461 | 0.177 | 0.575 |
| RPL19 | 0.780 | 0.726 | 0.032 |
| RPL32 | 0.242 | 0.805 | 0.050 |
| RPS7 | 0.775 | 0.635 | 0.057 |
| RPS8 | 0.524 | 0.636 | 0.017 |
| SDHA | 0.829 | 0.401 | 0.258 |
| TBP | 0.916 | 0.322 | 0.155 |
| YWHAZ | 0.893 | 0.631 | 0.182 |
| Liver | Brain | Muscle | |
|---|---|---|---|
| Gene combination | GAPDH and RPS8 | PGK1 and RPL32 | PGK1 and ACTB |
| Stability value | 0.026 | 0.022 | 0.047 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vitorino Carvalho, A.; Couroussé, N.; Crochet, S.; Coustham, V. Identification of Reference Genes for Quantitative Gene Expression Studies in Three Tissues of Japanese Quail. Genes 2019, 10, 197. https://doi.org/10.3390/genes10030197
Vitorino Carvalho A, Couroussé N, Crochet S, Coustham V. Identification of Reference Genes for Quantitative Gene Expression Studies in Three Tissues of Japanese Quail. Genes. 2019; 10(3):197. https://doi.org/10.3390/genes10030197
Chicago/Turabian StyleVitorino Carvalho, Anaïs, Nathalie Couroussé, Sabine Crochet, and Vincent Coustham. 2019. "Identification of Reference Genes for Quantitative Gene Expression Studies in Three Tissues of Japanese Quail" Genes 10, no. 3: 197. https://doi.org/10.3390/genes10030197
APA StyleVitorino Carvalho, A., Couroussé, N., Crochet, S., & Coustham, V. (2019). Identification of Reference Genes for Quantitative Gene Expression Studies in Three Tissues of Japanese Quail. Genes, 10(3), 197. https://doi.org/10.3390/genes10030197





