RNA 2′-O-Methylation (Nm) Modification in Human Diseases
Abstract
:1. Introduction
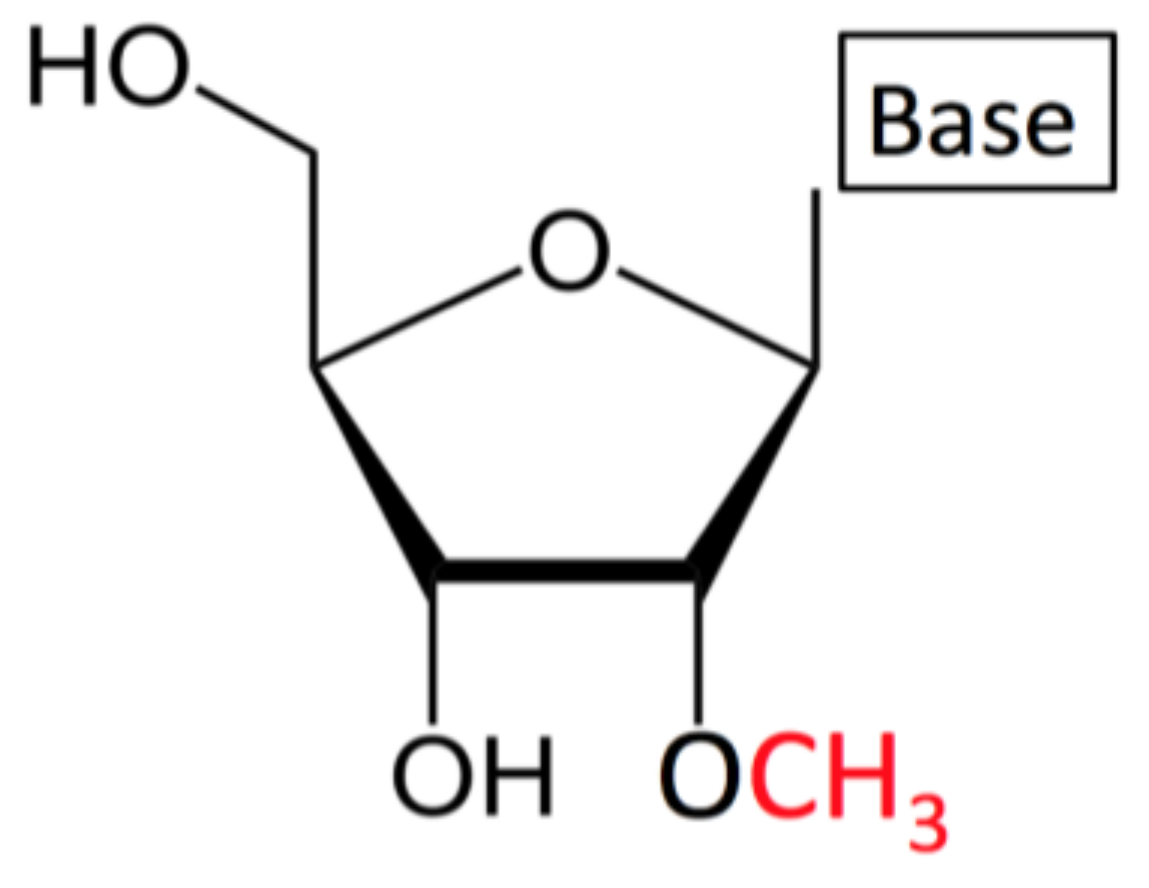
1.1. What is Nm/2′-O-Methylation?
1.2. Nm Detection
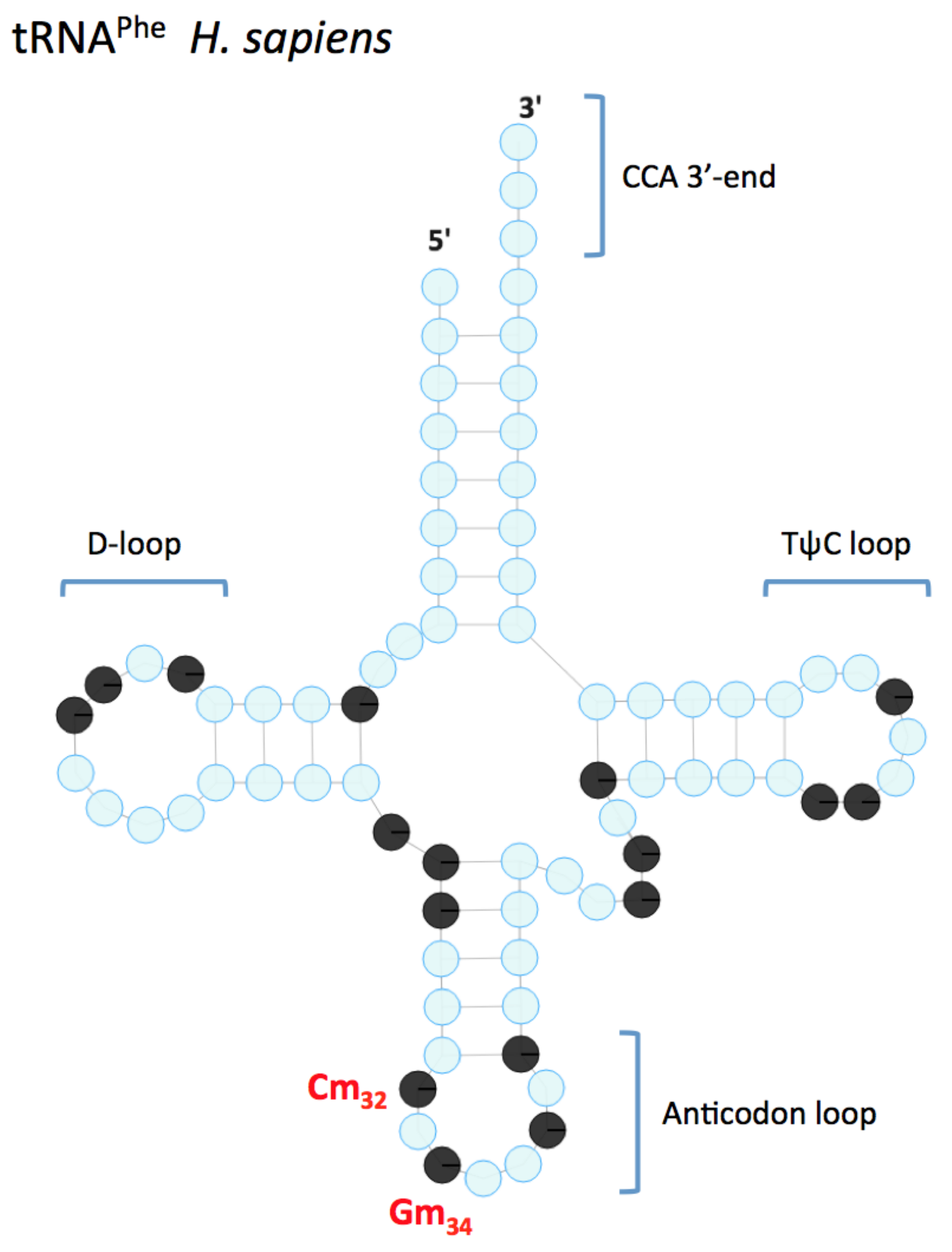
2. Nm in tRNA
2.1. FTSJ1 Identity and Conservation
2.2. FTSJ1 Link to Intellectual Disability
2.3. Discussion on FTSJ1
2.4. TRMT44 and Idiopathic Epilepsy
3. Nm in rRNAs
3.1. snoRNAs and Prader-Willi Syndrome
3.2. Fibrillarin and Diseases: Identity and Conservation
3.3. Fibrillarin and Cancer
3.4. Fibrillarin and Autoimmune Diseases
3.5. Fibrillarin: Possible Link to Other Diseases
3.6. FTSJ2 and Cancer
4. Nm in mRNAs
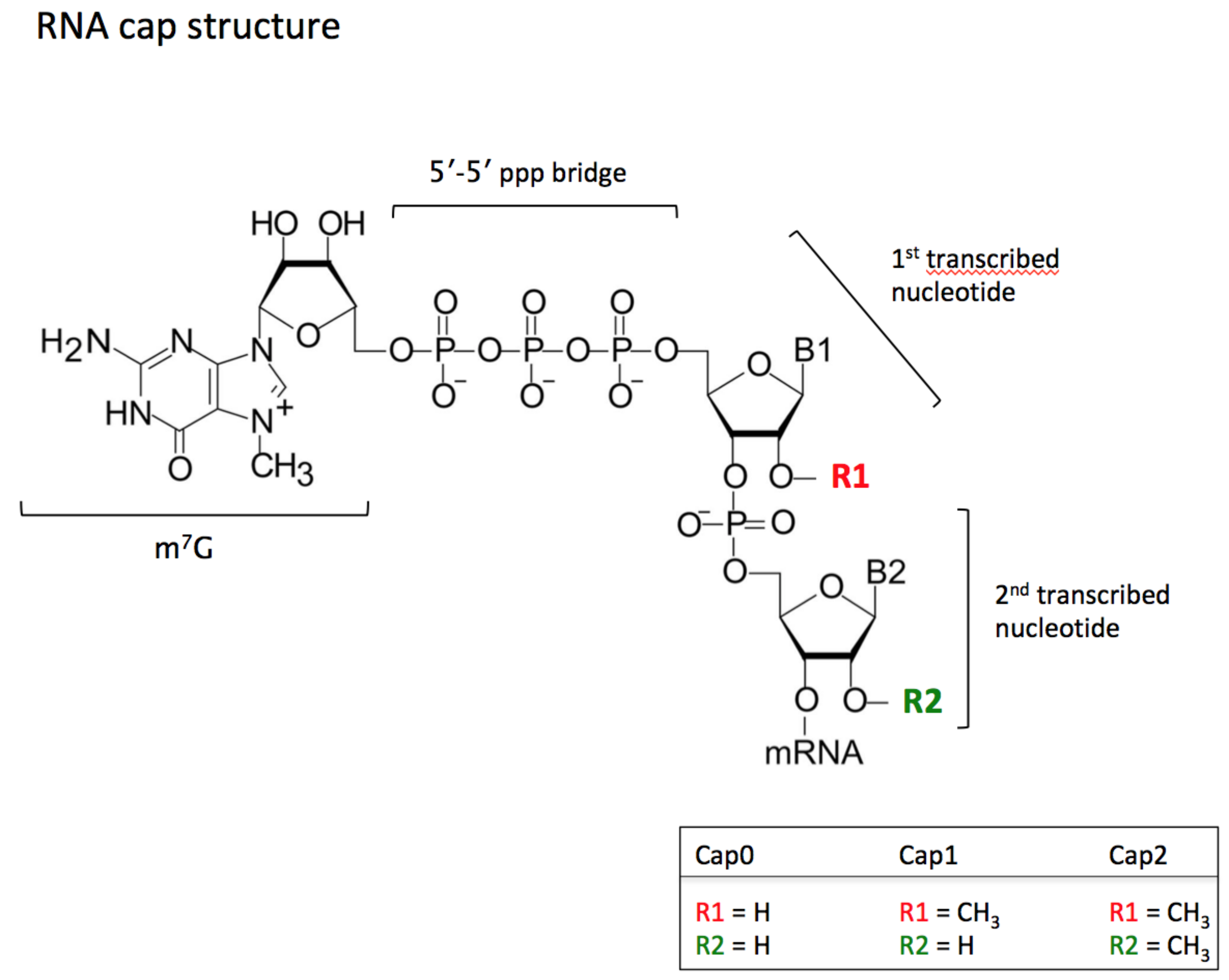
4.1. mRNAs and Cap Protection
4.2. CMTR1 and Diseases
4.3. CMTR1 and Asthma
4.4. CMTR1 and Alzheimer’s Disease
4.5. CMTR1 and Cancer
4.6. mRNA and Splicing
4.7. snRNAs
4.8. scaRNAs
4.9. mRNAs and Internal Nm Modifications
5. Nm in Small RNAs Silencing Pathways
HENMT1
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Darzacq, X.; Jády, B.E.; Verheggen, C.; Kiss, A.M.; Bertrand, E.; Kiss, T. Cajal body-specific small nuclear RNAs: A novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002, 21, 2746–2756. [Google Scholar] [CrossRef] [PubMed]
- Rebane, A.; Roomere, H.; Metspalu, A. Locations of several novel 2′-O-methylated nucleotides in human 28S rRNA. BMC Mol. Biol. 2002, 3, 1. [Google Scholar] [CrossRef]
- Somme, J.; Van Laer, B.; Roovers, M.; Steyaert, J.; Versées, W.; Droogmans, L. Characterization of two homologous 2′-O-methyltransferases showing different specificities for their tRNA substrates. RNA 2014, 20, 1257–1271. [Google Scholar] [CrossRef] [PubMed]
- Dai, Q.; Moshitch-Moshkovitz, S.; Han, D.; Kol, N.; Amariglio, N.; Rechavi, G.; Dominissini, D.; He, C. Nm-seq maps 2′-O-methylation sites in human mRNA with base precision. Nat. Methods 2017, 14, 695–698. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yang, Z.; Yu, B.; Liu, J.; Chen, X. Methylation Protects miRNAs and siRNAs from a 3′-End Uridylation Activity in Arabidopsis. Curr. Biol. 2005, 15, 1501–1507. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Yang, Z.; Li, J.; Minakhina, S.; Yang, M.; Padgett, R.W.; Steward, R.; Chen, X. Methylation as a crucial step in plant microRNA biogenesis. Science 2005, 307, 932–935. [Google Scholar] [CrossRef] [PubMed]
- Kurth, H.M.; Mochizuki, K. 2′-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. RNA 2009, 15, 675–685. [Google Scholar] [CrossRef]
- Zhao, Y.; Mo, B.; Chen, X. Mechanisms that impact microRNA stability in plants. RNA Biol. 2012, 9, 1218–1223. [Google Scholar] [CrossRef]
- Horwich, M.D.; Li, C.; Matranga, C.; Vagin, V.; Farley, G.; Wang, P.; Zamore, P.D. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 2007, 17, 1265–1272. [Google Scholar] [CrossRef]
- Saito, K.; Sakaguchi, Y.; Suzuki, T.; Suzuki, T.; Siomi, H.; Siomi, M.C. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes Dev. 2007, 21, 1603–1608. [Google Scholar] [CrossRef]
- Sproat, B.S.; Lamond, A.I.; Beijer, B.; Neuner, P.; Ryder, U. Highly efficient chemical synthesis of 2′-O-methyloligoribonucleotides and tetrabiotinylated derivatives; novel probes that are resistant to degradation by RNA or DNA specific nucleases. Nucleic Acids Res. 1989, 17, 3373–3386. [Google Scholar] [CrossRef] [PubMed]
- Byszewska, M.; Śmietański, M.; Purta, E.; Bujnicki, J.M. RNA methyltransferases involved in 5′ cap biosynthesis. RNA Biol. 2014, 11, 1597–1607. [Google Scholar] [CrossRef]
- Kumar, S.; Mapa, K.; Maiti, S. Understanding the effect of locked nucleic acid and 2′-O-methyl modification on the hybridization thermodynamics of a miRNA-mRNA pair in the presence and absence of AfPiwi protein. Biochemistry 2014, 53, 1607–1615. [Google Scholar] [CrossRef]
- Yildirim, I.; Kierzek, E.; Kierzek, R.; Schatz, G.C. Interplay of LNA and 2′-O-methyl RNA in the structure and thermodynamics of RNA hybrid systems: A molecular dynamics study using the revised AMBER force field and comparison with experimental results. J. Phys. Chem. B 2014, 118, 14177–14187. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Hayase, Y.; Imura, A.; Iwai, S.; Miura, K.; Ohtsuka, E. Synthesis and hybridization studies on two complementary nona (2′-O-methyl) ribonucleotides. Nucleic Acids Res. 1987, 15, 6131–6148. [Google Scholar] [CrossRef] [PubMed]
- Kawai, G.; Yamamoto, Y.; Kamimura, T.; Masegi, T.; Sekine, M.; Hata, T.; Iimori, T.; Watanabe, T.; Miyazawa, T.; Yokoyama, S. Conformational rigidity of specific pyrimidine residues in tRNA arises from posttranscriptional modifications that enhance steric interaction between the base and the 2′-hydroxyl group. Biochemistry 1992, 31, 1040–1046. [Google Scholar] [CrossRef] [PubMed]
- Majlessi, M.; Nelson, N.C.; Becker, M.M. Advantages of 2′-O-methyl oligoribonucleotide probes for detecting RNA targets. Nucleic Acids Res. 1998, 26, 2224–2229. [Google Scholar] [CrossRef]
- Tsourkas, A.; Behlke, M.A.; Bao, G. Hybridization of 2′-O-methyl and 2′-deoxy molecular beacons to RNA and DNA targets. Nucleic Acids Res. 2002, 30, 5168–5174. [Google Scholar] [CrossRef]
- Lebars, I.; Legrand, P.; Aimé, A.; Pinaud, N.; Fribourg, S.; Di Primo, C. Exploring TAR–RNA aptamer loop–loop interaction by X-ray crystallography, UV spectroscopy and surface plasmon resonance. Nucleic Acids Res. 2008, 36, 7146–7156. [Google Scholar] [CrossRef]
- Hou, Y.-M.; Zhang, X.; Holland, J.A.; Davis, D.R. An important 2′-OH group for an RNA--protein interaction. Nucleic Acids Res. 2001, 29, 976–985. [Google Scholar] [CrossRef]
