Core Element Cloning, Cis-Element Mapping and Serum Regulation of the Human EphB4 Promoter: A Novel TATA-Less Inr/MTE/DPE-Like Regulated Gene
Abstract
1. Introduction
2. Materials and Methods
2.1. Molecular Biology Reagents
2.2. Human EphB4 Promoter Sequencing and Bioinformatic Analysis
2.3. Human EphB4 Core Promoter and Luc-Reporter Construct Cloning
2.4. Cell Cultures, Transient Transfections and EphB4 Promoter Reporter Assay
3. Results
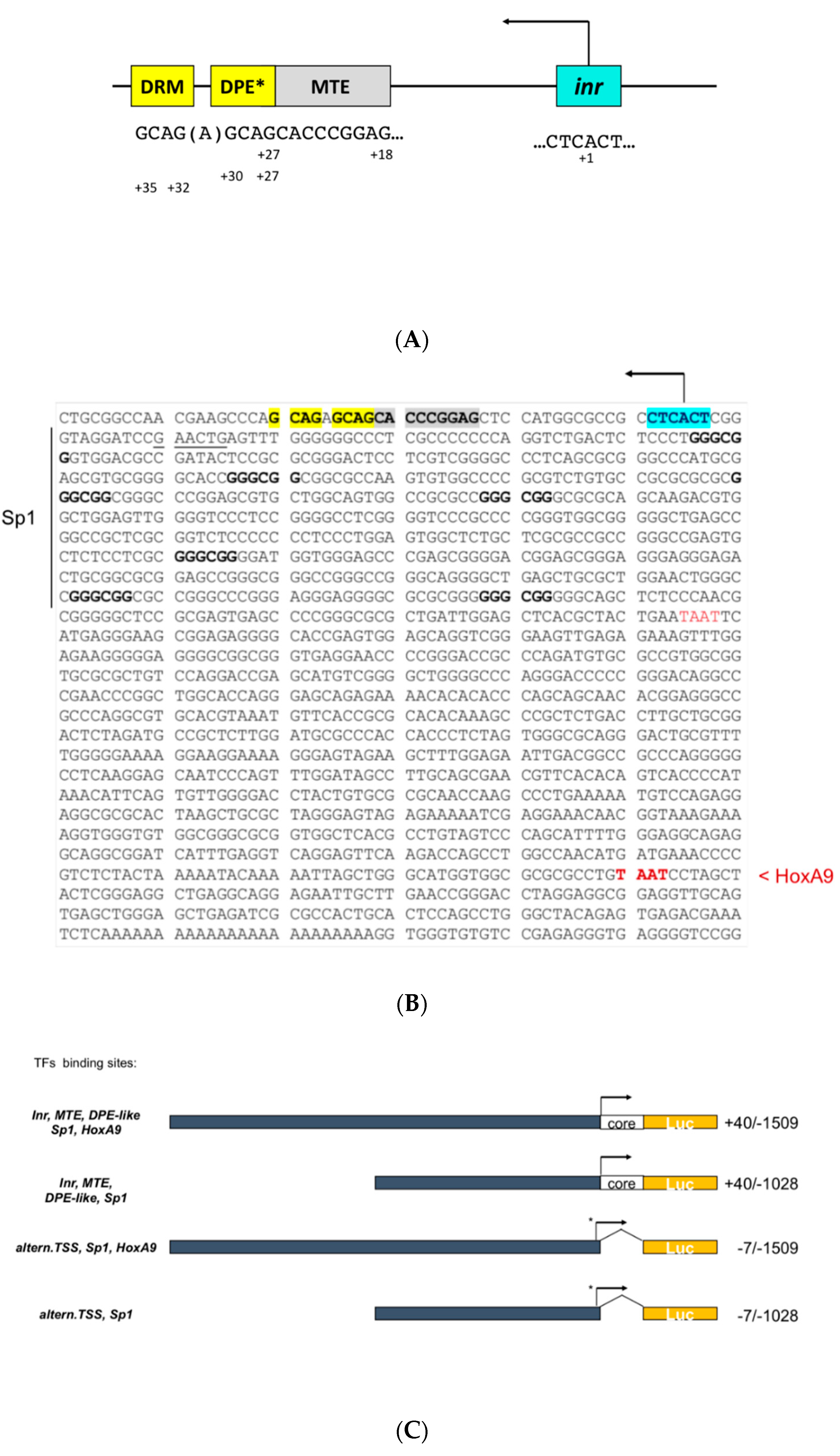
3.1. Cloning of the Human EphB4 Core Promoter Reveals a TATA-less Promoter Regulated by Inr/MTE/DPE-Like Elements
3.2. The EphB4 Inr/MTE/DPE-Like Core Promoter
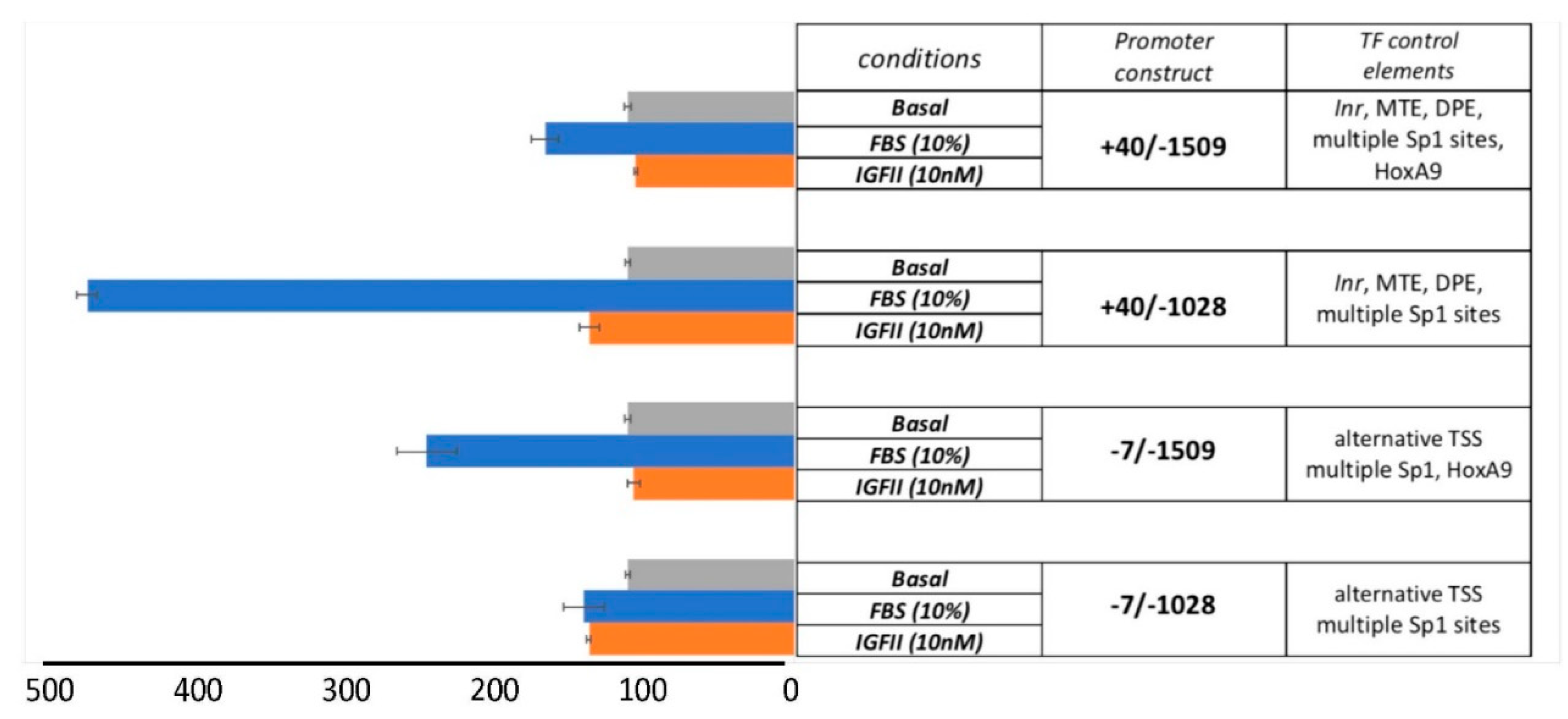
3.3. The Serum-Dependent Transcriptional Response of EphB4 TATA-Less Inr/MTE/DPE-Like Downstream Promoter Is Enhanced by Multiple Proximal Sp1 Consensus Motifs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wang, H.U.; Chen, Z.F.; Anderson, D.J. Molecular distinction and angiogenic interaction between embryonic arteries and veins revealed by ephrin-B2 and its receptor Eph-B4. Cell 1998, 93, 741–753. [Google Scholar] [CrossRef]
- Gerety, S.S.; Wang, H.U.; Chen, Z.F.; Anderson, D.J. Symmetrical mutant phenotypes of the receptor EphB4 and its specific transmembrane ligand ephrin-B2 in cardiovascular development. Mol. Cell 1999, 4, 403–414. [Google Scholar] [CrossRef]
- Bruhl, T.; Urbich, C.; Aicher, D.; Acker-Palmer, A.; Zeiher, A.M.; Dimmeler, S. Homeobox A9 transcriptionally regulates the EphB4 receptor to modulate endothelial cell migration and tube formation. Circ. Res. 2004, 94, 743–751. [Google Scholar] [CrossRef] [PubMed]
- Pasquale, E.B. Eph receptors and ephrins in cancer: Bidirectional signalling and beyond. Nat. Rev. Cancer 2010, 10, 165–180. [Google Scholar] [CrossRef] [PubMed]
