Identification of Genes Involved in Low Temperature-Induced Senescence of Panicle Leaf in Litchi chinensis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. RNA Isolation, cDNA Synthesis, and Library Construction for Transcriptome
2.3. RNA-Seq Data Analysis and Gene Functional Annotation
2.4. Quantitative Real-Time PCR Analysis
2.5. Statistical Analysis
3. Results
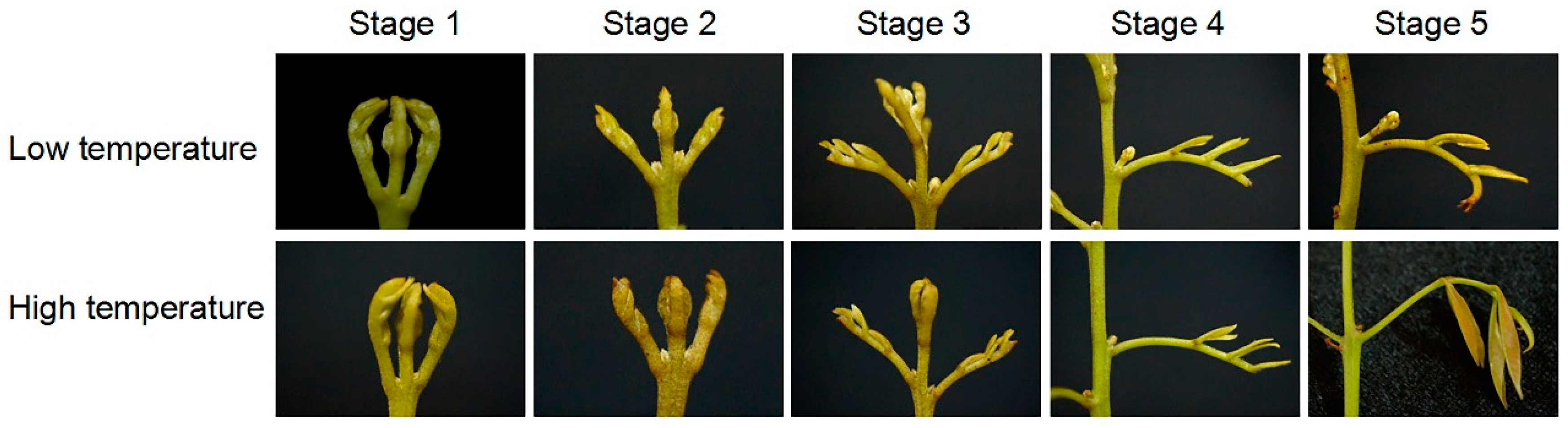
3.1. Morphology of Senescing and Developing Panicle Leaves
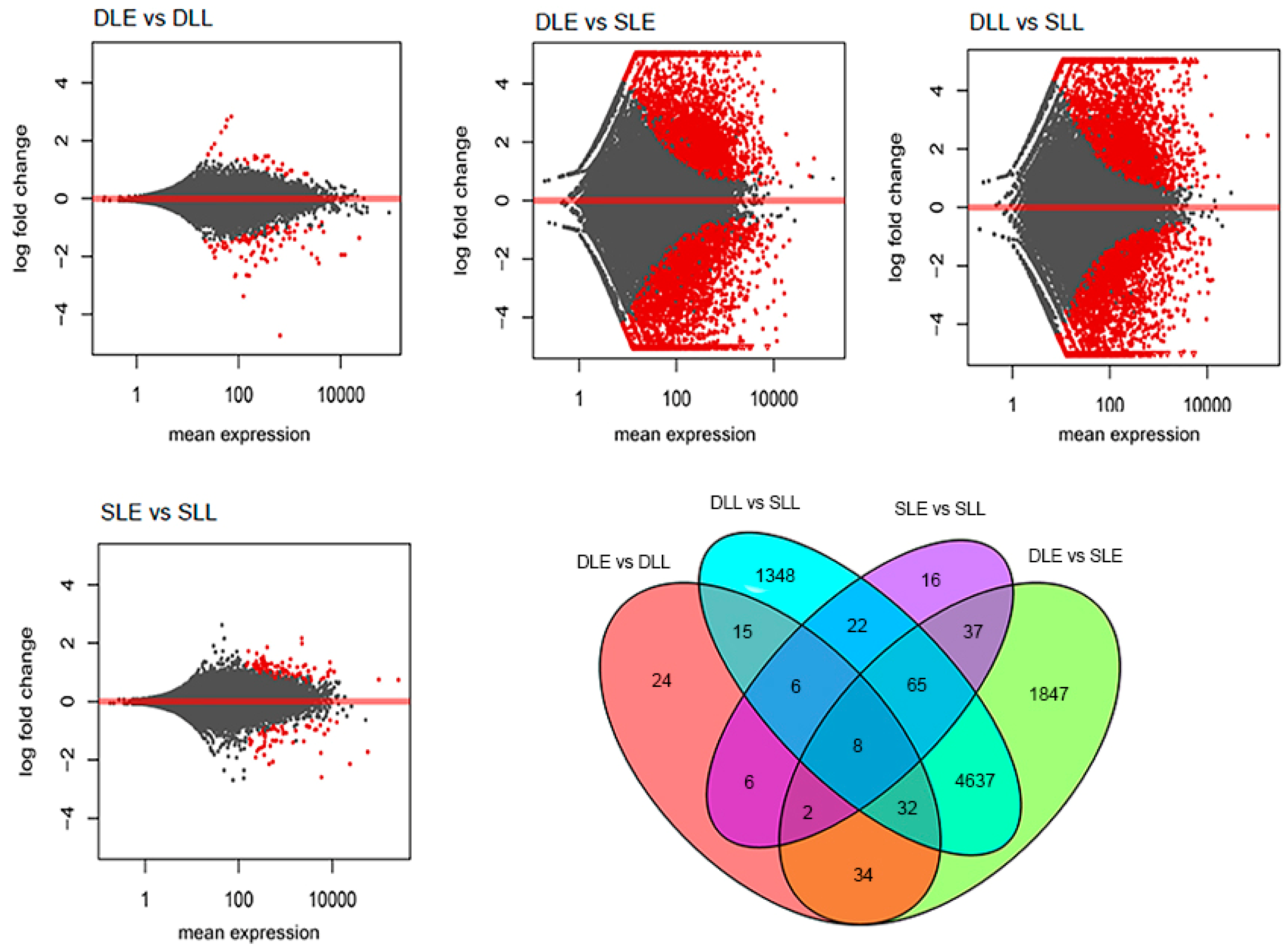
3.2. Digital Transcriptome Analysis
3.3. DEG Identification and GO-Term Analysis
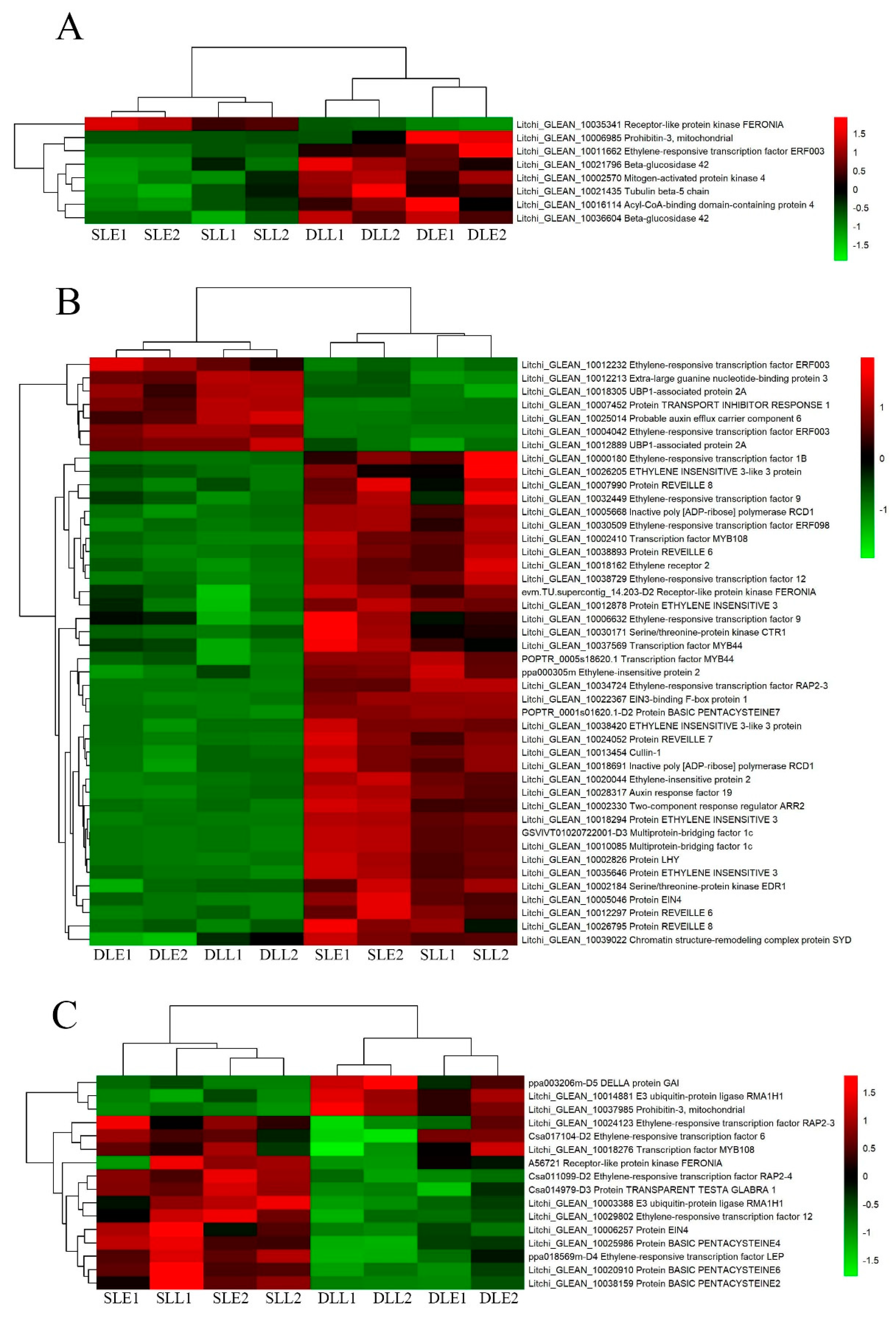
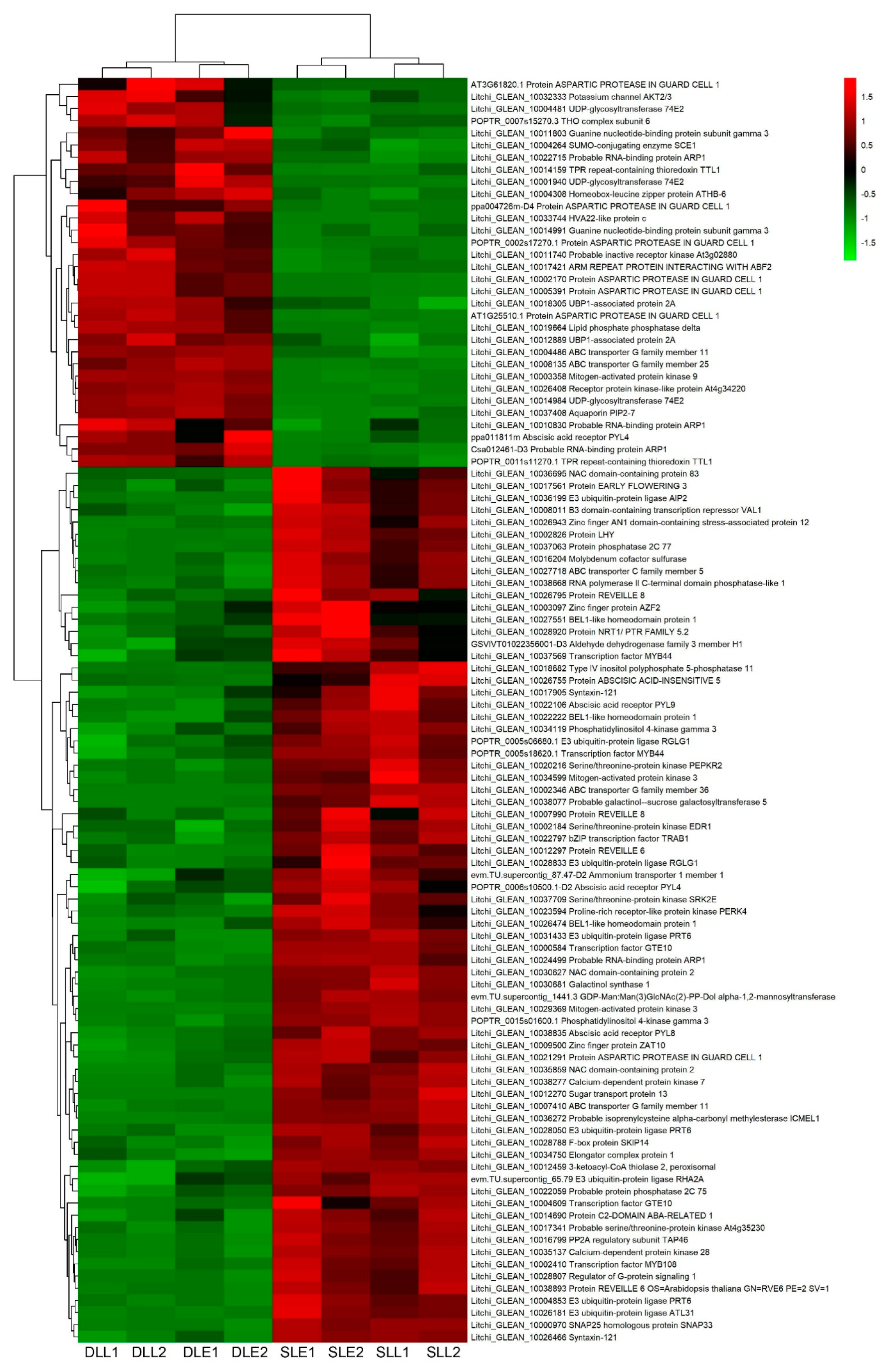
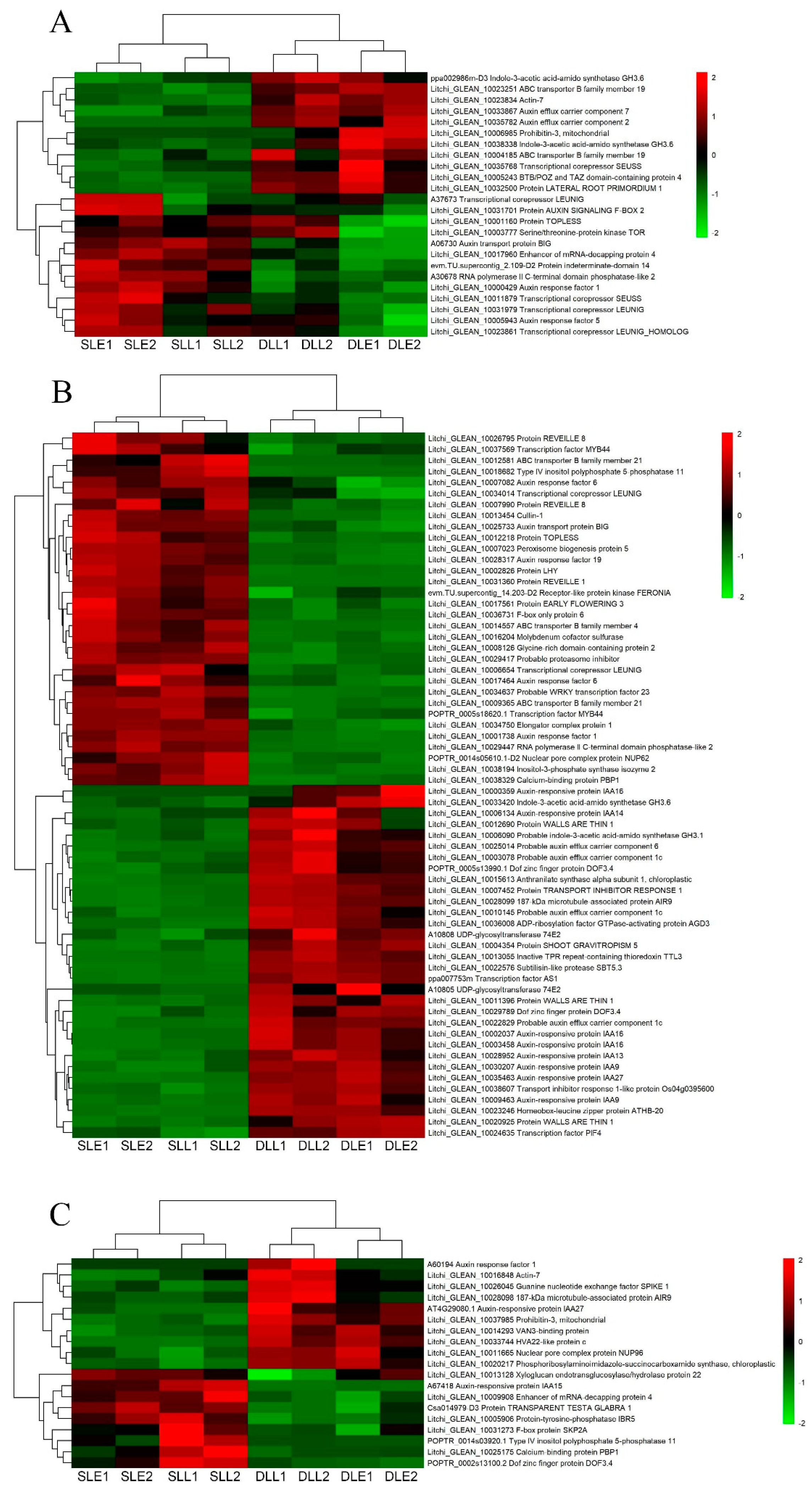
3.4. Identification of the Plant Hormone-Related Genes Potentially Involved in LT-Induced Panicle Leaf Senescence
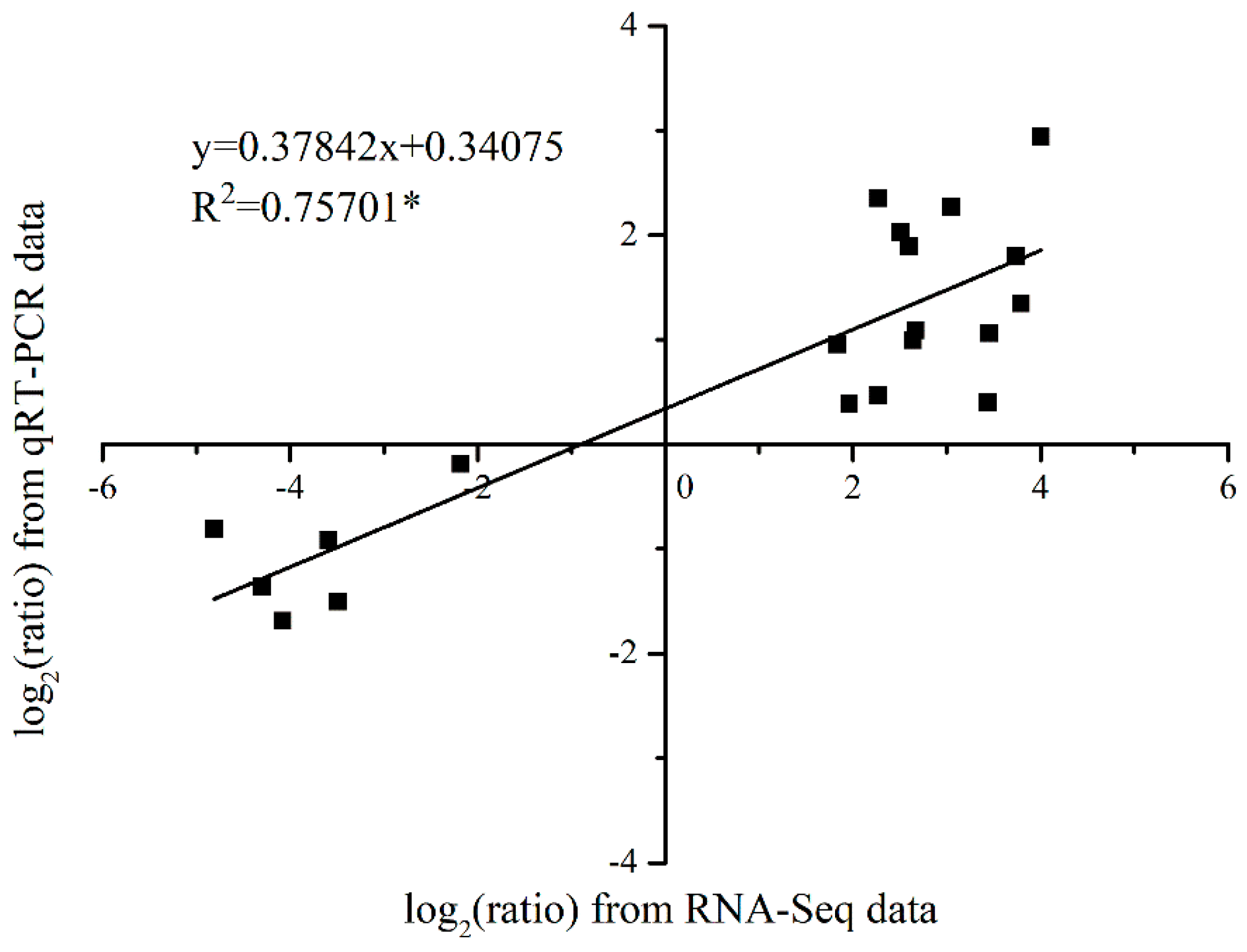
3.5. Validation of the Expression Pattern of DEGs by qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Menzel, C.M.; Simpson, D.X. Effect of temperature on growth and flowering of litchi (Litchi chinensis Sonn.) cultivars. J. Hortic. Sci. 1988, 63, 349–360. [Google Scholar] [CrossRef]
- Chen, H.B.; Huang, H.B. Low temperature requirements for floral induction in lychee. Acta Hortic. 2005, 665, 195–202. [Google Scholar] [CrossRef]