- Lacoux, C.; Di Marino, D.; Pilo Boyl, P.; Zalfa, F.; Yan, B.; Ciotti, M.T.; Falconi, M.; Urlaub, H.; Achsel, T.; Mougin, A.; et al. BC1-FMRP interaction is modulated by 2′-O-methylation: RNA-binding activity of the tudor domain and translational regulation at synapses. Nucleic Acids Res. 2012, 40, 4086–4096. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.; Daley, D.T.A.; Luscombe, N.M.; Berman, H.M.; Thornton, J.M. Protein–RNA interactions: A structural analysis. Nucleic Acids Res. 2001, 29, 943–954. [Google Scholar] [CrossRef]
- Treger, M.; Westhof, E. Statistical analysis of atomic contacts at RNA–protein interfaces. J. Mol. Recognit. 2001, 14, 199–214. [Google Scholar] [CrossRef] [PubMed]
- Douthwaite, S.; Kirpekar, F. Identifying modifications in RNA by MALDI mass spectrometry. Methods Enzymol. 2007, 425, 3–20. [Google Scholar]
- Grosjean, H.; Keith, G.; Droogmans, L. Detection and quantification of modified nucleotides in RNA using thin-layer chromatography. Methods Mol. Biol. 2004, 265, 357–391. [Google Scholar] [PubMed]
- Grosjean, H.; Droogmans, L.; Roovers, M.; Keith, G. Detection of enzymatic activity of transfer RNA modification enzymes using radiolabeled tRNA substrates. Methods Enzymol. 2007, 425, 55–101. [Google Scholar] [PubMed]
- Maden, B.E. Mapping 2′-O-methyl groups in ribosomal RNA. Methods 2001, 25, 374–382. [Google Scholar] [CrossRef]
- Deryusheva, S.; Gall, J.G.; Matera, A.G. Small Cajal Body–specific RNAs of Drosophila Function in the Absence of Cajal Bodies. MBoC 2009, 20, 5250–5259. [Google Scholar] [CrossRef]
- Belin, S.; Beghin, A.; Solano-Gonzàlez, E.; Bezin, L.; Brunet-Manquat, S.; Textoris, J.; Prats, A.-C.; Mertani, H.C.; Dumontet, C.; Diaz, J.-J. Dysregulation of ribosome biogenesis and translational capacity is associated with tumor progression of human breast cancer cells. PLoS ONE 2009, 4, e7147. [Google Scholar] [CrossRef]
- Yu, Y.T.; Shu, M.D.; Steitz, J.A. A new method for detecting sites of 2′-O-methylation in RNA molecules. RNA 1997, 3, 324–331. [Google Scholar]
- Saikia, M.; Dai, Q.; Decatur, W.A.; Fournier, M.J.; Piccirilli, J.A.; Pan, T. A systematic, ligation-based approach to study RNA modifications. RNA 2006, 12, 2025–2033. [Google Scholar] [CrossRef] [PubMed]
- Buchhaupt, M.; Peifer, C.; Entian, K.-D. Analysis of 2′-O-methylated nucleosides and pseudouridines in ribosomal RNAs using DNAzymes. Anal. Biochem. 2007, 361, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.-W.; Shao, P.; Diao, L.-T.; Zhou, H.; Yu, C.-H.; Qu, L.-H. RTL-P: A sensitive approach for detecting sites of 2′-O-methylation in RNA molecules. Nucleic Acids Res. 2012, 40, e157. [Google Scholar] [CrossRef]
- Marchand, V.; Blanloeil-Oillo, F.; Helm, M.; Motorin, Y. Illumina-based RiboMethSeq approach for mapping of 2′-O-Me residues in RNA. Nucleic Acids Res. 2016, 44, e135. [Google Scholar] [CrossRef]
- Marchand, V.; Ayadi, L.; El Hajj, A.; Blanloeil-Oillo, F.; Helm, M.; Motorin, Y. High-Throughput Mapping of 2′-O-Me Residues in RNA Using Next-Generation Sequencing (Illumina RiboMethSeq Protocol). In RNA Methylation: Methods and Protocols; Lusser, A., Ed.; Springer New York: New York, NY, USA, 2017; pp. 171–187. ISBN 9781493968077. [Google Scholar]
- Marchand, V.; Pichot, F.; Thüring, K.; Ayadi, L.; Freund, I.; Dalpke, A.; Helm, M.; Motorin, Y. Next-Generation Sequencing-Based RiboMethSeq Protocol for Analysis of tRNA 2′-O-Methylation. Biomolecules 2017, 7, 13. [Google Scholar] [CrossRef] [PubMed]
- Ayadi, L.; Motorin, Y.; Marchand, V. Quantification of 2′-O-Me Residues in RNA Using Next-Generation Sequencing (Illumina RiboMethSeq Protocol). In RNA Detection: Methods and Protocols; Gaspar, I., Ed.; Springer New York: New York, NY, USA, 2018; ISBN 9781493972135. [Google Scholar]
- Wang, N.; Qu, S.; Sun, W.; Zeng, Z.; Liang, H.; Zhang, C.-Y.; Chen, X.; Zen, K. Direct quantification of 3′ terminal 2′-O-methylation of small RNAs by RT-qPCR. RNA 2018, 24, 1520–1529. [Google Scholar] [CrossRef]
- Zhu, Y.; Pirnie, S.P.; Carmichael, G.G. High-throughput and site-specific identification of 2′-O-methylation sites using ribose oxidation sequencing (RibOxi-seq). RNA 2017, 23, 1303–1314. [Google Scholar] [CrossRef]
- Motorin, Y.; Marchand, V. Detection and Analysis of RNA Ribose 2′-O-Methylations: Challenges and Solutions. Genes 2018, 9, 642. [Google Scholar] [CrossRef]
- Towns, W.L.; Begley, T.J. Transfer RNA methytransferases and their corresponding modifications in budding yeast and humans: Activities, predications, and potential roles in human health. DNA Cell Biol. 2012, 31, 434–454. [Google Scholar] [CrossRef]
- Torres, A.G.; Batlle, E.; Ribas de Pouplana, L. Role of tRNA modifications in human diseases. Trends Mol. Med. 2014, 20, 306–314. [Google Scholar] [CrossRef]
- Chou, H.-J.; Donnard, E.; Gustafsson, H.T.; Garber, M.; Rando, O.J. Transcriptome-wide Analysis of Roles for tRNA Modifications in Translational Regulation. Mol. Cell 2017, 68, 978–992. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, S.S.; Guenther, R.H.; Ansari, G.; Malkiewicz, A.; Sochacka, E.; Agris, P.F. Role of modified nucleosides of yeast tRNA(Phe) in ribosomal binding. Cell Biochem. Biophys. 2000, 33, 241–252. [Google Scholar] [CrossRef]
- Pintard, L.; Lecointe, F.; Bujnicki, J.M.; Bonnerot, C.; Grosjean, H.; Lapeyre, B. Trm7p catalyses the formation of two 2′-O-methylriboses in yeast tRNA anticodon loop. EMBO J. 2002, 21, 1811–1820. [Google Scholar] [CrossRef] [PubMed]