- Xia, G.; Kumar, S.R.; Masood, R.; Zhu, S.; Reddy, R.; Krasnoperov, V.; Quinn, D.I.; Henshall, S.M.; Sutherland, R.L.; Pinski, J.K.; et al. EphB4 expression and biological significance in prostate cancer. Cancer Res. 2005, 65, 4623–4632. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Ferguson, B.D.; Zhou, Y.; Naga, K.; Salgia, R.; Gill, P.S.; Krasnoperov, V. EphB4 as a therapeutic target in mesothelioma. BMC Cancer 2013, 13, 269. [Google Scholar] [CrossRef] [PubMed]
- Scalia, P.; Pandini, G.; Carnevale, V.; Giordano, A.; Williams, S.J. Identification of a novel EphB4 phosphodegron regulated by the autocrine IGFII/IRA axis in malignant mesothelioma. Oncogene 2019, 38, 5987–6001. [Google Scholar] [CrossRef] [PubMed]
- Juven-Gershon, T.; Kadonaga, J.T. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev. Biol. 2010, 339, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.Y.; Santoso, B.; Boulay, T.; Dong, E.; Ohler, U.; Kadonaga, J.T. The MTE, a new core promoter element for transcription by RNA polymerase II. Genes Dev. 2004, 18, 1606–1617. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Theisen, J.W.; Lim, C.Y.; Kadonaga, J.T. Three key subregions contribute to the function of the downstream RNA polymerase II core promoter. Mol. Cell. Biol. 2010, 30, 3471–3479. [Google Scholar] [CrossRef] [PubMed]
- Kadonaga, J.T. The DPE, a core promoter element for transcription by RNA polymerase II. Exp. Mol. Med. 2002, 34, 259–264. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Scalia, P.; Heart, E.; Comai, L.; Vigneri, R.; Sung, C.K. Regulation of the Akt/Glycogen synthase kinase-3 axis by insulin-like growth factor-II via activation of the human insulin receptor isoform-A. J. Cell. Biochem. 