- Zhou, B.; Chen, H.; Huang, X.; Li, N.; Hu, Z.; Gao, Z.; Lu, Y. Rudimentary leaf abortion with the development of panicle in litchi: Changes in ultrastructure, antioxidant enzymes and phytohormones. Sci. Hortic. 2008, 117, 288–296. [Google Scholar] [CrossRef]
- Zhou, B.; Li, N.; Zhang, Z.; Huang, X.; Chen, H.; Hu, Z.; Pang, X.; Liu, W.; Lu, Y. Hydrogen peroxide and nitric oxide promote reproductive growth in Litchi chinensis. Biol. Plant. 2012, 56, 321–329. [Google Scholar] [CrossRef]
- Zhou, B.; Huang, X.; Chen, H.; Shu, W.; Hu, Z.; Liu, W.; Xiang, C.; Zhang, S. Effects of ethylene on rudimentary leaf and panicle primordium in litchi: Antioxidant enzymes, hydrogen peroxide and nitric oxide. Acta Hortic. 2013, 975, 247–254. [Google Scholar] [CrossRef]
- Jing, H.; Hebeler, R.; Oeljeklaus, S.; Sitek, B.; Stuehler, K.; Meyer, H.; Sturre, M.; Hille, J.; Warscheid, B.; Dijkwel, P.; et al. Early leaf senescence is associated with an altered cellular redox balance in Arabidopsis cpr5/old1 mutants. Plant Biol. 2008, 10, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.Y.; Kuai, B.K.; Jia, J.Z.; Jing, H.C. Regulation of leaf senescence and crop genetic improvement. J. Integr. Plant Biol. 2012, 54, 936–952. [Google Scholar] [CrossRef] [PubMed]