- Benítez-Páez, A.; Villarroya, M.; Douthwaite, S.; Gabaldón, T.; Armengod, M.-E. YibK is the 2′-O-methyltransferase TrmL that modifies the wobble nucleotide in Escherichia coli tRNA(Leu) isoacceptors. RNA 2010, 16, 2131–2143. [Google Scholar] [CrossRef]
- Bügl, H.; Fauman, E.B.; Staker, B.L.; Zheng, F.; Kushner, S.R.; Saper, M.A.; Bardwell, J.C.; Jakob, U. RNA methylation under heat shock control. Mol. Cell 2000, 6, 349–360. [Google Scholar] [CrossRef]
- Feder, M.; Pas, J.; Wyrwicz, L.S.; Bujnicki, J.M. Molecular phylogenetics of the RrmJ/fibrillarin superfamily of ribose 2′-O-methyltransferases. Gene 2003, 302, 129–138. [Google Scholar] [CrossRef]
- Guy, M.P.; Phizicky, E.M. Conservation of an intricate circuit for crucial modifications of the tRNAPhe anticodon loop in eukaryotes. RNA 2015, 21, 61–74. [Google Scholar] [CrossRef]
- Guy, M.P.; Podyma, B.M.; Preston, M.A.; Shaheen, H.H.; Krivos, K.L.; Limbach, P.A.; Hopper, A.K.; Phizicky, E.M. Yeast Trm7 interacts with distinct proteins for critical modifications of the tRNAPhe anticodon loop. RNA 2012, 18, 1921–1933. [Google Scholar] [CrossRef]
- Noma, A.; Kirino, Y.; Ikeuchi, Y.; Suzuki, T. Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J. 2006, 25, 2142–2154. [Google Scholar] [CrossRef]
- Pintard, L.; Bujnicki, J.M.; Lapeyre, B.; Bonnerot, C. MRM2 encodes a novel yeast mitochondrial 21S rRNA methyltransferase. EMBO J. 2002, 21, 1139–1147. [Google Scholar] [CrossRef]
- Ching, Y.-P.; Zhou, H.-J.; Yuan, J.-G.; Qiang, B.-Q.; Kung Hf, H.-F.; Jin, D.-Y. Identification and characterization of FTSJ2, a novel human nucleolar protein homologous to bacterial ribosomal RNA methyltransferase. Genomics 2002, 79, 2–6. [Google Scholar] [CrossRef] [PubMed]
- Pintard, L.; Kressler, D.; Lapeyre, B. Spb1p Is a Yeast Nucleolar Protein Associated with Nop1p and Nop58p That Is Able to BindS-Adenosyl-l-Methionine In Vitro. Mol. Cell. Biol. 2000, 20, 1370–1381. [Google Scholar] [CrossRef] [PubMed]
- Simabuco, F.M.; Morello, L.G.; Aragão, A.Z.B.; Paes Leme, A.F.; Zanchin, N.I.T. Proteomic characterization of the human FTSJ3 preribosomal complexes. J. Proteome Res. 2012, 11, 3112–3126. [Google Scholar] [CrossRef] [PubMed]
- Maulik, P.K.; Mascarenhas, M.N.; Mathers, C.D.; Dua, T.; Saxena, S. Prevalence of intellectual disability: A meta-analysis of population-based studies. Res. Dev. Disabil. 2011, 32, 419–436. [Google Scholar] [CrossRef] [PubMed]
- Willems, P.; Vits, L.; Buntinx, I.; Raeymaekers, P.; Van Broeckhoven, C.; Ceulemans, B. Localization of a gene responsible for nonspecific mental retardation (MRX9) to the pericentromeric region of the X chromosome. Genomics 1993, 18, 290–294. [Google Scholar] [CrossRef]
- Hamel, B.C.J.; Smits, A.P.T.; van den Helm, B.; Smeets, D.F.; Knoers, N.V.; van Roosmalen, T.; Thoonen, G.H.J.; Assman-Hulsmans, C.F.; Ropers, H.-H.; Mariman, E.C.M.; et al. Four families (MRX43, MRX44, MRX45, MRX52) with nonspecific X-linked mental retardation: Clinical and psychometric data and results of linkage analysis. Am. J. Med. Genet. 1999, 85, 290–304. [Google Scholar] [CrossRef]
- Freude, K.; Hoffmann, K.; Jensen, L.-R.; Delatycki, M.B.; des Portes, V.; Moser, B.; Hamel, B.; van Bokhoven, H.; Moraine, C.; Fryns, J.-P.; et al. Mutations in the FTSJ1 Gene Coding for a Novel S-Adenosylmethionine–Binding Protein Cause Nonsyndromic X-Linked Mental Retardation. Am. J. Hum. Genet. 2004, 75, 305–309. [Google Scholar] [CrossRef]
- Ramser, J.; Winnepenninckx, B.; Lenski, C.; Errijgers, V.; Platzer, M.; Schwartz, C.E.; Meindl, A.; Kooy, R.F. A splice site mutation in the methyltransferase gene FTSJ1 in Xp11.23 is associated with non-syndromic mental retardation in a large Belgian family (MRX9). J. Med. Genet. 2004, 41, 679–683. [Google Scholar] [CrossRef]
- Froyen, G.; Bauters, M.; Boyle, J.; Van Esch, H.; Govaerts, K.; van Bokhoven, H.; Ropers, H.-H.; Moraine, C.; Chelly, J.; Fryns, J.-P.; et al. Loss of SLC38A5 and FTSJ1 at Xp11.23 in three brothers with non-syndromic mental retardation due to a microdeletion in an unstable genomic region. Hum. Genet. 2007, 121, 539–547. [Google Scholar] [CrossRef]
- Dai, L.; Xing, L.; Gong, P.; Zhang, K.; Gao, X.; Zheng, Z.; Zhou, J.; Guo, Y.; Guo, S.; Zhang, F. Positive association of the FTSJ1 gene polymorphisms with nonsyndromic X-linked mental retardation in young Chinese male subjects. J. Hum. Genet. 2008, 53, 592–597. [Google Scholar] [CrossRef]
- Takano, K.; Nakagawa, E.; Inoue, K.; Kamada, F.; Kure, S.; Goto, Y.-I.; Consortium, J.M.R. A loss-of-function mutation in the FTSJ1 gene causes nonsyndromic X-linked mental retardation in a japanese family. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2008, 147, 479–484. [Google Scholar] [CrossRef]
- Guy, M.P.; Shaw, M.; Weiner, C.L.; Hobson, L.; Stark, Z.; Rose, K.; Kalscheuer, V.M.; Gecz, J.; Phizicky, E.M. Defects in tRNA Anticodon Loop 2′-O-Methylation Are Implicated in Nonsyndromic X-Linked Intellectual Disability due to Mutations in FTSJ1. Hum. Mutat. 2015, 36, 1176–1187. [Google Scholar] [CrossRef] [PubMed]
- Angelova, M.T.; Dimitrova, D.G.; Dinges, N.; Lence, T.; Worpenberg, L.; Carré, C.; Roignant, J.-Y. The Emerging Field of Epitranscriptomics in Neurodevelopmental and Neuronal Disorders. Front. Bioeng. Biotechnol. 2018, 6, 46. [Google Scholar] [CrossRef]
- FTSJ1 single nucleotide variant reported by Ambry Genetics at Clinvar NCBI. Available online: https://www.ncbi.nlm.nih.gov/clinvar/variation/208659/ (accessed on 27 July 2017).
