Rhizobia Isolated from the Relict Legume Vavilovia formosa Represent a Genetically Specific Group within Rhizobium leguminosarum biovar viciae
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Collection and DNA Isolation
2.2. Polymerase Chain Reaction (PCR) Analysis
2.3. Sequence Analysis
2.4. Group Separation Statistics
2.5. Sterile Tube Test Experiment
3. Results
3.1. Population Diversity in Symbionts of V. formosa
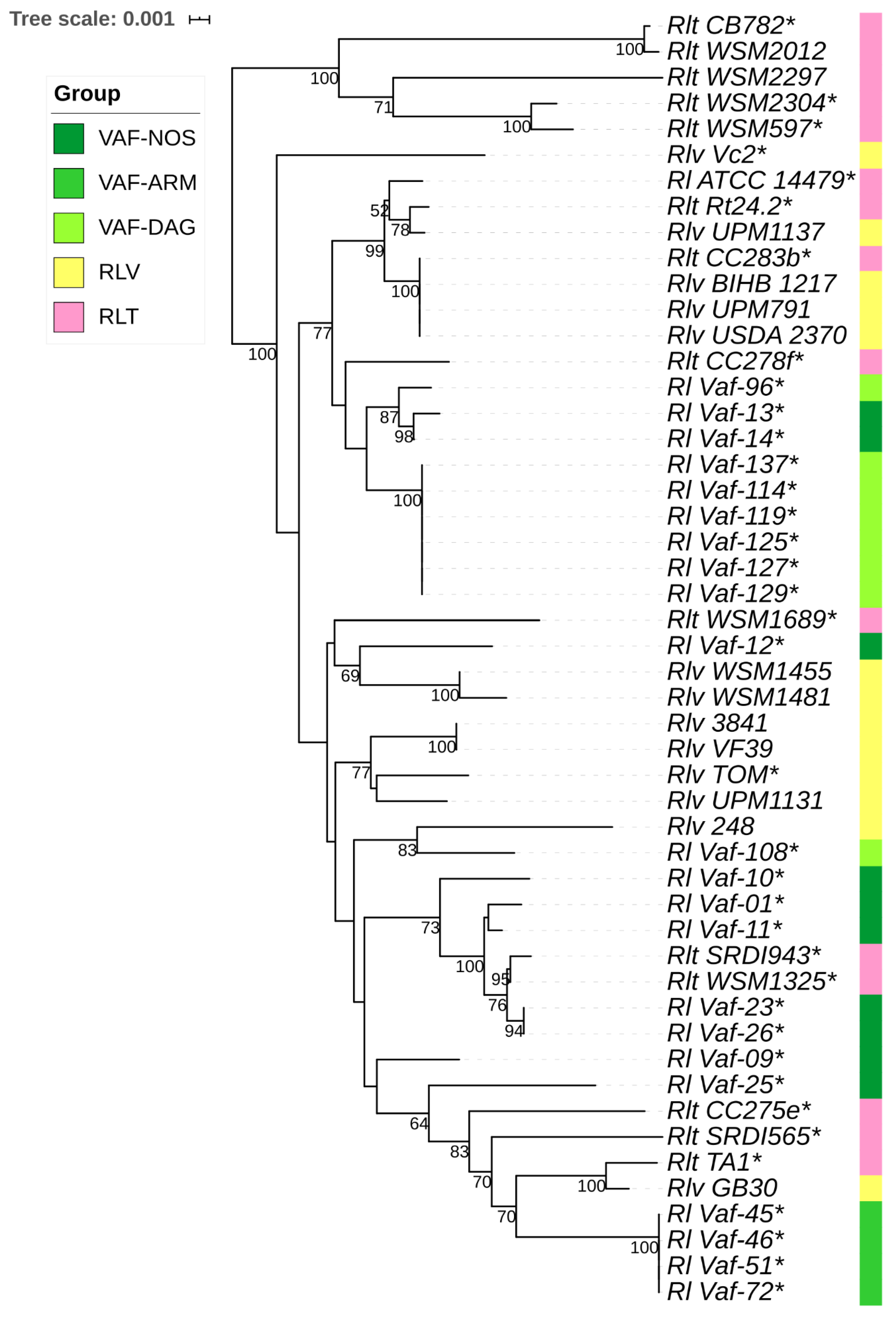
3.2. Phylogenetic Analysis of Symbionts of V. formosa
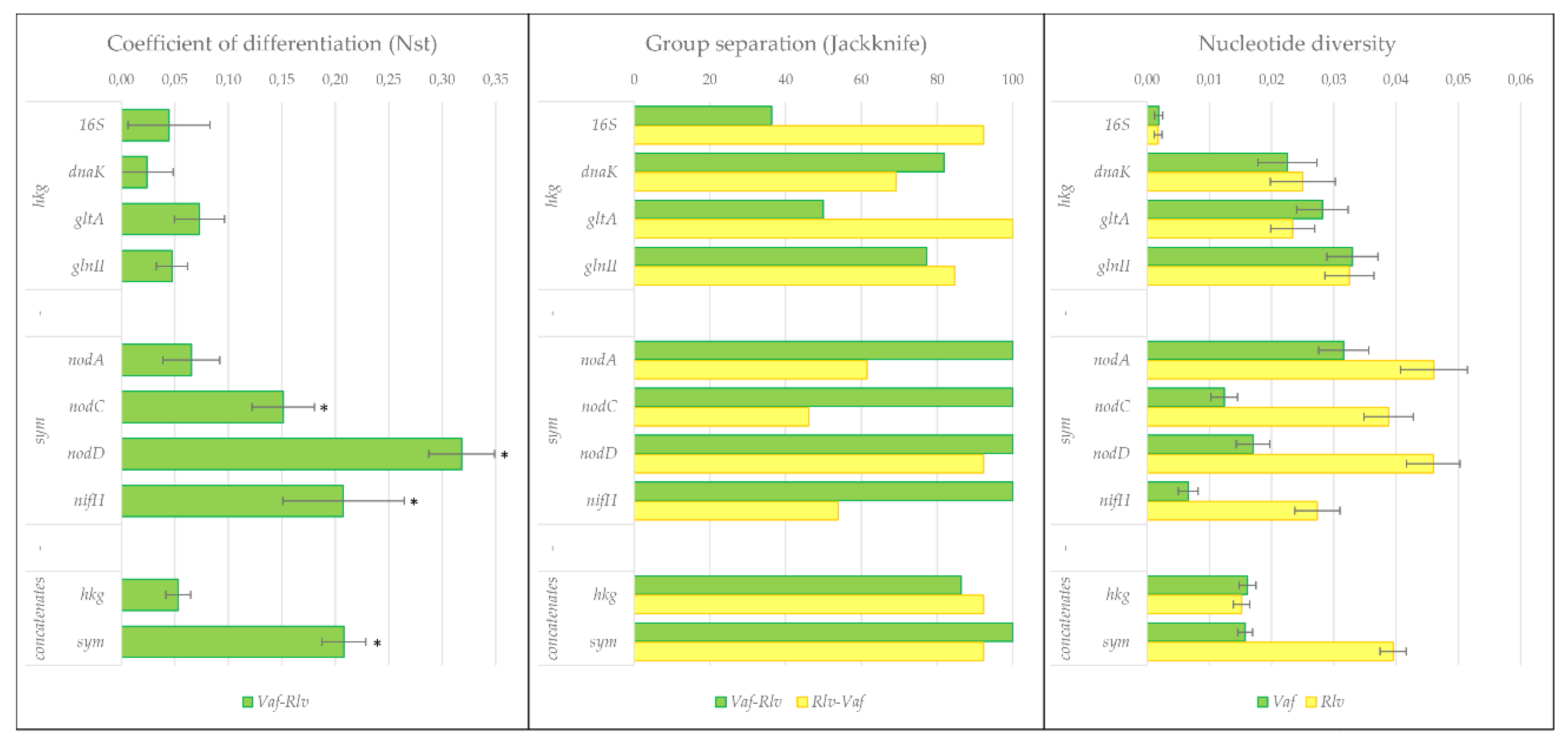
3.3. Separation Statistics of symbionts of V. formosa from Rlv
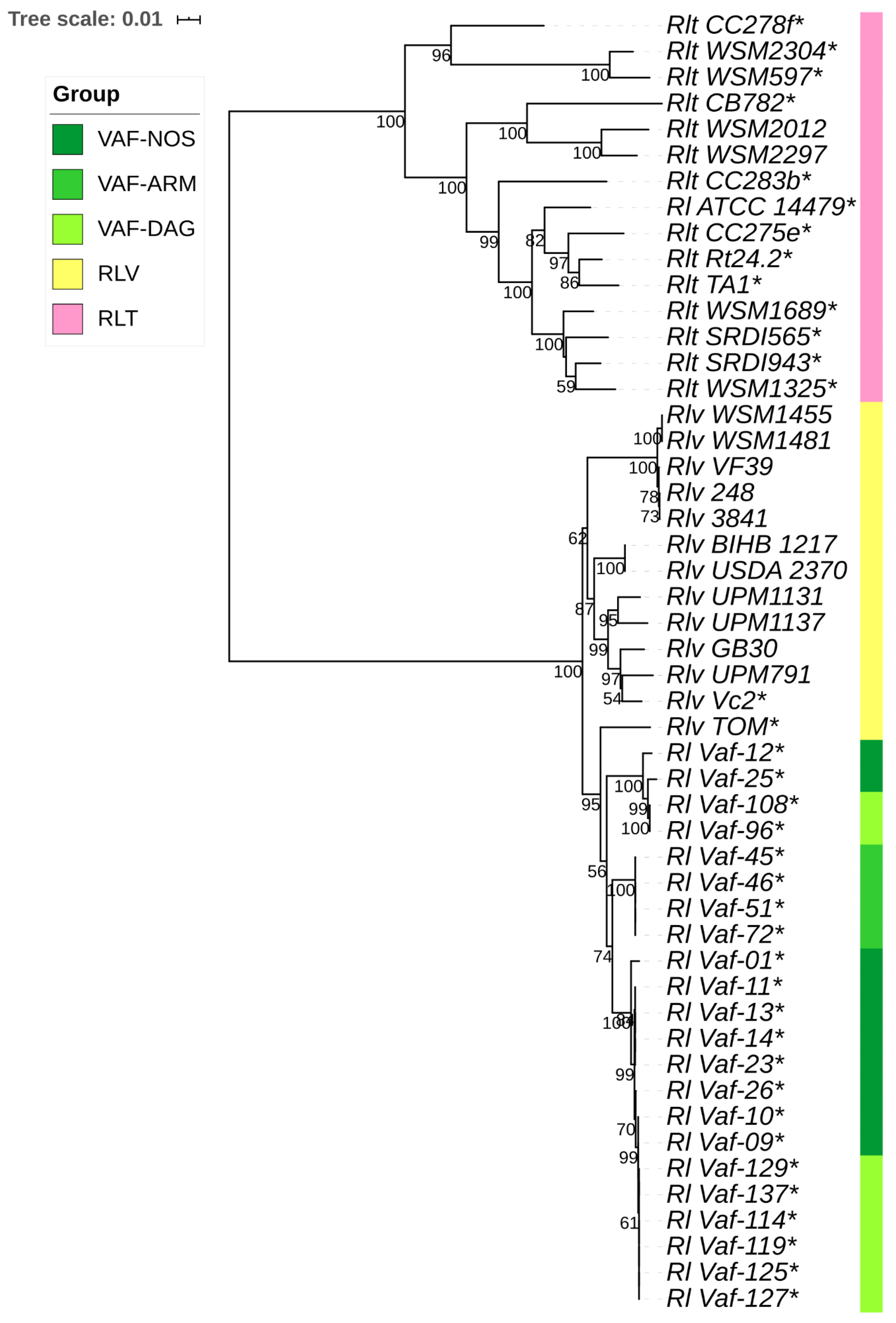
3.4. Divergence of Hkg and Sym Genes in R. leguminosarum
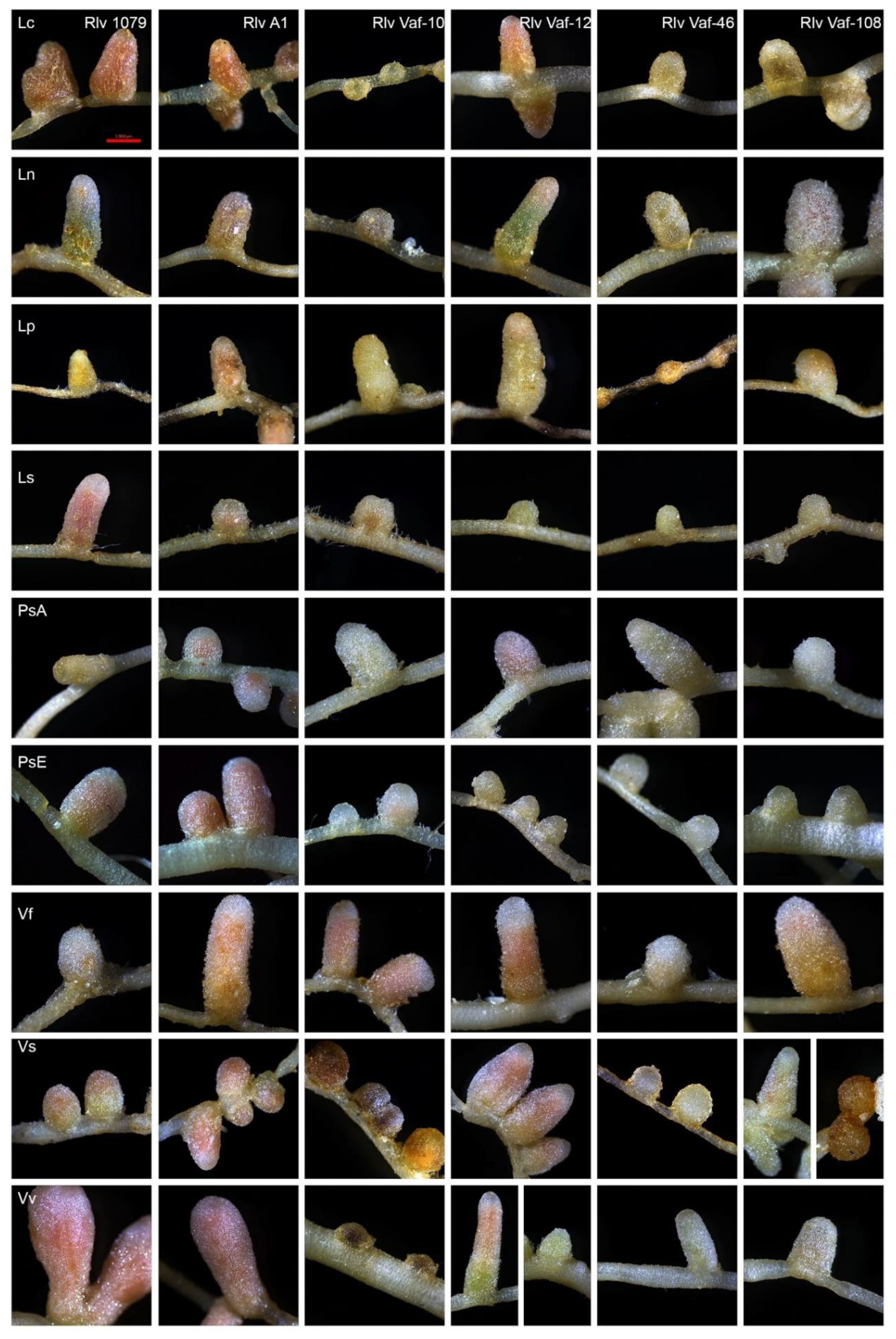
3.5. Sterile Tube Test Experiment
4. Discussion
4.1. Symbiotic Behavior of Isolates of V. formosa
4.2. Divergent Evolution within R. leguminosarum
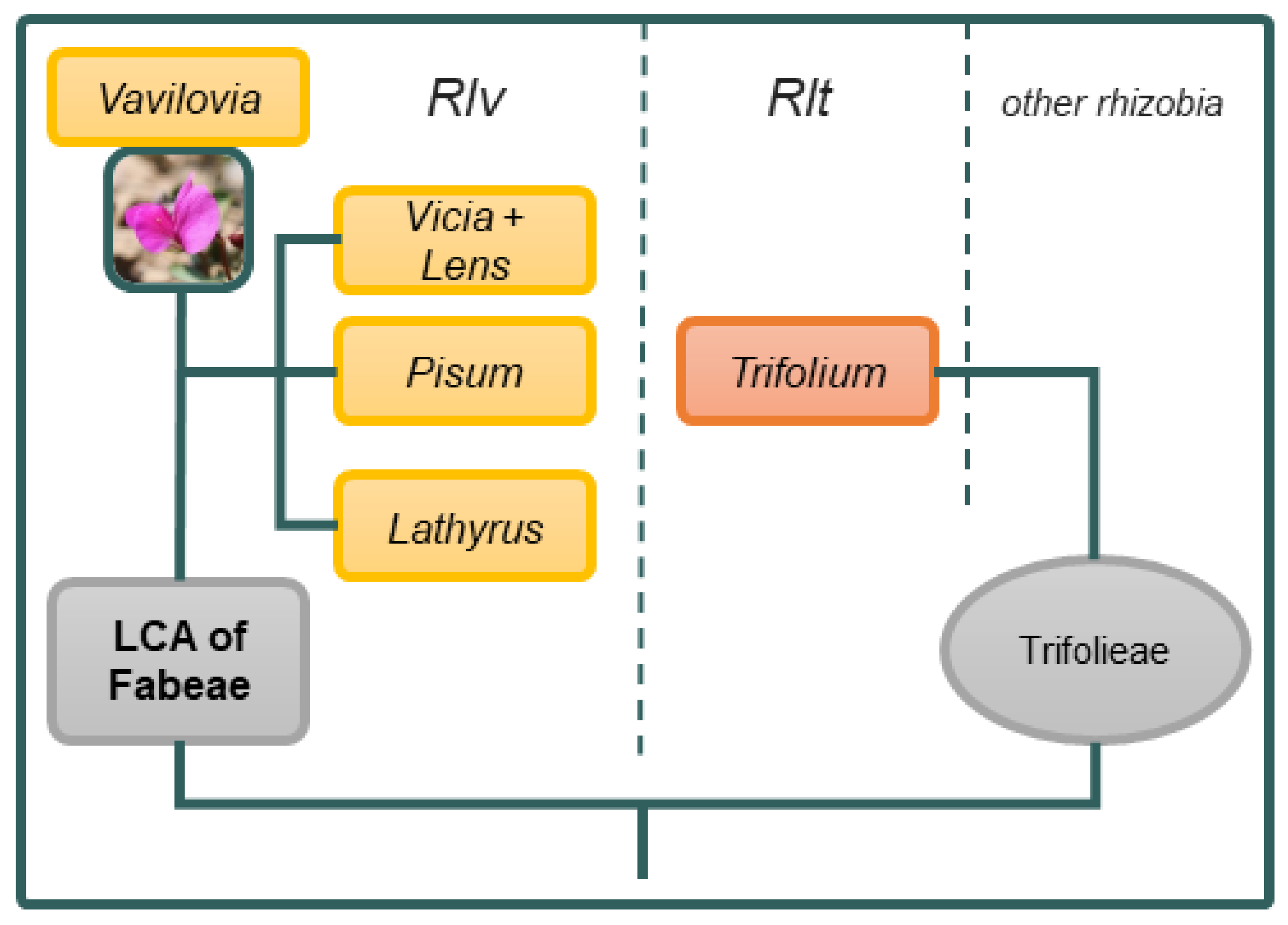
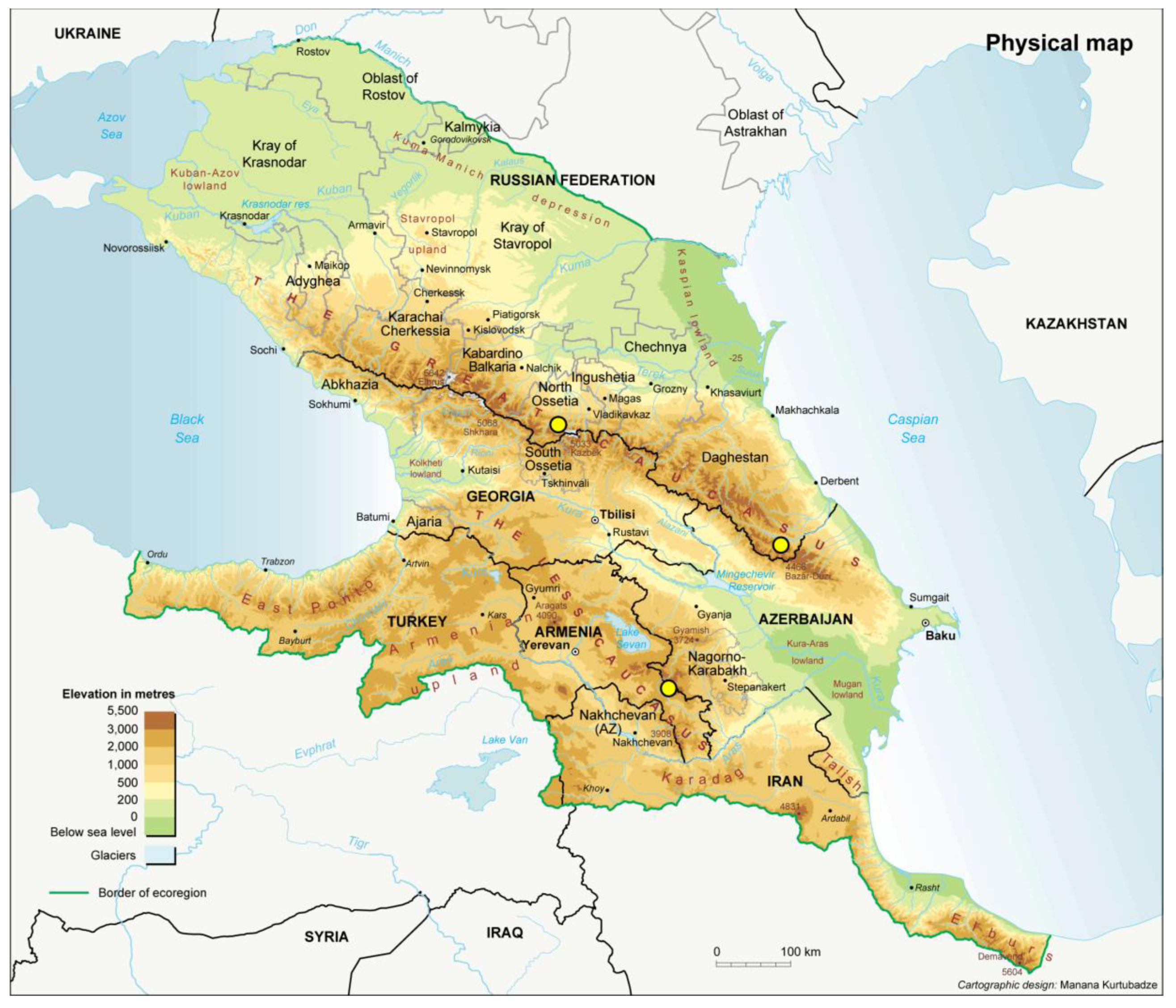
4.3. Role of Geographic Factors in the Diversification of Rhizobia
4.4. Evolutionary Status of Symbionts of V. formosa
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bailly, X.; Olivieri, I.; De Mita, S.; Cleyet-Marel, J.-C.; Béna, G. Recombination and selection shape the molecular diversity pattern of nitrogen-fixing Sinorhizobium sp. associated to Medicago. Mol. Ecol. 2006, 15, 2719–2734. [Google Scholar] [CrossRef] [PubMed]