- Schippers, J.H.M.; Schmidt, R.; Wagstaff, C.; Jing, H.C. Living to die and dying to live: The survival strategy behind leaf senescence. Plant Physiol. 2015, 169, 914–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, H.; Gong, Y.; Zhang, D. Effect of high temperature on leaf senescence and related enzymes of grain starch synthesis in stay-green wheat after anthesis. Chin. J. Plant Ecol. 2011, 35, 769–778. [Google Scholar] [CrossRef]
- Allu, A.D.; Soja, A.M.; Wu, A.; Szymanski, J.; Balazadeh, S. Salt stress and senescence: Identification of cross-talk regulatory components. J. Exp. Bot. 2014, 65, 3993–4008. [Google Scholar] [CrossRef]
- Naschitz, S.; Naor, A.; Wolf, S.; Goldschmidt, E. The effects of temperature and drought on autumnal senescence and leaf shed in apple under warm, east mediterranean climate. Trees-Struct. Funct. 2014, 28, 879–890. [Google Scholar] [CrossRef]
- Yang, H.F.; Lu, X.Y.; Chen, H.B.; Wang, C.C.; Zhou, B.Y. Low temperature-induced leaf senescence and the expression of senescence-related genes in the panicles of Litchi chinensis. Biol. Plant. 2017, 61, 315–322. [Google Scholar] [CrossRef]
- Yang, H.; Kim, H.J.; Chen, H.; Lu, Y.; Lu, X.; Wang, C.; Zhou, B. Reactive oxygen species and nitric oxide induce senescence of rudimentary leaves in Litchi chinensis and the expression profiles of the related genes. Hortic. Res. 2018, 5, 23. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhou, C. Signal transduction in leaf senescence. Plant Mol. Biol. 2013, 82, 539–545. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Rozhon, W.; Poppenberger, B. The role of hormones in the aging of plants—A mini-review. Gerontology 2014, 60, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Jirage, D.; Tootle, T.L.; Reuber, T.L.; Frost, L.N.; Feys, B.J.; Parker, J.E.; Ausubel, F.M.; Glazebrook, J. Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. USA 1999, 96, 1358–13588. [Google Scholar] [CrossRef]
- Caarls, L.; Pieterse, C.M.J.; Van Wees, S.C.M. How salicylic acid takes transcriptional control over jasmonic acid signaling. Front. Plant Sci. 2015, 6, 170. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Kim, H.; Zhong, S.; Chen, H.; Hu, Z.; Zhou, B. De novo transcriptome assembly for rudimentary leaves in Litchi chinesis Sonn. and identification of differentially expressed genes in response to reactive oxygen species. BMC Genom. 2014, 15, 805–819. [Google Scholar] [CrossRef] [PubMed]
- Saha, G.; Park, J.I.; Jung, H.J.; Ahmed, N.U.; Kayum, M.A.; Chung, M.Y.; Hur, Y.; Cho, Y.G.; Watanabe, M.; Nou, I.S. Genome-wide identification and characterization of MADS-box family genes related to organ development and stress resistance in Brassica rapa. BMC Genom. 2015, 16, 178. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Guo, J.J.; He, C.T.; Shen, C.; Huang, Y.Y.; Chen, J.X.; Gao, J.H.; Yuan, J.G.; Zhang, Z.Y. Comparative transcriptome analysis between low- and high-Cadmium-accumulating genotypes of pakchoi (Brassica chinensis L.) in response to cadmium stress. Environ. Sci. Technol. 2016, 50, 6485–6494. [Google Scholar] [CrossRef] [PubMed]