- Gong, P.; Li, J.; Dai, L.; Zhang, K.; Zheng, Z.; Gao, X.; Zhang, F. Genetic variations in FTSJ1 influence cognitive ability in young males in the Chinese Han population. J. Neurogenet. 2008, 22, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Chishty, M.; Reichel, A.; Abbott, N.J.; Begley, D.J. S-adenosylmethionine is substrate for carrier mediated transport at the blood–brain barrier in vitro. Brain Res. 2002, 942, 46–50. [Google Scholar] [CrossRef]
- Han, L.; Guy, M.P.; Kon, Y.; Phizicky, E.M. Lack of 2′-O-methylation in the tRNA anticodon loop of two phylogenetically distant yeast species activates the general amino acid control pathway. PLoS Genet. 2018, 14, e1007288. [Google Scholar] [CrossRef] [PubMed]
- Honda, S.; Hayashi, S.; Imoto, I.; Toyama, J.; Okazawa, H.; Nakagawa, E.; Goto, Y.-I.; Inazawa, J. Copy-number variations on the X chromosome in Japanese patients with mental retardation detected by array-based comparative genomic hybridization analysis. J. Hum. Genet. 2010, 55, 590–599. [Google Scholar] [CrossRef]
- Giorda, R.; Bonaglia, M.C.; Beri, S.; Fichera, M.; Novara, F.; Magini, P.; Urquhart, J.; Sharkey, F.H.; Zucca, C.; Grasso, R.; et al. Complex Segmental Duplications Mediate a Recurrent dup(X)(p11.22-p11.23) Associated with Mental Retardation, Speech Delay, and EEG Anomalies in Males and Females. Am. J. Hum. Genet. 2009, 85, 419. [Google Scholar] [CrossRef]
- Leschziner, G.D.; Coffey, A.J.; Andrew, T.; Gregorio, S.P.; Dias-Neto, E.; Calafato, M.; Bentley, D.R.; Kinton, L.; Sander, J.W.; Johnson, M.R. Q8IYL2 is a candidate gene for the familial epilepsy syndrome of Partial Epilepsy with Pericentral Spikes (PEPS). Epilepsy Res. 2011, 96, 109–115. [Google Scholar] [CrossRef]
- Polikanov, Y.S.; Melnikov, S.V.; Söll, D.; Steitz, T.A. Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat. Struct. Mol. Biol. 2015, 22, 342–344. [Google Scholar] [CrossRef]
- Monaco, P.L.; Marcel, V.; Diaz, J.-J.; Catez, F. 2′-O-Methylation of Ribosomal RNA: Towards an Epitranscriptomic Control of Translation? Biomolecules 2018, 8, 106. [Google Scholar] [CrossRef]
- Sloan, K.E.; Warda, A.S.; Sharma, S.; Entian, K.-D.; Lafontaine, D.L.J.; Bohnsack, M.T. Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2017, 14, 1138–1152. [Google Scholar] [CrossRef] [PubMed]
- Erales, J.; Marchand, V.; Panthu, B.; Gillot, S.; Belin, S.; Ghayad, S.E.; Garcia, M.; Laforêts, F.; Marcel, V.; Baudin-Baillieu, A.; et al. Evidence for rRNA 2′-O-methylation plasticity: Control of intrinsic translational capabilities of human ribosomes. Proc. Natl. Acad. Sci. USA 2017, 114, 12934–12939. [Google Scholar] [CrossRef]
- Cavaillé, J.; Nicoloso, M.; Bachellerie, J.P. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature 1996, 383, 732–735. [Google Scholar] [CrossRef] [PubMed]
- Bachellerie, J.P.; Cavaillé, J.; Hüttenhofer, A. The expanding snoRNA world. Biochimie 2002, 84, 775–790. [Google Scholar] [CrossRef]
- Reichow, S.L.; Hamma, T.; Ferré-D’Amaré, A.R.; Varani, G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007, 35, 1452–1464. [Google Scholar] [CrossRef] [PubMed]
- Henras, A.K.; Soudet, J.; Gérus, M.; Lebaron, S.; Caizergues-Ferrer, M.; Mougin, A.; Henry, Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 2008, 65, 2334–2359. [Google Scholar] [CrossRef]
- Higa-Nakamine, S.; Suzuki, T.; Uechi, T.; Chakraborty, A.; Nakajima, Y.; Nakamura, M.; Hirano, N.; Suzuki, T.; Kenmochi, N. Loss of ribosomal RNA modification causes developmental defects in zebrafish. Nucleic Acids Res. 2012, 40, 391–398. [Google Scholar] [CrossRef]
- Sridhar, P.; Gan, H.H.; Schlick, T. A computational screen for C/D box snoRNAs in the human genomic region associated with Prader-Willi and Angelman syndromes. J. Biomed. Sci. 2008, 15, 697–705. [Google Scholar] [CrossRef]
- Doe, C.M.; Relkovic, D.; Garfield, A.S.; Dalley, J.W.; Theobald, D.E.H.; Humby, T.; Wilkinson, L.S.; Isles, A.R. Loss of the imprinted snoRNA mbii-52 leads to increased 5htr2c pre-RNA editing and altered 5HT2CR-mediated behaviour. Hum. Mol. Genet. 2009, 18, 2140–2148. [Google Scholar] [CrossRef]
- Cavaillé, J. Box C/D small nucleolar RNA genes and the Prader-Willi syndrome: A complex interplay. Wiley Interdiscip. Rev. RNA 2017, 8, e1417. [Google Scholar] [CrossRef] [PubMed]
- Peters, J. Prader-Willi and snoRNAs. Nat. Genet. 2008, 40, 688–689. [Google Scholar] [CrossRef] [PubMed]
- Belmonte, M.K.; Cook, E.H., Jr; Anderson, G.M.; Rubenstein, J.L.R.; Greenough, W.T.; Beckel-Mitchener, A.; Courchesne, E.; Boulanger, L.M.; Powell, S.B.; Levitt, P.R.; et al. Autism as a disorder of neural information processing: Directions for research and targets for therapy. Mol. Psychiatry 2004, 9, 646. [Google Scholar] [CrossRef] [PubMed]
- Bolton, P.F.; Veltman, M.W.M.; Weisblatt, E.; Holmes, J.R.; Thomas, N.S.; Youings, S.A.; Thompson, R.J.; Roberts, S.E.; Dennis, N.R.; Browne, C.E.; et al. Chromosome 15q11-13 abnormalities and other medical conditions in individuals with autism spectrum disorders. Psychiatr. Genet. 2004, 14, 131–137. [Google Scholar] [CrossRef]
- Cook, E.H., Jr; Scherer, S.W. Copy-number variations associated with neuropsychiatric conditions. Nature 2008, 455, 919–923. [Google Scholar] [CrossRef] [PubMed]
- Cavaillé, J.; Buiting, K.; Kiefmann, M.; Lalande, M.; Brannan, C.I.; Horsthemke, B.; Bachellerie, J.-P.; Brosius, J.; Hüttenhofer, A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl. Acad. Sci. USA 2000, 97, 14311–14316. [Google Scholar] [CrossRef] [PubMed]
- Bortolin-Cavaillé, M.-L.; Cavaillé, J. The SNORD115 (H/MBII-52) and SNORD116 (H/MBII-85) gene clusters at the imprinted Prader-Willi locus generate canonical box C/D snoRNAs. Nucleic Acids Res. 2012, 40, 6800–6807. [Google Scholar] [CrossRef]
- Vitali, P.; Basyuk, E.; Le Meur, E.; Bertrand, E.; Muscatelli, F.; Cavaillé, J.; Huttenhofer, A. ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. J. Cell Biol. 2005, 169, 745–753. [Google Scholar] [CrossRef]
- Kishore, S.; Stamm, S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science 2006, 311, 230–232. [Google Scholar] [CrossRef]
- Sahoo, T.; del Gaudio, D.; German, J.R.; Shinawi, M.; Peters, S.U.; Person, R.E.; Garnica, A.; Cheung, S.W.; Beaudet, A.L. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat. Genet. 2008, 40, 719–721. [Google Scholar] [CrossRef]
- Duker, A.L.; Ballif, B.C.; Bawle, E.V.; Person, R.E.; Mahadevan, S.; Alliman, S.; Thompson, R.; Traylor, R.; Bejjani, B.A.; Shaffer, L.G.; et al. Paternally inherited microdeletion at 15q11.2 confirms a significant role for the SNORD116 C/D box snoRNA cluster in Prader-Willi syndrome. Eur. J. Hum. Genet. 2010, 18, 1196–1201. [Google Scholar] [CrossRef] [PubMed]
- Tollervey, D.; Lehtonen, H.; Jansen, R.; Kern, H.; Hurt, E.C. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell 1993, 72, 443–457. [Google Scholar] [CrossRef]
- Jansen, R.P.; Hurt, E.C.; Kern, H.; Lehtonen, H.; Carmo-Fonseca, M.; Lapeyre, B.; Tollervey, D. Evolutionary conservation of the human nucleolar protein fibrillarin and its functional expression in yeast. J. Cell Biol. 1991, 113, 715–729. [Google Scholar] [CrossRef] [PubMed]
- Amiri, K.A. Fibrillarin-like proteins occur in the domain Archaea. J. Bacteriol. 1994, 176, 2124–2127. [Google Scholar] [CrossRef] [PubMed]
- Narcisi, E.M.; Glover, C.V.C.; Fechheimer, M. Fibrillarin, A Conserved Pre-ribosomal RNA Processing Protein of Giardia. J. Eukaryot. Microbiol. 1998, 45, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Corona, U.; Sobol, M.; Rodriguez-Zapata, L.C.; Hozak, P.; Castano, E. Fibrillarin from Archaea to human. Biol. Cell 2015, 107, 159–174. [Google Scholar] [CrossRef] [PubMed]