- Burghardt, L.T.; Epstein, B.; Tiffin, P. Legacy of prior host and soil selection on rhizobial fitness in planta. Evolution 2019, 73, 2013–2023. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.; Vinuesa, P.; Eguiarte, L.E.; Souza, V.; Martínez-Romero, E. Evolutionary genetics and biogeographic structure of Rhizobium gallicum sensu lato, a widely distributed bacterial symbiont of diverse legumes. Mol. Ecol. 2005, 14, 4033–4050. [Google Scholar] [CrossRef] [PubMed]
- Young, J.P.W.; Crossman, L.C.; Johnston, A.W.; Thomson, N.R.; Ghazoui, Z.F.; Hull, K.H.; Wexler, M.; Curson, A.R.; Todd, J.D.; Poole, P.S.; et al. The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol. 2006, 7, R34. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, A.; Lum, M.R.; Downie, J.A. What Makes the Rhizobia-Legume Symbiosis So Special? Plant Physiol. 2002, 127, 1484–1492. [Google Scholar] [CrossRef]
- Mutch, L.A.; Young, J.P.W. Diversity and specificity of Rhizobium leguminosarum biovar viciae on wild and cultivated legumes. Mol. Ecol. 2004, 13, 2435–2444. [Google Scholar] [CrossRef]
- Young, J.P.W.; Johnston, A.W.B. The evolution of specificity in the legume-rhizobium symbiosis. Trends Ecol. Evol. 1989, 4, 341–349. [Google Scholar] [CrossRef]
- Laguerre, G.; Fernandez, M.P.; Edel, V.; Normand, P.; Amarger, N. Genomic heterogeneity among French Rhizobium strains isolated from Phaseolus vulgaris L. Int. J. Syst. Bacteriol. 1993, 43, 761–767. [Google Scholar] [CrossRef][Green Version]
- Jordan, D.C. Rhizobiaceae. In Bergey’s Manual of Systematic Bacteriology; Krieg, N.R., Holt, J.R., Eds.; Williams Wilkins: Baltimore, MD, USA, 1984; pp. 234–244. [Google Scholar]
