CLOCK Polymorphisms in Attention-Deficit/Hyperactivity Disorder (ADHD): Further Evidence Linking Sleep and Circadian Disturbances and ADHD
Abstract
:1. Introduction
2. Material and Methods
2.1. Sample
2.2. DNA and Genotyping
2.3. Genetic Markers
2.4. Statistical Analysis
2.5. Secondary Analyses
2.6. In Silico Functionality Analysis
2.7. Research Ethics
3. Results
3.1. Secondary Analysis
3.2. In Silico Functionality Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Polanczyk, G.V.; Willcutt, E.G.; Salum, G.A.; Kieling, C.; Rohde, L.A. ADHD prevalence estimates across three decades: An updated systematic review and meta-regression analysis. Int. J. Epidemiol. 2014, 43, 434–442. [Google Scholar] [CrossRef]
- Banaschewski, T.; Becker, K.; Döpfner, M.; Holtmann, M.; Rösler, M.; Romanos, M. Attention-deficit/hyperactivity disorder. Dtsch. Arztebl. Int. 2017, 114, 149–159. [Google Scholar] [CrossRef]
- Um, Y.H.; Hong, S.-C.; Jeong, J.-H. Sleep problems as predictors in attention-deficit hyperactivity disorder: Causal mechanisms, consequences and treatment. Clin. Psychopharmacol. Neurosci. 2017, 15, 9–18. [Google Scholar] [CrossRef]
- Hvolby, A. Associations of sleep disturbance with ADHD: Implications for treatment. ADHD Atten. Deficit Hyperact. Disord. 2015, 7, 1–18. [Google Scholar] [CrossRef]
- Coogan, A.N.; McGowan, N.M. A systematic review of circadian function, chronotype and chronotherapy in attention deficit hyperactivity disorder. Atten. DeficIT Hyperact. Disord. 2017, 9, 129–147. [Google Scholar] [CrossRef]
- Weiss, M.D.; Salpekar, J. Sleep problems in the child with attention-deficit hyperactivity disorder. CNS Drugs 2010, 24, 811–828. [Google Scholar] [CrossRef]
- Noble, G.S.; O’Laughlin, L.; Brubaker, B. Attention deficit hyperactivity disorder and sleep disturbances: Consideration of parental influence. Behav. Sleep Med. 2012, 10, 41–53. [Google Scholar] [CrossRef]
- Sung, V.; Hiscock, H.; Sciberras, E.; Efron, D. Sleep problems in children with attention-deficit/hyperactivity disorder: Prevalence and the effect on the child and family. Arch. Pediatr. Adoles. Med. 2008, 162, 336–342. [Google Scholar] [CrossRef]
- Dibner, C.; Schibler, U. Circadian timing of metabolism in animal models and humans. J. Intern. Med. 2015, 277, 513–527. [Google Scholar] [CrossRef] [Green Version]
- Waddington Lamont, E.; Legault-Coutu, D.; Cermakian, N.; Boivin, D.B. The role of circadian clock genes in mental disorders. Dialogues Clin. Neurosci. 2007, 9, 333–342. [Google Scholar]
- Schuch, J.B.; Genro, J.P.; Bastos, C.R.; Ghisleni, G.; Tovo-Rodrigues, L. The role of CLOCK gene in psychiatric disorders: Evidence from human and animal research. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2017, 177, 181–198. [Google Scholar] [CrossRef]