- Bowtie2. unpublished work. Available online: http://litchidb.genomics.cn (accessed on 6 November 2018).
- eXpress. Available online: https://pachterlab.github.io/eXpress/index.html (accessed on 10 March 2017).
- National Center for Biotechnology Information (NCBI). Available online: http://www.ncbi.nlm.nih.gov/guide/ (accessed on 6 November 2018).
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- UniProtKB Swiss-Prot. Available online: http://www.uniprot.org/uniprot/ (accessed on 4 November 2018).
- Falcon, S.; Gentleman, R. Using GOstats to test gene lists for GO term association. Bioinformatics 2006, 23, 257–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-DELTADELTACT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Janda, M.; Planchais, S.; Djafi, N.; Martinec, J.; Burketova, L.; Valentova, O.; Zachowski, A.; Ruelland, E. Phosphoglycerolipids are master players in plant hormone signal transduction. Plant Cell Rep. 2013, 32, 839–851. [Google Scholar] [CrossRef] [PubMed]
- Bennett, E.J.; Roberts, J.A.; Wagstaff, C. Use of mutants to dissect the role of ethylene signalling in organ senescence and the regulation of yield in Arabidopsis thaliana. J. Plant Growth. Regul. 2014, 33, 56–65. [Google Scholar] [CrossRef]