- Kumar, N.; Lad, G.; Giuntini, E.; Kaye, M.E.; Udomwong, P.; Shamsani, N.J.; Young, J.P.; Bailly, X. Bacterial genospecies that are not ecologically coherent: Population genomics of Rhizobium leguminosarum. Open Biol. 2015, 5, 140133. [Google Scholar] [CrossRef]
- Rogel, M.A.; Ormerino-Orrillo, E.; Martinez-Romero, E. Symbiovars in rhizobia reflect bacterial adaptation to legumes. Syst. Appl. Microbiol. 2011, 34, 96–104. [Google Scholar] [CrossRef]
- Akopian, J.; Sarukhanyan, N.; Gabrielyan, I.; Vanyan, A.; Mikić, A.; Smýkal, P.; Kenicer, G.; Vishnyakova, M.; Sinjushin, A.; Demidenko, N.; et al. Reports on establishing an ex situ site for ‘beautiful’ vavilovia (Vavilovia formosa) in Armenia. Genet. Resour. Crop Evol. 2010, 57, 1127–1134. [Google Scholar] [CrossRef]
- Mikić, A.; Smýkal, P.; Kenicer, G.; Vishnyakova, M.; Sarukhanyan, N.; Akopian, J.; Vanyan, A.; Gabrielyan, I.; Smýkalová, I.; Sherbakova, E.; et al. Beauty will save the world, but will the world save beauty? The case of the highly endangered Vavilovia formosa (Stev.) Fed. Planta 2014, 240, 1139–1146. [Google Scholar] [CrossRef]
- Ochatt, S.; Conreux, C.; Smykalova, I.; Smykal, P.; Mikić, A. Developing biotechnology tools for ‘beautiful’ vavilovia (Vavilovia formosa), a legume crop wild relative with taxonomic and agronomic potential. PCTOC 2016, 127, 637–648. [Google Scholar] [CrossRef]
- Vishnyakova, M.; Burlyaeva, M.; Akopian, J.; Murtazaliev, R.; Mikič, A. Reviewing and updating the detected locations of beautiful vavilovia (Vavilovia formosa) on the Caucasus sensu stricto. Genet. Resour. Crop Evol. 2016, 63, 1085–1102. [Google Scholar] [CrossRef]