- Dela Pena, I.J.I.; Dela Pena, I.; de la Pena, J.B.; Kim, H.J.; Sohn, A.; Shin, C.Y.; Han, D.H.; Kim, B.-N.; Ryu, J.H.; Cheong, J.H. Transcriptional profiling of SHR/NCrl prefrontal cortex shows hyperactivity-associated genes responsive to amphetamine challenge. Genes Brain Behav. 2017, 16, 664–674. [Google Scholar] [CrossRef]
- Huang, J.; Zhong, Z.; Wang, M.; Chen, X.; Tan, Y.; Zhang, S.; He, W.; He, X.; Huang, G.; Lu, H.; et al. Circadian modulation of dopamine levels and dopaminergic neuron development contributes to attention deficiency and hyperactive behavior. J. Neurosci. 2015, 35, 2572–2587. [Google Scholar] [CrossRef]
- Baird, A.L.; Coogan, A.N.; Kaufling, J.; Barrot, M.; Thome, J. Daily methylphenidate and atomoxetine treatment impacts on clock gene protein expression in the mouse brain. Brain Res. 2013, 1513, 61–71. [Google Scholar] [CrossRef]
- O’Keeffe, S.M.; Thome, J.; Coogan, A.N. The noradrenaline reuptake inhibitor atomoxetine phase-shifts the circadian clock in mice. Neuroscience 2012, 201, 219–230. [Google Scholar] [CrossRef]
- McClung, C.A.; Sidiropoulou, K.; Vitaterna, M.; Takahashi, J.S.; White, F.J.; Cooper, D.C.; Nestler, E.J. Regulation of dopaminergic transmission and cocaine reward by the Clock gene. Proc. Natl. Acad. Sci. USA 2005, 102, 9377–9381. [Google Scholar] [CrossRef]
- Jeong, S.H.; Yu, J.-C.; Lee, C.H.; Choi, K.-S.; Choi, J.-E.; Kim, S.H.; Joo, E.-J. Human CLOCK gene-associated attention deficit hyperactivity disorder-related features in healthy adults: Quantitative association study using Wender Utah Rating Scale. Eur. Arch. Psychiatry Clin. Neurosci. 2014, 264, 71–81. [Google Scholar] [CrossRef]
- Kissling, C.; Retz, W.; Wiemann, S.; Coogan, A.N.; Clement, R.M.; Hunnerkopf, R.; Conner, A.C.; Freitag, C.M.; Rosler, M.; Thome, J. A polymorphism at the 3′-untranslated region of the CLOCK gene is associated with adult attention-deficit hyperactivity disorder. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2008, 147, 333–338. [Google Scholar] [CrossRef]
- Xu, X.; Breen, G.; Chen, C.-K.; Huang, Y.-S.; Wu, Y.-Y.; Asherson, P. Association study between a polymorphism at the 3’-untranslated region of CLOCK gene and attention deficit hyperactivity disorder. Behav. Brain Funct. 2010, 6, 48. [Google Scholar] [CrossRef]
- Zheng, J.; Baird, D.; Borges, M.-C.; Bowden, J.; Hemani, G.; Haycock, P.; Evans, D.M.; Smith, G.D. Recent developments in Mendelian randomization studies. Curr. Epidemiol. Rep. 2017, 4, 330–345. [Google Scholar] [CrossRef]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 4th ed.; American Psychiatric Association: Washington, DC, USA, 1994; ISBN 978-0-89042-559-6. [Google Scholar]