- Guo, Y.; Gan, S.S. Convergence and divergence in gene expression profiles induced by leaf senescence and 27 senescence-promoting hormonal, pathological and environmental stress treatments. Plant Cell Environ. 2012, 35, 644–655. [Google Scholar] [CrossRef]
- Cui, Z.; Zhou, B.; Zhang, Z.; Hu, Z. Abscisic acid promotes flowering and enhances LcAP1 expression in Litchi chinensis Sonn. S. Afr. J. Bot. 2013, 88, 76–79. [Google Scholar] [CrossRef]
- Hönig, M.; Plíhalová, L.; Husičková, A.; Nisler, J.; Doležal, K. Role of cytokinins in senescence, antioxidant defence and photosynthesis. Int. J. Mol. Sci. 2018, 19, 4045. [Google Scholar] [CrossRef]
- Luo, Y.L.; Tang, Y.H.; Zhang, X.; Li, W.Q.; Chang, Y.L.; Pang, D.W.; Xu, X.; Li, Y.; Wang, Z.L. Interactions between cytokinin and nitrogen contribute to grain mass in wheat cultivars by regulating the flag leaf senescence process. Crop J. 2018, 6, 538–551. [Google Scholar] [CrossRef]
- Hu, Y.R.; Jiang, Y.J.; Han, X.; Wang, H.P.; Pan, J.J.; Yu, D.Q. Jasmonate regulates leaf senescence and tolerance to cold stress: Crosstalk with other phytohormones. J. Exp. Bot. 2017, 68, 1361–1369. [Google Scholar] [CrossRef]