- Oskoueiyan, R.; Osaloo, S.K.; Maassoumi, A.A.; Nejadsattari, T.; Mozaffarian, V. Phylogenetic status of Vavilovia formosa (Fabaceae-Fabeae) based on nrDNA ITS and cpDNA sequences. Biochem. Syst. Ecol. 2010, 38, 313–319. [Google Scholar] [CrossRef]
- Mikić, A.; Smýkal, P.; Kenicer, G.; Vishnyakova, M.; Sarukhanyan, N.; Akopian, J.; Vanyan, A.; Gabrielyan, I.; Smýkalová, I.; Sherbakova, E.; et al. The bicentenary of the research on ‘beautiful’ vavilovia (Vavilovia formosa), a legume crop wild relative with taxonomic and agronomic potential. Bot. J. Linn. Soc. 2013, 172, 524–531. [Google Scholar] [CrossRef]
- Safronova, V.I.; Kimeklis, A.K.; Chizhevskaya, E.P.; Belimov, A.A.; Andronov, E.E.; Pinaev, A.G.; Pukhaev, A.R.; Popov, K.P.; Tikhonovich, I.A. Genetic diversity of rhizobia isolated from nodules of the relic species Vavilovia formosa (Stev.) Fed. Antonie Van Leeuwenhoek 2014, 105, 389–399. [Google Scholar] [CrossRef]
- Ovtsyna, A.O.; Rademaker, G.-J.; Esser, E.; Weinman, J.; Rolfe, B.G.; Tikhonovich, I.A.; Lugtenberg, B.J.J.; Thomas-Oates, J.E.; Spaink, H.P. Comparison of Characteristics of the nodX Genes from Various Rhizobium leguminosarum Strains. MPMI 1999, 12, 252–258. [Google Scholar] [CrossRef]
- Novikova, N.; Safronova, V. Transconjugants of Agrobacterium radiobacter harbouring sym genes of Rhizobium galegae can form an effective symbiosis with Medicago sativa. FEMS Microbiol. Lett. 1992, 93, 261–268. [Google Scholar] [CrossRef]
- Safronova, V.; Tikhonovich, I. Automated cryobank of microorganisms: Unique possibilities for long-term authorized depositing of commercial microbial strains. In Microbes in Applied Research: Current Advances and Challenges; Mendez-Vilas, A., Ed.; Formatex Research Center: Badajoz, Spain, 2012; pp. 331–334. [Google Scholar] [CrossRef]