- Salatino-Oliveira, A.; Genro, J.P.; Chazan, R.; Zeni, C.; Schmitz, M.; Polanczyk, G.; Roman, T.; Rohde, L.A.; Hutz, M.H. Association study of GIT1 gene with attention-deficit hyperactivity disorder in Brazilian children and adolescents. Genes Brain Behav. 2012, 11, 864–868. [Google Scholar] [CrossRef]
- Bruxel, E.M.; Salatino-Oliveira, A.; Akutagava-Martins, G.C.; Tovo-Rodrigues, L.; Genro, J.P.; Zeni, C.P.; Polanczyk, G.V.; Chazan, R.; Schmitz, M.; Arcos-Burgos, M. LPHN3 and attention-deficit/hyperactivity disorder: A susceptibility and pharmacogenetic study. Genes Brain Behav. 2015, 14, 419–427. [Google Scholar] [CrossRef]
- Salatino-Oliveira, A.; Akutagava-Martins, G.C.; Bruxel, E.M.; Genro, J.P.; Polanczyk, G.V.; Zeni, C.; Kieling, C.; Karam, R.G.; Rovaris, D.L.; Contini, V. NOS1 and SNAP25 polymorphisms are associated with Attention-Deficit/Hyperactivity Disorder symptoms in adults but not in children. J. Psychiatr. Res. 2016, 75, 75–81. [Google Scholar] [CrossRef]
- Costa, D.S.; de Paula, J.J.; Malloy-Diniz, L.F.; Romano-Silva, M.A.; Miranda, D.M. Parent SNAP-IV rating of attention-deficit/hyperactivity disorder: Accuracy in a clinical sample of ADHD, validity, and reliability in a Brazilian sample. J. Pediatr. 2018, in press. [Google Scholar] [CrossRef]
- Lahiri, D.K.; Nurnberger, J.I., Jr. A rapid non-enzymatic method for the preparation of HMW DNA from blood for RFLP studies. Nucleic Acids Res. 1991, 19, 5444. [Google Scholar] [CrossRef]
- Schwender, H.; Taub, M.A.; Beaty, T.H.; Marazita, M.L.; Ruczinski, I. Rapid testing of SNPs and gene–environment interactions in case–parent trio data based on exact analytic parameter estimation. Biometrics 2012, 68, 766–773. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.; Daly, M.J. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Li, M.-X.; Yeung, J.M.; Cherny, S.S.; Sham, P.C. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum. Genet. 2012, 131, 747–756. [Google Scholar] [CrossRef]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2004, 21, 263–265. [Google Scholar] [CrossRef]
- Dudbridge, F. Likelihood-based association analysis for nuclear families and unrelated subjects with missing genotype data. Hum. Hered. 2008, 66, 87–98. [Google Scholar] [CrossRef]
- Ward, L.D.; Kellis, M. HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2011, 40, D930–D934. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.; Thormann, A.; Flicek, P.; Cunningham, F. The ensembl variant effect predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef]