- Tessarz, P.; Santos-Rosa, H.; Robson, S.C.; Sylvestersen, K.B.; Nelson, C.J.; Nielsen, M.L.; Kouzarides, T. Glutamine methylation in histone H2A is an RNA-polymerase-I-dedicated modification. Nature 2014, 505, 564–568. [Google Scholar] [CrossRef]
- Loza-Muller, L.; Rodríguez-Corona, U.; Sobol, M.; Rodríguez-Zapata, L.C.; Hozak, P.; Castano, E. Fibrillarin methylates H2A in RNA polymerase I trans-active promoters in Brassica oleracea. Front. Plant Sci. 2015, 6, 976. [Google Scholar] [CrossRef]
- Newton, K.; Petfalski, E.; Tollervey, D.; Cáceres, J.F. Fibrillarin Is Essential for Early Development and Required for Accumulation of an Intron-Encoded Small Nucleolar RNA in the Mouse. Mol. Cell. Biol. 2003, 23, 8519–8527. [Google Scholar] [CrossRef]
- Bouffard, S.; Dambroise, E.; Brombin, A.; Lempereur, S.; Hatin, I.; Simion, M.; Corre, R.; Bourrat, F.; Joly, J.-S.; Jamen, F. Fibrillarin is essential for S-phase progression and neuronal differentiation in zebrafish dorsal midbrain and retina. Dev. Biol. 2018, 437, 1–16. [Google Scholar] [CrossRef]
- Watanabe-Susaki, K.; Takada, H.; Enomoto, K.; Miwata, K.; Ishimine, H.; Intoh, A.; Ohtaka, M.; Nakanishi, M.; Sugino, H.; Asashima, M.; et al. Biosynthesis of ribosomal RNA in nucleoli regulates pluripotency and differentiation ability of pluripotent stem cells. Stem Cells 2014, 32, 3099–3111. [Google Scholar] [CrossRef] [PubMed]
- Shubina, M.Y.; Musinova, Y.R.; Sheval, E.V. Proliferation, cancer, and aging-novel functions of the nucleolar methyltransferase fibrillarin? Cell Biol. Int. 2018, 42, 1463–1466. [Google Scholar] [CrossRef] [PubMed]
- Ruggero, D.; Pandolfi, P.P. Does the ribosome translate cancer? Nat. Rev. Cancer 2003, 3, 179–192. [Google Scholar] [CrossRef]
- White, R.J. RNA polymerases I and III, growth control and cancer. Nat. Rev. Mol. Cell Biol. 2005, 6, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Koh, C.M.; Gurel, B.; Sutcliffe, S.; Aryee, M.J.; Schultz, D.; Iwata, T.; Uemura, M.; Zeller, K.I.; Anele, U.; Zheng, Q.; et al. Alterations in nucleolar structure and gene expression programs in prostatic neoplasia are driven by the MYC oncogene. Am. J. Pathol. 2011, 178, 1824–1834. [Google Scholar] [CrossRef]
- Marcel, V.; Ghayad, S.E.; Belin, S.; Therizols, G.; Morel, A.-P.; Solano-Gonzàlez, E.; Vendrell, J.A.; Hacot, S.; Mertani, H.C.; Albaret, M.A.; et al. p53 acts as a safeguard of translational control by regulating fibrillarin and rRNA methylation in cancer. Cancer Cell 2013, 24, 318–330. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Xu, T.; Ganapathy, S.; Shadfan, M.; Long, M.; Huang, T.H.-M.; Thompson, I.; Yuan, Z.-M. Elevated snoRNA biogenesis is essential in breast cancer. Oncogene 2014, 33, 1348–1358. [Google Scholar] [CrossRef]
- Derenzini, M.; Trerè, D.; Pession, A.; Montanaro, L.; Sirri, V.; Ochs, R.L. Nucleolar function and size in cancer cells. Am. J. Pathol. 1998, 152, 1291–1297. [Google Scholar]
- Coller, H.A.; Grandori, C.; Tamayo, P.; Colbert, T.; Lander, E.S.; Eisenman, R.N.; Golub, T.R. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc. Natl. Acad. Sci. USA 2000, 97, 3260–3265. [Google Scholar] [CrossRef]
- Schlosser, I.; Hölzel, M.; Mürnseer, M.; Burtscher, H.; Weidle, U.H.; Eick, D. A role for c-Myc in the regulation of ribosomal RNA processing. Nucleic Acids Res. 2003, 31, 6148–6156. [Google Scholar] [CrossRef]
- Imai, H.; Ochs, R.L.; Kiyosawa, K.; Furuta, S.; Nakamura, R.M.; Tan, E.M. Nucleolar antigens and autoantibodies in hepatocellular carcinoma and other malignancies. Am. J. Pathol. 1992, 140, 859–870. [Google Scholar] [PubMed]
- Imai, H.; Kiyosawa, K.; Chan, E.K.; Tan, E.M. Autoantibodies in viral hepatitis-related hepatocellular carcinoma. Intervirology 1993, 35, 73–85. [Google Scholar] [CrossRef]
- Covini, G.; von Mühlen, C.A.; Pacchetti, S.; Colombo, M.; Chan, E.K.; Tan, E.M. Diversity of antinuclear antibody responses in hepatocellular carcinoma. J. Hepatol. 1997, 26, 1255–1265. [Google Scholar] [CrossRef]
- El Hassouni, B.; Sarkisjan, D.; Vos, J.C.; Giovannetti, E.; Peters, G.J. Targeting the ribosome biogenesis key molecule fibrillarin to avoid chemoresistance. Curr. Med. Chem. 2018. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Tan, F.K.; Xiong, M.; Milewicz, D.M.; Feghali, C.A.; Fritzler, M.J.; Reveille, J.D.; Arnett, F.C. Systemic Sclerosis (Scleroderma): Specific Autoantigen Genes Are Selectively Overexpressed in Scleroderma Fibroblasts. J. Immunol. 2001, 167, 7126–7133. [Google Scholar] [CrossRef] [PubMed]
- Lischwe, M.A.; Ochs, R.L.; Reddy, R.; Cook, R.G.; Yeoman, L.C.; Tan, E.M.; Reichlin, M.; Busch, H. Purification and partial characterization of a nucleolar scleroderma antigen (Mr = 34,000; pI, 8.5) rich in NG,NG-dimethylarginine. J. Biol. Chem. 1985, 260, 14304–14310. [Google Scholar] [PubMed]
- Ochs, R.L.; Lischwe, M.A.; Spohn, W.H.; Busch, H. Fibrillarin: A new protein of the nucleolus identified by autoimmune sera. Biol. Cell 1985, 54, 123–133. [Google Scholar] [CrossRef]
- Busch, H.; Busch, R.K.; Black, A.; Chan, P.K.; Chatterjee, A.; Durban, E.; Freeman, J.; Ochs, R.; Reichlin, M.; Tan, E.M. Novel nucleolar antigens in autoimmune disease. J. Rheumatol. Suppl. 1987, 14 (Suppl. 13), 70–77. [Google Scholar]
- Reimer, G.; Steen, V.D.; Penning, C.A.; Medsger, T.A., Jr; Tan, E.M. Correlates between autoantibodies to nucleolar antigens and clinical features in patients with systemic sclerosis (scleroderma). Arthritis Rheum. 1988, 31, 525–532. [Google Scholar] [CrossRef]
- Pollard, K.M.; Reimer, G.; Tan, E.M. Autoantibodies in scleroderma. Clin. Exp. Rheumatol. 1989, 7 (Suppl. 3), S57–S62. [Google Scholar]
- Kühn, G.; Jarzabek-Chorzelska, M.; Blaszczyk, M.; Chorzelski, T.P.; Jablonska, S. [The fibrillarin (Scl-34) autoantibody in systemic scleroderma]. Dermatol. Monatsschr. 1990, 176, 19–26. [Google Scholar]
- Arnett, F.C.; Reveille, J.D.; Goldstein, R.; Pollard, K.M.; Leaird, K.; Smith, E.A.; Leroy, E.C.; Fritzler, M.J. Autoantibodies to fibrillarin in systemic sclerosis (scleroderma). An immunogenetic, serologic, and clinical analysis. Arthritis Rheum. 1996, 39, 1151–1160. [Google Scholar] [CrossRef] [PubMed]
- von Mühlen, C.A.; Tan, E.M. Autoantibodies in the diagnosis of systemic rheumatic diseases. Semin. Arthritis Rheum. 1995, 24, 323–358. [Google Scholar] [CrossRef]
- Boonstra, M.; Mertens, B.J.A.; Bakker, J.A.; Ninaber, M.K.; Ajmone Marsan, N.; van der Helm-van Mil, A.H.M.; Scherer, H.U.; Huizinga, T.W.J.; de Vries-Bouwstra, J.K. To what extent do autoantibodies help to identify high-risk patients in systemic sclerosis? Clin. Exp. Rheumatol. 2018, 36 (Suppl. 113), 109–117. [Google Scholar]
- Ferri, C.; Emdin, M.; Storino, F.A.; Giuggioli, D.; Longombardo, G.; Greco, F.; Fertig, N.; Medsger, T.A. Isolated pulmonary hypertension in diffuse cutaneous systemic sclerosis successfully treated with long-term plasma exchange. Scand. J. Rheumatol. 2000, 29, 198–200. [Google Scholar] [PubMed]
- Kasturi, K.N.; Hatakeyama, A.; Spiera, H.; Bona, C.A. Antifibrillarin autoantibodies present in systemic sclerosis and other connective tissue diseases interact with similar epitopes. J. Exp. Med. 1995, 181, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.-C.; Lee, Y.H. Meta-analysis of gene expression profiles of peripheral blood cells in systemic lupus erythematosus. Cell. Mol. Biol. 2018, 64, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Pellizzoni, L.; Baccon, J.; Charroux, B.; Dreyfuss, G. The survival of motor neurons (SMN) protein interacts with the snoRNP proteins fibrillarin and GAR1. Curr. Biol. 2001, 11, 1079–1088. [Google Scholar] [CrossRef]