- Fred, E.; Waksman, S.A. Laboratory Manual of General Microbiology; McGraw-Hill Book Company: New York, NY, USA, 1928; p. 33. [Google Scholar]
- Somasegaran, P.; Hoben, H.J. Isolating and Purifying Genomic DNA of Rhizobia Using a Large-Scale Method. In Handbook for Rhizobia; Garber, R.C., Ed.; Springer: New York, NY, USA, 1994; pp. 279–283. [Google Scholar]
- Malferrati, G.; Monferinin, P.; DeBlasio, P.; Diaferia, G.; Saltini, G.; Del Vecchio, E.; Rossi-Bernardi, L.; Biunno, I. High-quality genomic DNA from Human whole blood and mononuclear cells. BioTechniques 2002, 33, 1228–1230. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; Wiley: Chichester, UK, 1991; pp. 115–175. [Google Scholar]
- Stepkowski, T.; Czaplinska, M.; Miedzinska, K.; Moulin, L. The variable part of the dnaK gene as an alternative marker for phylogenetic studies of rhizobia and related alpha Proteobacteria. Syst. Appl. Microbiol. 2003, 26, 483–494. [Google Scholar] [CrossRef] [PubMed]
- Martens, M.; Delaere, M.; Coopman, R.; de Vos, P.; Gillis, M.; Willems, A. Multilocus sequence analysis of Ensifer and related taxa. Int. J. Syst. Evol. Microbiol. 2007, 57, 489–503. [Google Scholar] [CrossRef] [PubMed]
- Turner, S.L.; Young, J.P.W. The glutamine synthetases of rhizobia: Phylogenetics and evolutionary implications. Mol. Biol. Evol. 2000, 17, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Haukka, K.; Lindstrom, K.; Young, J.P. Three phylogenetic groups of nodA and nifH genes in Sinorhizobium and Mesorhizobium isolates from leguminous trees growing in Africa and Latin America. Appl. Environ. Microbiol. 1998, 64, 419–426. [Google Scholar]