- Rentzsch, P.; Witten, D.; Cooper, G.M.; Shendure, J.; Kircher, M. CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2018, 47, D886–D891. [Google Scholar] [CrossRef]
- Boyle, A.P.; Hong, E.L.; Hariharan, M.; Cheng, Y.; Schaub, M.A.; Kasowski, M.; Karczewski, K.J.; Park, J.; Hitz, B.C.; Weng, S. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012, 22, 1790–1797. [Google Scholar] [CrossRef] [Green Version]
- Fontenot, M.R.; Berto, S.; Liu, Y.; Werthmann, G.; Douglas, C.; Usui, N.; Gleason, K.; Tamminga, C.A.; Takahashi, J.S.; Konopka, G. Novel transcriptional networks regulated by CLOCK in human neurons. Genes Dev. 2017. [Google Scholar] [CrossRef]
- Coogan, A.N.; Baird, A.L.; Popa-Wagner, A.; Thome, J. Circadian rhythms and attention deficit hyperactivity disorder: The what, the when and the why. Prog. Neuropsychopharmacol. Biol. Psychiatry 2016, 67, 74–81. [Google Scholar] [CrossRef]
- Cassoff, J.; Wiebe, S.T.; Gruber, R. Sleep patterns and the risk for ADHD: A review. Nat. Sci. Sleep 2012, 4, 73–80. [Google Scholar] [CrossRef]
- Sadeh, A.; Tikotzky, L.; Kahn, M. Sleep in infancy and childhood: Implications for emotional and behavioral difficulties in adolescence and beyond. Curr. Opin. Psychiatry 2014, 27, 453–459. [Google Scholar] [CrossRef]
- Consortium, W.T.C.C. Genome-wide association study of 14,000 cases of seven common diseases and 3000 shared controls. Nature 2007, 447, 661. [Google Scholar] [CrossRef]
- Shi, J.; Wittke-Thompson, J.K.; Badner, J.A.; Hattori, E.; Potash, J.B.; Willour, V.L.; McMahon, F.J.; Gershon, E.S.; Liu, C. Clock genes may influence bipolar disorder susceptibility and dysfunctional circadian rhythm. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2008, 147, 1047–1055. [Google Scholar] [CrossRef]
- Ozburn, A.R.; Purohit, K.; Parekh, P.K.; Kaplan, G.N.; Falcon, E.; Mukherjee, S.; Cates, H.M.; McClung, C.A. Functional implications of the CLOCK 3111T/C single-nucleotide polymorphism. Front. Psychiatry 2016, 7, 67. [Google Scholar] [CrossRef]
- Saus, E.; Soria, V.; Escaramís, G.; Vivarelli, F.; Crespo, J.M.; Kagerbauer, B.; Menchón, J.M.; Urretavizcaya, M.; Gratacòs, M.; Estivill, X. Genetic variants and abnormal processing of pre-miR-182, a circadian clock modulator, in major depression patients with late insomnia. Hum. Mol. Genet. 2010, 19, 4017–4025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griggs, E.M.; Young, E.J.; Rumbaugh, G.; Miller, C.A. MicroRNA-182 regulates amygdala-dependent memory formation. J. Neurosci. 2013, 33, 1734–1740. [Google Scholar] [CrossRef]
- Wang, W.M.; Lu, G.; Su, X.W.; Lyu, H.; Poon, W.S. MicroRNA-182 regulates neurite outgrowth involving the PTEN/AKT pathway. Front. Cell. Neurosci. 2017, 11, 96. [Google Scholar] [CrossRef]