- Jones, K.W.; Gorzynski, K.; Hales, C.M.; Fischer, U.; Badbanchi, F.; Terns, R.M.; Terns, M.P. Direct interaction of the spinal muscular atrophy disease protein SMN with the small nucleolar RNA-associated protein fibrillarin. J. Biol. Chem. 2001, 276, 38645–38651. [Google Scholar] [CrossRef]
- Lefebvre, S.; Bürglen, L.; Reboullet, S.; Clermont, O.; Burlet, P.; Viollet, L.; Benichou, B.; Cruaud, C.; Millasseau, P.; Zeviani, M. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 1995, 80, 155–165. [Google Scholar] [CrossRef]
- Lefebvre, S.; Burlet, P.; Liu, Q.; Bertrandy, S.; Clermont, O.; Munnich, A.; Dreyfuss, G.; Melki, J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 1997, 16, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Dreyfuss, G. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 1996, 15, 3555–3565. [Google Scholar] [CrossRef] [PubMed]
- Béchade, C.; Rostaing, P.; Cisterni, C.; Kalisch, R.; La Bella, V.; Pettmann, B.; Triller, A. Subcellular distribution of survival motor neuron (SMN) protein: Possible involvement in nucleocytoplasmic and dendritic transport. Eur. J. Neurosci. 1999, 11, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Young, P.J.; Le, T.T.; Dunckley, M.; Nguyen, T.M.; Burghes, A.H.; Morris, G.E. Nuclear gems and Cajal (coiled) bodies in fetal tissues: Nucleolar distribution of the spinal muscular atrophy protein, SMN. Exp. Cell Res. 2001, 265, 252–261. [Google Scholar] [CrossRef]
- Todd, A.G.; Morse, R.; Shaw, D.J.; Stebbings, H.; Young, P.J. Analysis of SMN-neurite granules: Core Cajal body components are absent from SMN-cytoplasmic complexes. Biochem. Biophys. Res. Commun. 2010, 397, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Pellizzoni, L.; Charroux, B.; Rappsilber, J.; Mann, M.; Dreyfuss, G. A functional interaction between the survival motor neuron complex and RNA polymerase II. J. Cell Biol. 2001, 152, 75–85. [Google Scholar] [CrossRef]
- Calle, E.; Berciano, M.T.; Fernández, R.; Lafarga, M. Activation of the autophagy, c-FOS and ubiquitin expression, and nucleolar alterations in Schwann cells precede demyelination in tellurium-induced neuropathy. Acta Neuropathol. 1999, 97, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Sornjai, W.; Lithanatudom, P.; Erales, J.; Joly, P.; Francina, A.; Hacot, S.; Fucharoen, S.; Svasti, S.; Diaz, J.J.; Mertani, H.C.; et al. Hypermethylation of 28S ribosomal RNA in β-thalassemia trait carriers. Int. J. Biol. Macromol. 2017, 94, 728–734. [Google Scholar] [CrossRef]
- Lai, C.-W.; Chen, H.-L.; Lin, K.-Y.; Liu, F.-C.; Chong, K.-Y.; Cheng, W.T.K.; Chen, C.-M. FTSJ2, a heat shock-inducible mitochondrial protein, suppresses cell invasion and migration. PLoS ONE 2014, 9, e90818. [Google Scholar] [CrossRef]
- Campbell, J.M.; Lockwood, W.W.; Buys, T.P.H.; Chari, R.; Coe, B.P.; Lam, S.; Lam, W.L. Integrative genomic and gene expression analysis of chromosome 7 identified novel oncogene loci in non-small cell lung cancer. Genome 2008, 51, 1032–1039. [Google Scholar] [CrossRef] [PubMed]
- Reddy, R.; Singh, R.; Shimba, S. Methylated cap structures in eukaryotic RNAs: Structure, synthesis and functions. Pharmacol. Ther. 1992, 54, 249–267. [Google Scholar] [CrossRef]
- Kuge, H.; Brownlee, G.G.; Gershon, P.D.; Richter, J.D. Cap ribose methylation of c-mos mRNA stimulates translation and oocyte maturation in Xenopus laevis. Nucleic Acids Res. 1998, 26, 3208–3214. [Google Scholar] [CrossRef] [PubMed]
- Shuman, S. What messenger RNA capping tells us about eukaryotic evolution. Nat. Rev. Mol. Cell Biol. 2002, 3, 619–625. [Google Scholar] [CrossRef]
- Gu, M.; Lima, C.D. Processing the message: Structural insights into capping and decapping mRNA. Curr. Opin. Struct. Biol. 2005, 15, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, A.; Robb, G.B.; Chan, S.-H. mRNA capping: Biological functions and applications. Nucleic Acids Res. 2016, 44, 7511–7526. [Google Scholar] [CrossRef] [PubMed]
- Inesta-Vaquera, F.; Chaugule, V.K.; Galloway, A.; Chandler, L.; Rojas-Fernandez, A.; Weidlich, S.; Peggie, M.; Cowling, V.H. DHX15 regulates CMTR1-dependent gene expression and cell proliferation. Life Sci Alliance 2018, 1, e201800092. [Google Scholar] [CrossRef] [PubMed]
- Aregger, M.; Cowling, V.H. Regulation of mRNA capping in the cell cycle. RNA Biol. 2017, 14, 11–14. [Google Scholar] [CrossRef]
- Bélanger, F.; Stepinski, J.; Darzynkiewicz, E.; Pelletier, J. Characterization of hMTr1, a human Cap1 2′-O-ribose methyltransferase. J. Biol. Chem. 2010, 285, 33037–33044. [Google Scholar] [CrossRef]
- Perry, R.P.; Kelley, D.E. Kinetics of formation of 5′ terminal caps in mRNA. Cell 1976, 8, 433–442. [Google Scholar] [CrossRef]
- Werner, M.; Purta, E.; Kaminska, K.H.; Cymerman, I.A.; Campbell, D.A.; Mittra, B.; Zamudio, J.R.; Sturm, N.R.; Jaworski, J.; Bujnicki, J.M. 2′-O-ribose methylation of cap2 in human: Function and evolution in a horizontally mobile family. Nucleic Acids Res. 2011, 39, 4756–4768. [Google Scholar] [CrossRef]
- Banerjee, A.K. 5′-terminal cap structure in eucaryotic messenger ribonucleic acids. Microbiol. Rev. 1980, 44, 175–205. [Google Scholar] [PubMed]
- Smietanski, M.; Werner, M.; Purta, E.; Kaminska, K.H.; Stepinski, J.; Darzynkiewicz, E.; Nowotny, M.; Bujnicki, J.M. Structural analysis of human 2′-O-ribose methyltransferases involved in mRNA cap structure formation. Nat. Commun. 2014, 5, 3004. [Google Scholar] [CrossRef]
- Dahlin, A.; Denny, J.; Roden, D.M.; Brilliant, M.H.; Ingram, C.; Kitchner, T.E.; Linneman, J.G.; Shaffer, C.M.; Weeke, P.; Xu, H.; et al. CMTR1 is associated with increased asthma exacerbations in patients taking inhaled corticosteroids. Immun. Inflamm. Dis. 2015, 3, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Daffis, S.; Szretter, K.J.; Schriewer, J.; Li, J.; Youn, S.; Errett, J.; Lin, T.-Y.; Schneller, S.; Zust, R.; Dong, H.; et al. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 2010, 468, 452–456. [Google Scholar] [CrossRef] [PubMed]
- Züst, R.; Cervantes-Barragan, L.; Habjan, M.; Maier, R.; Neuman, B.W.; Ziebuhr, J.; Szretter, K.J.; Baker, S.C.; Barchet, W.; Diamond, M.S.; et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011, 12, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Sweeney, T.R.; Skabkin, M.A.; Skabkina, O.V.; Hellen, C.U.T.; Pestova, T.V. Inhibition of translation by IFIT family members is determined by their ability to interact selectively with the 5′-terminal regions of cap0-, cap1- and 5′ppp- mRNAs. Nucleic Acids Res. 2014, 42, 3228–3245. [Google Scholar] [CrossRef] [PubMed]
- Diamond, M.S. IFIT1: A dual sensor and effector molecule that detects non-2′-O methylated viral RNA and inhibits its translation. Cytokine Growth Factor Rev. 2014, 25, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Abbas, Y.M.; Laudenbach, B.T.; Martínez-Montero, S.; Cencic, R.; Habjan, M.; Pichlmair, A.; Damha, M.J.; Pelletier, J.; Nagar, B. Structure of human IFIT1 with capped RNA reveals adaptable mRNA binding and mechanisms for sensing N1 and N2 ribose 2′-O methylations. Proc. Natl. Acad. Sci. USA 2017, 114, E2106–E2115. [Google Scholar] [CrossRef]
- Jackson, D.J.; Sykes, A.; Mallia, P.; Johnston, S.L. Asthma exacerbations: Origin, effect, and prevention. J. Allergy Clin. Immunol. 2011, 128, 1165–1174. [Google Scholar] [CrossRef]