2001, 82, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Wierstra, I. Sp1: Emerging roles--beyond constitutive activation of TATA-less housekeeping genes. Biochem. Biophys. Res. Commun. 2008, 372, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kugel, J.F.; Goodrich, J.A. Finding the start site: Redefining the human initiator element. Genes Dev. 2017, 31, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Kadonaga, J.T. Perspectives on the RNA polymerase II core promoter. Wiley Interdiscip. Rev. Dev. Biol. 2012, 1, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Gershenzon, N.I.; Ioshikhes, I.P. Synergy of human Pol II core promoter elements revealed by statistical sequence analysis. Bioinformatics 2005, 21, 1295–1300. [Google Scholar] [CrossRef] [PubMed]
- Carninci, P.; Sandelin, A.; Lenhard, B.; Katayama, S.; Shimokawa, K.; Ponjavic, J.; Semple, C.A.; Taylor, M.S.; Engstrom, P.G.; Frith, M.C.; et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006, 38, 626–635. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Bolotin, E.; Jiang, T.; Sladek, F.M.; Martinez, E. Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene 2007, 389, 52–65. [Google Scholar] [CrossRef] [PubMed]
- Burke, T.W.; Willy, P.J.; Kutach, A.K.; Butler, J.E.; Kadonaga, J.T. The DPE, a conserved downstream core promoter element that is functionally analogous to the TATA box. Cold Spring Harb. Symp. Quant. Biol. 1998, 63, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Kutach, A.K.; Kadonaga, J.T. The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol. Cell. Biol. 2000, 20, 4754–4764. [Google Scholar] [CrossRef] [PubMed]
| Nt from TSS | +18 | +19 | +20 | +21 | +22 | +23 | +24 | +25 | +26 | +27 | +28 | +29 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MTE consensus | C | S | A | R | C | S | S | A | A | C | G | S |