- Laguerre, G.; Nour, S.M.; Macheret, V.; Sanjuan, J.; Drouin, P.; Amarger, N. Classification of rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts. Microbiology 2001, 147, 981–993. [Google Scholar] [CrossRef]
- Laguerre, G.; Mavingui, P.; Allard, M.R.; Charnay, M.P.; Louvrier, P.; Mazurier, S.I.; Rigottier-Gois, L.; Amarger, N. Typing of rhizobia by PCR DNA fingerprinting and PCR-restriction fragment length polymorphism analysis of chromosomal and symbiotic gene regions: Application to Rhizobium leguminosarum and its different biovars. Appl. Environ. Microbiol. 1996, 62, 2029–2036. [Google Scholar]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; The UGENE Team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- StatSoft, Inc. STATISTICA (Data Analysis Software System), Version 12, 2014. Available online: http://www.statsoft.com (accessed on 25 July 2017).
- RStudio Team. RStudio: Integrated Development for R; RStudio, Inc.: Boston, MA, USA, 2016; Available online: http://www.rstudio.com (accessed on 10 March 2019).
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- BioNumerics: BioNumerics Version 7.6.3 Created by Applied Maths NV. Available online: http://www.applied-maths.com (accessed on 5 February 2018).
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000; pp. 240–297. [Google Scholar]
- Chetkova, S.A.; Tikhonovich, I.A. Isolation and study of Rhizobium leguminosarum strains effective on peas from Afghanistan. Mikrobiologiya 1986, 55, 143–147. [Google Scholar]