- Ye, Y.; Xu, H.; Su, X.; He, X. Role of microRNA in governing synaptic plasticity. Neural Plast. 2016, 2016, 4959523. [Google Scholar] [CrossRef]
- Yuan, H.; Mischoulon, D.; Fava, M.; Otto, M.W. Circulating microRNAs as biomarkers for depression: Many candidates, few finalists. J. Affect. Disord. 2018, 233, 68–78. [Google Scholar] [CrossRef]
- Kohen, R.; Dobra, A.; Tracy, J.H.; Haugen, E. Transcriptome profiling of human hippocampus dentate gyrus granule cells in mental illness. Transl. Psychiatry 2014, 4, e366. [Google Scholar] [CrossRef]
- Jones, S.E.; Tyrrell, J.; Wood, A.R.; Beaumont, R.N.; Ruth, K.S.; Tuke, M.A.; Yaghootkar, H.; Hu, Y.; Teder-Laving, M.; Hayward, C.; et al. Genome-wide association analyses in 128,266 individuals identifies new morningness and sleep duration loci. PLoS Genet. 2016, 12, e1006125. [Google Scholar] [CrossRef]
- Lane, J.M.; Liang, J.; Vlasac, I.; Anderson, S.G.; Bechtold, D.A.; Bowden, J.; Emsley, R.; Gill, S.; Little, M.A.; Luik, A.I.; et al. Genome-wide association analyses of sleep disturbance traits identify new loci and highlight shared genetics with neuropsychiatric and metabolic traits. Nat. Genet. 2017, 49, 274–281. [Google Scholar] [CrossRef]
- Hammerschlag, A.R.; Stringer, S.; de Leeuw, C.A.; Sniekers, S.; Taskesen, E.; Watanabe, K.; Blanken, T.F.; Dekker, K.; Te Lindert, B.H.; Wassing, R. Genome-wide association analysis of insomnia complaints identifies risk genes and genetic overlap with psychiatric and metabolic traits. Nat. Genet. 2017, 49, 1584. [Google Scholar] [CrossRef]
- Marinelli, M.; Pappa, I.; Bustamante, M.; Bonilla, C.; Bonilla, C.; Suarez, A.; Tiesler, C.M.; Vilor-Tejedor, N.; Zafarmand, M.H.; Alvarez-Pedrerol, M. Heritability and genome-wide association analyses of sleep duration in children: The EAGLE consortium. Sleep 2016, 39, 1859–1869. [Google Scholar] [CrossRef] [PubMed]
- Boyle, E.A.; Li, Y.I.; Pritchard, J.K. An expanded view of complex traits: From polygenic to omnigenic. Cell 2017, 169, 1177–1186. [Google Scholar] [CrossRef]
- Faraone, S.V.; Larsson, H. Genetics of attention deficit hyperactivity disorder. Mol. Psychiatry 2018. [Google Scholar] [CrossRef]
- Durso, D.F.; Bydlowski, S.P.; Hutz, M.H.; Suarez-Kurtz, G.; Magalhães, T.R.; Pena, S.D.J. Association of genetic variants with self-assessed color categories in Brazilians. PLoS ONE 2014, 9, e83926. [Google Scholar] [CrossRef]
- Salzano, F.M.; Sans, M. Interethnic admixture and the evolution of Latin American populations. Genet. Mol. Biol. 2014, 37, 151–170. [Google Scholar] [CrossRef] [Green Version]