| H EphB4 | C | G | A | G | G | C | C | C | A | C | G | A |
| CG4427 | C | G | A | A | C | G | C | A | A | C | G | G |
| CG15312 | C | G | A | A | C | G | C | A | A | C | G | G |
| CG14720 | C | G | A | A | C | C | A | A | A | C | G | G |
| Hedgehog | C | G | A | G | C | G | C | A | A | C | G | G |
| CG15695 | C | G | A | G | C | G | G | A | G | C | G | G |
| CG10479 | C | G | A | A | C | C | G | A | T | C | G | C |
| Tollo | C | G | A | G | C | C | G | A | G | C | G | G |
| Side | C | A | A | G | C | C | A | A | G | C | G | C |
| CG12537 | C | A | A | G | C | G | G | A | G | C | G | G |
| % consensus | 100% | 80% | 100% | 60% | 0 | 50% | 40% | 0 | 50% | 100% | 100% | 0 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scalia, P.; Williams, S.J.; Giordano, A. Core Element Cloning, Cis-Element Mapping and Serum Regulation of the Human EphB4 Promoter: A Novel TATA-Less Inr/MTE/DPE-Like Regulated Gene. Genes 2019, 10, 997. https://doi.org/10.3390/genes10120997
Scalia P, Williams SJ, Giordano A. Core Element Cloning, Cis-Element Mapping and Serum Regulation of the Human EphB4 Promoter: A Novel TATA-Less Inr/MTE/DPE-Like Regulated Gene. Genes. 2019; 10(12):997. https://doi.org/10.3390/genes10120997
Chicago/Turabian StyleScalia, Pierluigi, Stephen J. Williams, and Antonio Giordano. 2019. "Core Element Cloning, Cis-Element Mapping and Serum Regulation of the Human EphB4 Promoter: A Novel TATA-Less Inr/MTE/DPE-Like Regulated Gene" Genes 10, no. 12: 997. https://doi.org/10.3390/genes10120997
APA StyleScalia, P., Williams, S. J., & Giordano, A. (2019). Core Element Cloning, Cis-Element Mapping and Serum Regulation of the Human EphB4 Promoter: A Novel TATA-Less Inr/MTE/DPE-Like Regulated Gene. Genes, 10(12), 997. https://doi.org/10.3390/genes10120997