- Geiss, G.K.; Carter, V.S.; He, Y.; Kwieciszewski, B.K.; Holzman, T.; Korth, M.J.; Lazaro, C.A.; Fausto, N.; Bumgarner, R.E.; Katze, M.G. Gene expression profiling of the cellular transcriptional network regulated by alpha/beta interferon and its partial attenuation by the hepatitis C virus nonstructural 5A protein. J. Virol. 2003, 77, 6367–6375. [Google Scholar] [CrossRef]
- Haline-Vaz, T.; Silva, T.C.L.; Zanchin, N.I.T. The human interferon-regulated ISG95 protein interacts with RNA polymerase II and shows methyltransferase activity. Biochem. Biophys. Res. Commun. 2008, 372, 719–724. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.C. Viruses in asthma exacerbations. Curr. Opin. Pulm. Med. 2005, 11, 21–26. [Google Scholar] [CrossRef]
- World Health Organization. Bulletin of WHO Vol. 90 No. 02 2012; World Health Organization: Geneva, Switzerland, 2012; ISBN 9789240689718. [Google Scholar]
- Ardura-Fabregat, A.; Boddeke, E.W.G.M.; Boza-Serrano, A.; Brioschi, S.; Castro-Gomez, S.; Ceyzériat, K.; Dansokho, C.; Dierkes, T.; Gelders, G.; Heneka, M.T.; et al. Targeting Neuroinflammation to Treat Alzheimer’s Disease. CNS Drugs 2017, 31, 1057–1082. [Google Scholar] [CrossRef] [PubMed]
- Boza-Serrano, A.; Yang, Y.; Paulus, A.; Deierborg, T. Innate immune alterations are elicited in microglial cells before plaque deposition in the Alzheimer’s disease mouse model 5xFAD. Sci. Rep. 2018, 8, 1550. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Tian, T.; Wei, Z.; Peck, K.N.; Shih, N.; Chalian, A.A.; O’Malley, B.W.; Weinstein, G.S.; Feldman, M.D.; Alwine, J.; et al. Microbial Signatures Associated with Oropharyngeal and Oral Squamous Cell Carcinomas. Sci. Rep. 2017, 7, 4036. [Google Scholar] [CrossRef] [PubMed]
- Vicente, C.; Schwab, C.; Broux, M.; Geerdens, E.; Degryse, S.; Demeyer, S.; Lahortiga, I.; Elliott, A.; Chilton, L.; La Starza, R.; et al. Targeted sequencing identifies associations between IL7R-JAK mutations and epigenetic modulators in T-cell acute lymphoblastic leukemia. Haematologica 2015, 100, 1301–1310. [Google Scholar] [CrossRef]
- Degryse, S.; Cools, J. JAK kinase inhibitors for the treatment of acute lymphoblastic leukemia. J. Hematol. Oncol. 2015, 8, 91. [Google Scholar] [CrossRef] [PubMed]
- Degryse, S.; de Bock, C.E.; Demeyer, S.; Govaerts, I.; Bornschein, S.; Verbeke, D.; Jacobs, K.; Binos, S.; Skerrett-Byrne, D.A.; Murray, H.C.; et al. Mutant JAK3 phosphoproteomic profiling predicts synergism between JAK3 inhibitors and MEK/BCL2 inhibitors for the treatment of T-cell acute lymphoblastic leukemia. Leukemia 2018, 32, 788–800. [Google Scholar] [CrossRef]
- Jemal, A.; Center, M.M.; DeSantis, C.; Ward, E.M. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol. Biomark. Prev. 2010, 19, 1893–1907. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2015. CA Cancer J. Clin. 2015, 65, 5–29. [Google Scholar] [CrossRef]
- Devarakonda, S.; Morgensztern, D.; Govindan, R. Genomic alterations in lung adenocarcinoma. Lancet Oncol. 2015, 16, e342–e351. [Google Scholar] [CrossRef]
- Soda, M.; Choi, Y.L.; Enomoto, M.; Takada, S.; Yamashita, Y.; Ishikawa, S.; Fujiwara, S.-I.; Watanabe, H.; Kurashina, K.; Hatanaka, H.; et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007, 448, 561–566. [Google Scholar] [CrossRef]
- Wen, M.; Wang, X.; Sun, Y.; Xia, J.; Fan, L.; Xing, H.; Zhang, Z.; Li, X. Detection of EML4-ALK fusion gene and features associated with EGFR mutations in Chinese patients with non-small-cell lung cancer. Onco Targets Ther. 2016, 9, 1989–1995. [Google Scholar] [CrossRef]
- Du, X.; Shao, Y.; Qin, H.-F.; Tai, Y.-H.; Gao, H.-J. ALK-rearrangement in non-small-cell lung cancer (NSCLC). Thorac. Cancer 2018, 9, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Hoskins, A.A.; Moore, M.J. The spliceosome: A flexible, reversible macromolecular machine. Trends Biochem. Sci. 2012, 37, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Bohnsack, M.T.; Sloan, K.E. Modifications in small nuclear RNAs and their roles in spliceosome assembly and function. Biol. Chem. 2018, 399, 1265–1276. [Google Scholar] [CrossRef]
- Karijolich, J.; Yu, Y.-T. Spliceosomal snRNA modifications and their function. RNA Biol. 2010, 7, 192–204. [Google Scholar] [CrossRef]
- Dönmez, G.; Hartmuth, K.; Lührmann, R. Modified nucleotides at the 5′ end of human U2 snRNA are required for spliceosomal E-complex formation. RNA 2004, 10, 1925–1933. [Google Scholar] [CrossRef]
- Jia, Y.; Mu, J.C.; Ackerman, S.L. Mutation of a U2 snRNA gene causes global disruption of alternative splicing and neurodegeneration. Cell 2012, 148, 296–308. [Google Scholar] [CrossRef]
- Mao, Y.S.; Zhang, B.; Spector, D.L. Biogenesis and function of nuclear bodies. Trends Genet. 2011, 27, 295–306. [Google Scholar] [CrossRef]
- Massenet, S.; Bertrand, E.; Verheggen, C. Assembly and trafficking of box C/D and H/ACA snoRNPs. RNA Biol. 2017, 14, 680–692. [Google Scholar] [CrossRef]
- Meier, U.T. RNA modification in Cajal bodies. RNA Biol. 2017, 14, 693–700. [Google Scholar] [CrossRef] [PubMed]
- Jády, B.E.; Kiss, T. A small nucleolar guide RNA functions both in 2′-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J. 2001, 20, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Kishore, S.; Gruber, A.R.; Jedlinski, D.J.; Syed, A.P.; Jorjani, H.; Zavolan, M. Insights into snoRNA biogenesis and processing from PAR-CLIP of snoRNA core proteins and small RNA sequencing. Genome Biol. 2013, 14, R45. [Google Scholar] [CrossRef] [PubMed]
- Deryusheva, S.; Gall, J.G. scaRNAs and snoRNAs: Are they limited to specific classes of substrate RNAs? RNA 2018, 25, 17–22. [Google Scholar] [CrossRef]
- Raj, B.; Blencowe, B.J. Alternative Splicing in the Mammalian Nervous System: Recent Insights into Mechanisms and Functional Roles. Neuron 2015, 87, 14–27. [Google Scholar] [CrossRef]
- Patil, P.; Kibiryeva, N.; Uechi, T.; Marshall, J.; O’Brien, J.E., Jr; Artman, M.; Kenmochi, N.; Bittel, D.C. scaRNAs regulate splicing and vertebrate heart development. Biochim. Biophys. Acta 2015, 1852, 1619–1629. [Google Scholar] [CrossRef]
- Nagasawa, C.; Ogren, A.; Kibiryeva, N.; Marshall, J.; O’Brien, J.E.; Kenmochi, N.; Bittel, D.C. The Role of scaRNAs in Adjusting Alternative mRNA Splicing in Heart Development. J. Cardiovasc. Dev. Dis. 2018, 5, 26. [Google Scholar] [CrossRef]
- Carlile, T.M.; Rojas-Duran, M.F.; Zinshteyn, B.; Shin, H.; Bartoli, K.M.; Gilbert, W.V. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 2014, 515, 143–146. [Google Scholar] [CrossRef]
- Lovejoy, A.F.; Riordan, D.P.; Brown, P.O. Transcriptome-wide mapping of pseudouridines: Pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS ONE 2014, 9, e110799. [Google Scholar] [CrossRef]
- Schwartz, S.; Bernstein, D.A.; Mumbach, M.R.; Jovanovic, M.; Herbst, R.H.; León-Ricardo, B.X.; Engreitz, J.M.; Guttman, M.; Satija, R.; Lander, E.S.; et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 2014, 159, 148–162. [Google Scholar] [CrossRef] [PubMed]
- Beckmann, B.M.; Horos, R.; Fischer, B.; Castello, A.; Eichelbaum, K.; Alleaume, A.-M.; Schwarzl, T.; Curk, T.; Foehr, S.; Huber, W.; et al. The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nat. Commun. 2015, 6, 10127. [Google Scholar] [CrossRef] [PubMed]
- Clouet d’Orval, B.; Bortolin, M.L.; Gaspin, C.; Bachellerie, J.P. Box C/D RNA guides for the ribose methylation of archaeal tRNAs. The tRNATrp intron guides the formation of two ribose-methylated nucleosides in the mature tRNATrp. Nucleic Acids Res. 2001, 29, 4518–4529. [Google Scholar] [CrossRef] [PubMed]