- De Neves Manta, F.S.; Pereira, R.; Vianna, R.; de Araújo, A.R.B.; Gitaí, D.L.G.; da Silva, D.A.; de Vargas Wolfgramm, E.; da Mota Pontes, I.; Aguiar, J.I.; Moraes, M.O. Revisiting the genetic ancestry of Brazilians using autosomal AIM-Indels. PLoS ONE 2013, 8, e75145. [Google Scholar] [CrossRef]
- Santos, N.P.; Ribeiro-Rodrigues, E.M.; Ribeiro-dos-Santos, Â.K.; Pereira, R.; Gusmão, L.; Amorim, A.; Guerreiro, J.F.; Zago, M.A.; Matte, C.; Hutz, M.H. Assessing individual interethnic admixture and population substructure using a 48–insertion-deletion (INSEL) ancestry-informative marker (AIM) panel. Hum. Mutat. 2010, 31, 184–190. [Google Scholar] [CrossRef]
- Grinde, K.E.; Qi, Q.; Thornton, T.A.; Liu, S.; Shadyab, A.H.; Chan, K.H.K.; Reiner, A.P.; Sofer, T. Generalizing genetic risk scores from europeans to Hispanics/Latinos. bioRxiv 2018, 242404. [Google Scholar] [CrossRef]
| Variable N | N (%) | ||
|---|---|---|---|
| Mean (Standard Deviation, SD) | |||
| Age(years), mean (SD) | 259 | 10.42 (3.2) | |
| IQ, mean (SD) | 255 | 93.52 (13.1) | |
| Gender, N male (%) | 259 | 198 (76.4) | |
| Skin color | 259 | ||
| White, N (%) | 216(83.4) | ||
| Brown or Black, N (%) | 43 (16.6) | ||
| ADHD presentations | 259 | ||
| Inattentive, N (%) | 114(44.0) | ||
| Hyperactive, N (%) | 13 (5.0) | ||
| Combined, N (%) | 122 (47.1) | ||
| Subthreshold N (%) | 10 (3.9) | ||
| Comorbid Conditions | |||
| Conduct Disorders, N (%) | 258 | 36 (13.9) | |
| Oppositional Defiant Disorders, N (%) | 258 | 92 (35.5) | |
| Mood disorders, N (%) | 241 | 23 (8.9) | |
| Anxiety disorders, N (%) | 241 | 72 (27.8) | |
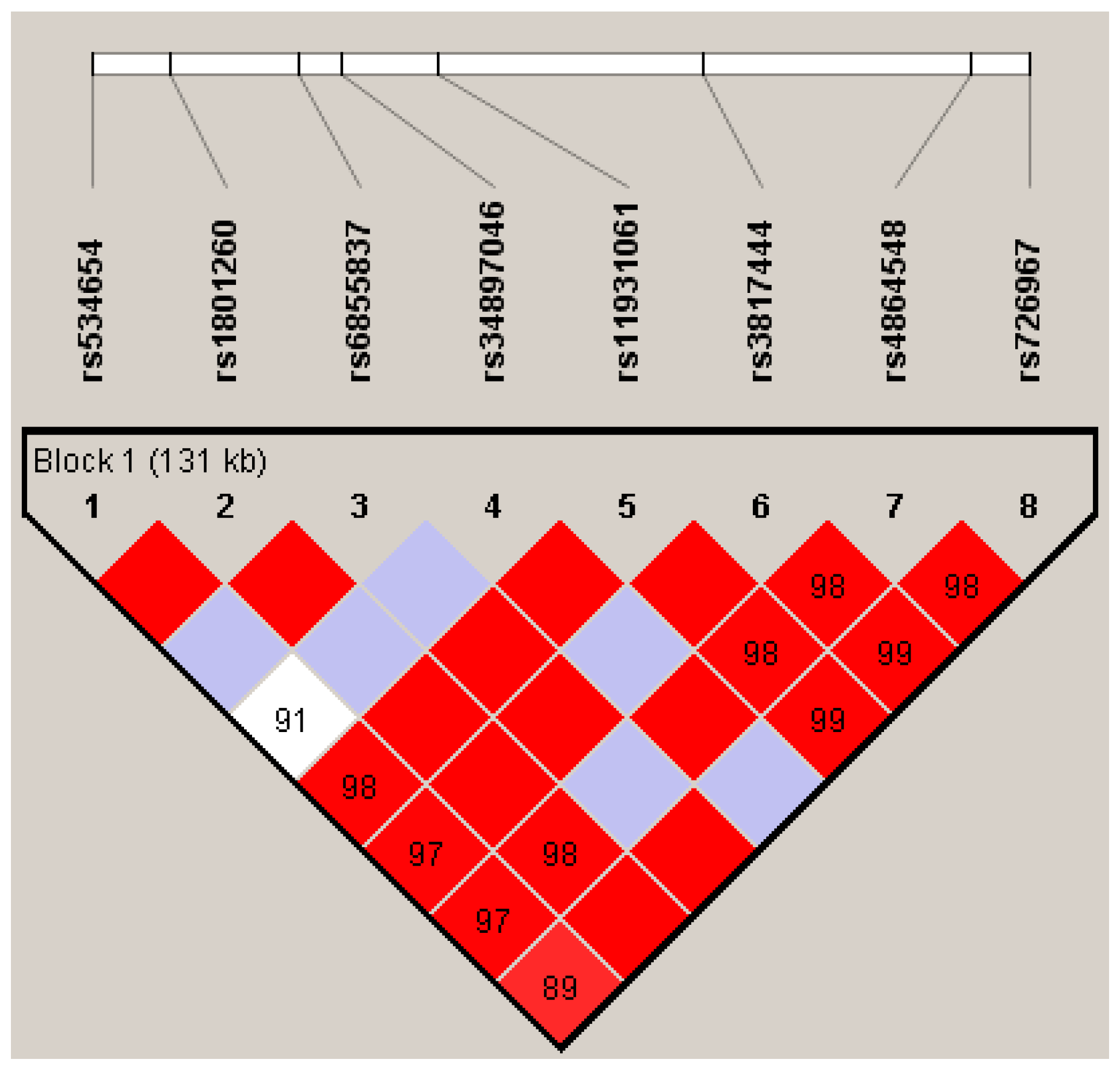
| SNP | Location | Position Regarding CLOCK Gene | EA | RA | FREQ EA * | HWE (p-value) | OR (95% CI) | p |
|---|---|---|---|---|---|---|---|---|
| rs534654 | 56290220 | 3848 bp downstream | A | G | 0.226 | 0.593 | 1.54 (1.11–2.12) | 0.010 |
| rs1801260 | 56301369 | exon 23 (3’UTR) | G | A | 0.253 | 0.252 | 0.87 (0.66–1.15) | 0.329 |
| rs6855837 | 56319244 | exon 15 | T | G | 0.048 | 0.456 | 1.96 (0.99–3.86) | 0.054 |
| rs34897046 | 56325365 | exon 10 | C | G | 0.029 | 0.188 | 0.79 (0.40–1.56) | 0.492 |
| rs11931061 | 56338793 | intron 7 | G | A | 0.402 | 0.120 | 1.180 (0.92–1.51) | 0.188 |
| rs3817444 | 56375981 | intron 2 | A | C | 0.344 | 0.075 | 1.059 (0.82–1.36) | 0.655 |
| rs4864548 | 56413803 | 727 bp upstream | A | G | 0.344 | 0.681 | 0.911 (0.71–1.18) | 0.473 |
| rs726967 | 56421713 | 8637 pb upstream | T | A | 0.334 | 0.094 | 1.068 (0.83–1.37) | 0.608 |
| Haplotype * | Haplotype Frequency ** | OR (95% CI) |
|---|---|---|
| G-A-G-G-A-C-A-A | 0.314 | 1 |
| G-G-G-G-A-C-G-A | 0.255 | 0.92 (0.66–1.27) |
| A-A-G-G-G-A-G-T | 0.213 | 1.43 (1.00–2.07) |