- Provorov, N.A.; Simarov, B.V. Genetic Variation in Alfalfa, Sweet Clover and Fenugreek for the Activity of Symbiosis with Rhizobium meliloti. Plant Breed. 1990, 105, 300–310. [Google Scholar] [CrossRef]
- Kimeklis, A.K.; Safronova, V.I.; Kuznetsova, I.G.; Sazanova, A.L.; Belimov, A.A.; Pinaev, A.G.; Chizhevskaya, E.P.; Pukhaev, A.R.; Popov, K.P.; Andronov, E.E.; et al. Phylogenetic analysis of Rhizobium strains, isolated from nodules of Vavilovia formosa (Stev.) Fed. Agric. Biol. 2015, 50, 655–664. [Google Scholar] [CrossRef]
- Geurts, R.; Heidstra, R.; Hadri, A.; Downie, J.; Franssen, H.; Kammen, A.; Bisseling, T. Sym2 of Pea Is Involved in a Nodulation Factor-Perception Mechanism That Controls the Infection Process in the Epidermis. Plant Physiol. 1997, 115, 351–359. [Google Scholar] [CrossRef]
- Kimeklis, A.K.; Kuznetsova, I.G.; Sazanova, A.L.; Safronova, V.I.; Belimov, A.A.; Onishchuk, O.P.; Kurchak, O.N.; Aksenova, T.S.; Pinaev, A.G.; Andronov, E.E.; et al. Divergent Evolution of Symbiotic Bacteria: Rhizobia of the Relic Legume Vavilovia formosa Form an Isolated Group within Rhizobium leguminosarum bv. viciae. Russ. J. Genet. 2018, 54, 866–870. [Google Scholar] [CrossRef]
- Chirak, E.R.; Kimeklis, A.K.; Kopat, V.V.; Karasev, E.S.; Safronova, V.I.; Belimov, A.A.; Aksenova, T.S.; Kabilov, M.R.; Provorov, N.A.; Andronov, E.E. Search for ancestral features in genomes of Rhizobium leguminosarum bv. viciae strains isolated from the relict legume Vavilovia formosa. Manuscript in preparation.