- Zhu, Z.; Lee, B. Friends or foes: New insights in jasmonate and ethylene co-actions. Plant Cell Physiol. 2015, 56, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Torres, D.P.; Proels, R.K.; Schempp, H.; Huckelhoven, R. Silencing of RBOHF2 causes leaf age-dependent accelerated senescence, salicylic acid accumulation, and powdery mildew resistance in barley. Mol. Plant-Microbe Interact. 2017, 30, 906–918. [Google Scholar] [CrossRef] [PubMed]
- Gomes, M.M.A. Physiological effects related to brassinosteroid application in plants. In Brassinosteroids: A Class of Plant Hormone; Hayat, S., Ahmad, A., Eds.; Springer: Dordrecht, The Netherlands, 2011; pp. 193–242. [Google Scholar]
- de Assis-Gomes, M.D.; Pinheiro, D.T.; Bressan-Smith, R.; Campostrini, E. Exogenous brassinosteroid application delays senescence and promotes hyponasty in Carica papaya L. leaves. Theor. Exp. Plant Physiol. 2018, 30, 193–201. [Google Scholar] [CrossRef]




| Sample | Raw Reads | Clean Reads | Clean Reads/Raw Reads (%) | Aligned Reads | Alignment Rate |
|---|---|---|---|---|---|
| DLE 1 | 6,521,358 | 6,395,241 | 98.07 | 3,825,633 | 59.82% |
| DLE 2 | 5,658,367 | 5,514,047 | 97.45 | 3,398,858 | 61.64% |
| DLL 1 | 7,370,305 | 6,518,556 | 88.44 | 3,761,206 | 57.70% |
| DLL 2 | 6,533,923 | 6,219,090 | 95.18 | 3,853,348 | 61.96% |
| SLE 1 | 9,081,562 | 8,998,048 | 99.08 | 5,485,210 | 60.96% |
| SLE 2 | 7,620,278 | 7,560,087 | 99.21 | 4,843,747 | 64.07% |
| SLL 1 | 4,825,239 | 4,781,295 | 99.09 | 3,219,724 | 67.34% |
| SLL 2 | 6,004,061 | 5,868,244 | 97.74 | 3,679,388 | 62.70% |