| G-A-G-G-G-A-G-T | 0.125 | 0.77 (0.52–1.15) |
| G-A-T-G-G-C-G-A | 0.050 | 2.15 (1.04–4.43) |
| G-A-G-C-A-C-A-A | 0.030 | 0.82 (0.40–1.66) |
| A-A-G-G-G-A-G-A | 0.012 | 1.24 (0.37–4.14) |
| Inattention | Hyperactivity | ADHD Total Symptoms | |||||
|---|---|---|---|---|---|---|---|
| SNP | EA | OR (95%CI95) * | p * | OR (95%CI95) * | p * | OR (95%CI95) * | p * |
| rs534654 | A | 1.53 (0.94–2.5) | 0.088 | 1.157 (0.71–1.90) | 0.565 | 1.16 (0.72; 1.88) | 0.534 |
| rs1801260 | G | 0.70 (0.45–1.10) | 0.122 | 0.774 (0.49–1.22) | 0.265 | 0.80 (0.52; 6.93) | 0.309 |
| rs6855837 | T | 1.51 (0.55–4.15) | 0.424 | 2.654 (0.94–7.47) | 0.064 | 2.37 (0.81; 6.93) | 0.117 |
| rs34897046 | C | 1.16 (0.39–3.43) | 0.836 | 0.955 (0.32–2.89) | 0.934 | 1.04 (0.36; 3.03) | 0.936 |
| rs11931061 | G | 1.56 (1.05–2.33) | 0.029 | 1.289 (0.87–1.20) | 0.210 | 1.469 (0.99; 2.17) | 0.054 |
| rs3817444 | A | 1.60 (1.06–2.42) | 0.026 | 1.117 (0.74–1.68) | 0.597 | 1.39 (0.93; 2.08) | 0.107 |
| rs4864548 | A | 0.84 (0.55–1.29) | 0.433 | 0.954 (0.63–1.46) | 0.829 | 0.816 (0.54; 1.23) | 0.335 |
| rs726967 | T | 1.57 (1.03–2.37) | 0.035 | 1.096 (0.73–1.66) | 0.663 | 1.38 (0.92; 2.07) | 0.117 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carpena, M.X.; Hutz, M.H.; Salatino-Oliveira, A.; Polanczyk, G.V.; Zeni, C.; Schmitz, M.; Chazan, R.; Genro, J.P.; Rohde, L.A.; Tovo-Rodrigues, L. CLOCK Polymorphisms in Attention-Deficit/Hyperactivity Disorder (ADHD): Further Evidence Linking Sleep and Circadian Disturbances and ADHD. Genes 2019, 10, 88. https://doi.org/10.3390/genes10020088
Carpena MX, Hutz MH, Salatino-Oliveira A, Polanczyk GV, Zeni C, Schmitz M, Chazan R, Genro JP, Rohde LA, Tovo-Rodrigues L. CLOCK Polymorphisms in Attention-Deficit/Hyperactivity Disorder (ADHD): Further Evidence Linking Sleep and Circadian Disturbances and ADHD. Genes. 2019; 10(2):88. https://doi.org/10.3390/genes10020088
Chicago/Turabian StyleCarpena, Marina Xavier, Mara H. Hutz, Angélica Salatino-Oliveira, Guilherme V. Polanczyk, Cristian Zeni, Marcelo Schmitz, Rodrigo Chazan, Julia P. Genro, Luis Augusto Rohde, and Luciana Tovo-Rodrigues. 2019. "CLOCK Polymorphisms in Attention-Deficit/Hyperactivity Disorder (ADHD): Further Evidence Linking Sleep and Circadian Disturbances and ADHD" Genes 10, no. 2: 88. https://doi.org/10.3390/genes10020088
APA StyleCarpena, M. X., Hutz, M. H., Salatino-Oliveira, A., Polanczyk, G. V., Zeni, C., Schmitz, M., Chazan, R., Genro, J. P., Rohde, L. A., & Tovo-Rodrigues, L. (2019). CLOCK Polymorphisms in Attention-Deficit/Hyperactivity Disorder (ADHD): Further Evidence Linking Sleep and Circadian Disturbances and ADHD. Genes, 10(2), 88. https://doi.org/10.3390/genes10020088