- Kristen M Bartoli, Cassandra Schaening, Thomas Carlile, Wendy V Gilbert Conserved Methyltransferase Spb1 Targets mRNAs for Regulated Modification with 2′-O-Methyl Ribose. bioRxiv 2018. [CrossRef]
- Baltz, A.G.; Munschauer, M.; Schwanhäusser, B.; Vasile, A.; Murakawa, Y.; Schueler, M.; Youngs, N.; Penfold-Brown, D.; Drew, K.; Milek, M.; et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 2012, 46, 674–690. [Google Scholar] [CrossRef] [PubMed]
- Castello, A.; Fischer, B.; Eichelbaum, K.; Horos, R.; Beckmann, B.M.; Strein, C.; Davey, N.E.; Humphreys, D.T.; Preiss, T.; Steinmetz, L.M.; et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 2012, 149, 1393–1406. [Google Scholar] [CrossRef] [PubMed]
- Ringeard, M.; Marchand, V.; Decroly, E.; Motorin, Y.; Bennasser, Y. FTSJ3 is an RNA 2′-O-methyltransferase recruited by HIV to avoid innate immune sensing. Nature 2019, 565, 500–504. [Google Scholar] [CrossRef]
- Choi, J.; Indrisiunaite, G.; DeMirci, H.; Ieong, K.-W.; Wang, J.; Petrov, A.; Prabhakar, A.; Rechavi, G.; Dominissini, D.; He, C.; et al. 2′-O-methylation in mRNA disrupts tRNA decoding during translation elongation. Nat. Struct. Mol. Biol. 2018, 25, 208–216. [Google Scholar] [CrossRef]
- Costa, M.C.; Leitão, A.L.; Enguita, F.J. Biogenesis and mechanism of action of small non-coding RNAs: Insights from the point of view of structural biology. Int. J. Mol. Sci. 2012, 13, 10268–10295. [Google Scholar] [CrossRef]
- Olina, A.V.; Kulbachinskiy, A.V.; Aravin, A.A.; Esyunina, D.M. Argonaute Proteins and Mechanisms of RNA Interference in Eukaryotes and Prokaryotes. Biochemistry 2018, 83, 483–497. [Google Scholar] [CrossRef]
- Cox, D.N.; Chao, A.; Baker, J.; Chang, L.; Qiao, D.; Lin, H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998, 12, 3715–3727. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Lin, H. Miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev. Cell 2002, 2, 819–830. [Google Scholar] [CrossRef]
- Vagin, V.V.; Sigova, A.; Li, C.; Seitz, H.; Gvozdev, V.; Zamore, P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 2006, 313, 320–324. [Google Scholar] [CrossRef]
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007, 128, 1089–1103. [Google Scholar] [CrossRef] [PubMed]
- Kirino, Y.; Mourelatos, Z. 2′-O-methyl modification in mouse piRNAs and its methylase. Nucleic Acids Symp. Ser. 2007, 417–418. [Google Scholar] [CrossRef]
- Juliano, C.; Wang, J.; Lin, H. Uniting germline and stem cells: The function of Piwi proteins and the piRNA pathway in diverse organisms. Annu. Rev. Genet. 2011, 45, 447–469. [Google Scholar] [CrossRef] [PubMed]
- Plotnikova, A.; Baranauskė, S.; Osipenko, A.; Klimašauskas, S.; Vilkaitis, G. Mechanistic insights into small RNA recognition and modification by the HEN1 methyltransferase. Biochem. J. 2013, 453, 281–290. [Google Scholar] [CrossRef]
- Lim, S.L.; Qu, Z.P.; Kortschak, R.D.; Lawrence, D.M.; Geoghegan, J.; Hempfling, A.-L.; Bergmann, M.; Goodnow, C.C.; Ormandy, C.J.; Wong, L.; et al. HENMT1 and piRNA Stability Are Required for Adult Male Germ Cell Transposon Repression and to Define the Spermatogenic Program in the Mouse. PLoS Genet. 2015, 11, e1005620. [Google Scholar]
- Hempfling, A.L.; Lim, S.L.; Adelson, D.L.; Evans, J.; O’Connor, A.E.; Qu, Z.P.; Kliesch, S.; Weidner, W.; O’Bryan, M.K.; Bergmann, M. Expression patterns of HENMT1 and PIWIL1 in human testis: Implications for transposon expression. Reproduction 2017, 154, 363–374. [Google Scholar] [CrossRef]
- Hoff, A.M.; Alagaratnam, S.; Zhao, S.; Bruun, J.; Andrews, P.W.; Lothe, R.A.; Skotheim, R.I. Identification of Novel Fusion Genes in Testicular Germ Cell Tumors. Cancer Res. 2016, 76, 108–116. [Google Scholar] [CrossRef]
- Peng, J.C.; Lin, H. Beyond transposons: The epigenetic and somatic functions of the Piwi-piRNA mechanism. Curr. Opin. Cell Biol. 2013, 25, 190–194. [Google Scholar] [CrossRef] [PubMed]
- Martinez, V.D.; Vucic, E.A.; Thu, K.L.; Hubaux, R.; Enfield, K.S.S.; Pikor, L.A.; Becker-Santos, D.D.; Brown, C.J.; Lam, S.; Lam, W.L. Unique somatic and malignant expression patterns implicate PIWI-interacting RNAs in cancer-type specific biology. Sci. Rep. 2015, 5, 10423. [Google Scholar] [CrossRef]
- Ponnusamy, M.; Yan, K.-W.; Liu, C.-Y.; Li, P.-F.; Wang, K. PIWI family emerging as a decisive factor of cell fate: An overview. Eur. J. Cell Biol. 2017, 96, 746–757. [Google Scholar] [CrossRef] [PubMed]
- Lim, R.S.M.; Anand, A.; Nishimiya-Fujisawa, C.; Kobayashi, S.; Kai, T. Analysis of Hydra PIWI proteins and piRNAs uncover early evolutionary origins of the piRNA pathway. Dev. Biol. 2014, 386, 237–251. [Google Scholar] [CrossRef] [PubMed]
- Lewis, S.H.; Quarles, K.A.; Yang, Y.; Tanguy, M.; Frézal, L.; Smith, S.A.; Sharma, P.P.; Cordaux, R.; Gilbert, C.; Giraud, I.; et al. Pan-arthropod analysis reveals somatic piRNAs as an ancestral defence against transposable elements. Nat. Ecol. Evol. 2018, 2, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Rajasethupathy, P.; Antonov, I.; Sheridan, R.; Frey, S.; Sander, C.; Tuschl, T.; Kandel, E.R. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell 2012, 149, 693–707. [Google Scholar] [CrossRef] [PubMed]
- Abe, M.; Naqvi, A.; Hendriks, G.-J.; Feltzin, V.; Zhu, Y.; Grigoriev, A.; Bonini, N.M. Impact of age-associated increase in 2′-O-methylation of miRNAs on aging and neurodegeneration in Drosophila. Genes Dev. 2014, 28, 44–57. [Google Scholar] [CrossRef]
- Pepper, A.S.-R.; Beerman, R.W.; Bhogal, B.; Jongens, T.A. Argonaute2 suppresses Drosophila fragile X expression preventing neurogenesis and oogenesis defects. PLoS ONE 2009, 4, e7618. [Google Scholar] [CrossRef]
| FTSJ1 Allele | Family | Mutation/Location | Effect | Reference |
|---|---|---|---|---|
| Ftsj1Δ | 6 (AU) | Deletion of FTSJ1 and SLC38A5 | Loss of FTSJ1 | [61] |
| FTSJ1-ss | A3 | c.121 + 1delG, p.Gly41Valfs*10 (IVS2, G DEL, + 1)/ Exon 2 | Significant reduction of FTSJ1 mRNA level (NMD) | [59] |
| 196C > T | P48 | c.196C>T, p.Gln66*/ Exon 4 | Almost undetectable FTSJ1 transcripts (NMD) | [59] |
| 655G > A | MRX44 | c.655G > A, p.Glu191_Tyr218del/ Exon 9 | Loss of exon 9, protein lacking 28 amino acids | [59] |
| A > G | MRX9 | c.192-2A>G, p.Gly65Cysfs*18 (IVS3AS, A-G, -2)/ Intron 3 | Truncated protein | [60] |
| G > A | MRW06 | c.571 + 1G > A, p.Glu191Glyfs*44/ Intron 8 | Significant reduction of FTSJ1 mRNA level (NMD) | [63] |
| p.A26P | 7 | c.76G > C; p.Ala26Pro/ Exon 2 | Altered FTSJ1 protein function | [64] |
| A > T | de novo variation | c.362-2A > T, p.?/ Intron 5 of trascrit NM_012280.3 | Unknown (probable loss of exon 6 of transcript NM_012280.3 causing a frameshift) | Amélie Piton & Elise Schaefer, HUS, personal communication |
| Y > N | de novo variation | c.34T > A; p.Tyr12Asn/ Exon 2 | Deposited as pathogenic | Ambry Genetics Clinvar NCBI [66] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dimitrova, D.G.; Teysset, L.; Carré, C. RNA 2′-O-Methylation (Nm) Modification in Human Diseases. Genes 2019, 10, 117. https://doi.org/10.3390/genes10020117
Dimitrova DG, Teysset L, Carré C. RNA 2′-O-Methylation (Nm) Modification in Human Diseases. Genes. 2019; 10(2):117. https://doi.org/10.3390/genes10020117
Chicago/Turabian StyleDimitrova, Dilyana G., Laure Teysset, and Clément Carré. 2019. "RNA 2′-O-Methylation (Nm) Modification in Human Diseases" Genes 10, no. 2: 117. https://doi.org/10.3390/genes10020117
APA StyleDimitrova, D. G., Teysset, L., & Carré, C. (2019). RNA 2′-O-Methylation (Nm) Modification in Human Diseases. Genes, 10(2), 117. https://doi.org/10.3390/genes10020117