| Sample 1 | Sample 2 | Total | Up (Sample 1 to 2) | Down (Sample 1 to 2) |
|---|---|---|---|---|
| DLE | DLL | 127 | 30 | 97 |
| SLE | SLL | 162 | 108 | 54 |
| DLE | SLE | 6662 | 3295 | 3367 |
| DLL | SLL | 6133 | 3182 | 2951 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Liu, H.; Yu, S.; Chen, H.; Hu, F.; Zhan, H.; Pan, X.; Lao, Y.; Zhong, S.; Zhou, B. Identification of Genes Involved in Low Temperature-Induced Senescence of Panicle Leaf in Litchi chinensis. Genes 2019, 10, 111. https://doi.org/10.3390/genes10020111
Wang C, Liu H, Yu S, Chen H, Hu F, Zhan H, Pan X, Lao Y, Zhong S, Zhou B. Identification of Genes Involved in Low Temperature-Induced Senescence of Panicle Leaf in Litchi chinensis. Genes. 2019; 10(2):111. https://doi.org/10.3390/genes10020111
Chicago/Turabian StyleWang, Congcong, Hao Liu, Sheng Yu, Houbin Chen, Fuchu Hu, Huiling Zhan, Xifen Pan, Yuhua Lao, Silin Zhong, and Biyan Zhou. 2019. "Identification of Genes Involved in Low Temperature-Induced Senescence of Panicle Leaf in Litchi chinensis" Genes 10, no. 2: 111. https://doi.org/10.3390/genes10020111
APA StyleWang, C., Liu, H., Yu, S., Chen, H., Hu, F., Zhan, H., Pan, X., Lao, Y., Zhong, S., & Zhou, B. (2019). Identification of Genes Involved in Low Temperature-Induced Senescence of Panicle Leaf in Litchi chinensis. Genes, 10(2), 111. https://doi.org/10.3390/genes10020111