- Karasev, E.S.; Andronov, E.E.; Aksenova, T.S.; Tupikin, A.E.; Provorov, N.A. Evolution of goat’s rue rhizobia (Neorhizobium galegae): An analysis of the polymorphism of the nitrogen fixation genes and the genes of nodule formation. Russ. J. Genet. 2019, 55, 234–238. [Google Scholar] [CrossRef]
| Genes | Primers | Sequences | T °C | References |
|---|---|---|---|---|
| 16S rDNA | 27F | AGAGTTTGATCMTGGCTCAG | 55 | [25] |
| 1525R | AAGGAGGTGWTCCARCC | |||
| dnaK | dnaK1466F | AAGGARCANCAGATCCGCATCCA | 62 | [26] |
| dnaK1777R | TASATSGCCTSRCCRAGCTTCAT | |||
| gltA | gltA428F | CSGCCTTCTAYCAYGACTC | 53 | [27] |
| gltA1111R | GGGAGCCSAKCGCCTTCAG | |||
| glnII | GSII-1 | AACGCAGATCAAGGAATTCG | 55 | [28] |
| GSII-2 | ATGCCCGAGCCGTTCCAGTC | |||
| nodA | nodA-1 | TGCRGTGGAARNTRNNCTGGGAAA | 49 | [29] |
| nodA-2 | GGNCCGTCRTCRAAWGTCARGTA | |||
| nodC | nodCF | AYGTHGTYGAYGACGGTTC | 57 | [30] |
| nodCI | CGYGACAGCCANTCKCTATTG | |||
| nodD | NBA12 | GGATSGCAATCATCTAYRGMRTGG | 57 | [31] |
| NBF12′ | GGATCRAAAGCATCCRCASTATGG | |||
| nodX | oMP199 | CCATGGGACCATCCAATGAAC | 53 | [19] |
| oMP196 | TTAAGCGACGGAAAGCCTTC | |||
| nifH | nifHF | TACGGNAARGGSGGNATCGGCAA | 62 | [30] |
| nifHI | AGCATGTCYTCSAGYTCNTCCA |
| Gene | Mantel Statistic r | p-Value |
|---|---|---|
| 16S rRNA | 0.0192 | 0.2978 |
| dnaK | 0.3379 | 0.0004 |
| glnII | 0.4051 | 0.0001 |
| gltA | 0.4494 | 0.0001 |
| nodA | 0.0173 | 0.2807 |
| nodC | 0.1266 | 0.039 |
| nodD | 0.2553 | 0.0037 |
| nodX | 0.3705 | 0.0007 |
| nifH | 0.1899 | 0.0077 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kimeklis, A.K.; Chirak, E.R.; Kuznetsova, I.G.; Sazanova, A.L.; Safronova, V.I.; Belimov, A.A.; Onishchuk, O.P.; Kurchak, O.N.; Aksenova, T.S.; Pinaev, A.G.; et al. Rhizobia Isolated from the Relict Legume Vavilovia formosa Represent a Genetically Specific Group within Rhizobium leguminosarum biovar viciae. Genes 2019, 10, 991. https://doi.org/10.3390/genes10120991
Kimeklis AK, Chirak ER, Kuznetsova IG, Sazanova AL, Safronova VI, Belimov AA, Onishchuk OP, Kurchak ON, Aksenova TS, Pinaev AG, et al. Rhizobia Isolated from the Relict Legume Vavilovia formosa Represent a Genetically Specific Group within Rhizobium leguminosarum biovar viciae. Genes. 2019; 10(12):991. https://doi.org/10.3390/genes10120991
Chicago/Turabian StyleKimeklis, Anastasiia K., Elizaveta R. Chirak, Irina G. Kuznetsova, Anna L. Sazanova, Vera I. Safronova, Andrey A. Belimov, Olga P. Onishchuk, Oksana N. Kurchak, Tatyana S. Aksenova, Alexander G. Pinaev, and et al. 2019. "Rhizobia Isolated from the Relict Legume Vavilovia formosa Represent a Genetically Specific Group within Rhizobium leguminosarum biovar viciae" Genes 10, no. 12: 991. https://doi.org/10.3390/genes10120991
APA StyleKimeklis, A. K., Chirak, E. R., Kuznetsova, I. G., Sazanova, A. L., Safronova, V. I., Belimov, A. A., Onishchuk, O. P., Kurchak, O. N., Aksenova, T. S., Pinaev, A. G., Andronov, E. E., & Provorov, N. A. (2019). Rhizobia Isolated from the Relict Legume Vavilovia formosa Represent a Genetically Specific Group within Rhizobium leguminosarum biovar viciae. Genes, 10(12), 991. https://doi.org/10.3390/genes10120991






