Characterization and Functional Divergence of a Novel DUF668 Gene Family in Rice Based on Comprehensive Expression Patterns
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of the DUF668 Genes
2.2. Phylogenetic Analysis, Gene Structure, and Conserved Motifs
2.3. Chromosomal Locations, Gene Duplication Events, and Cis-acting Elements Analysis
2.4. Plant Materials and Treatments
2.5. RNA Isolation and RT-PCR Analysis
3. Results
3.1. Genome-Wide Identification and Characterization of DUF668 Genes in Rice
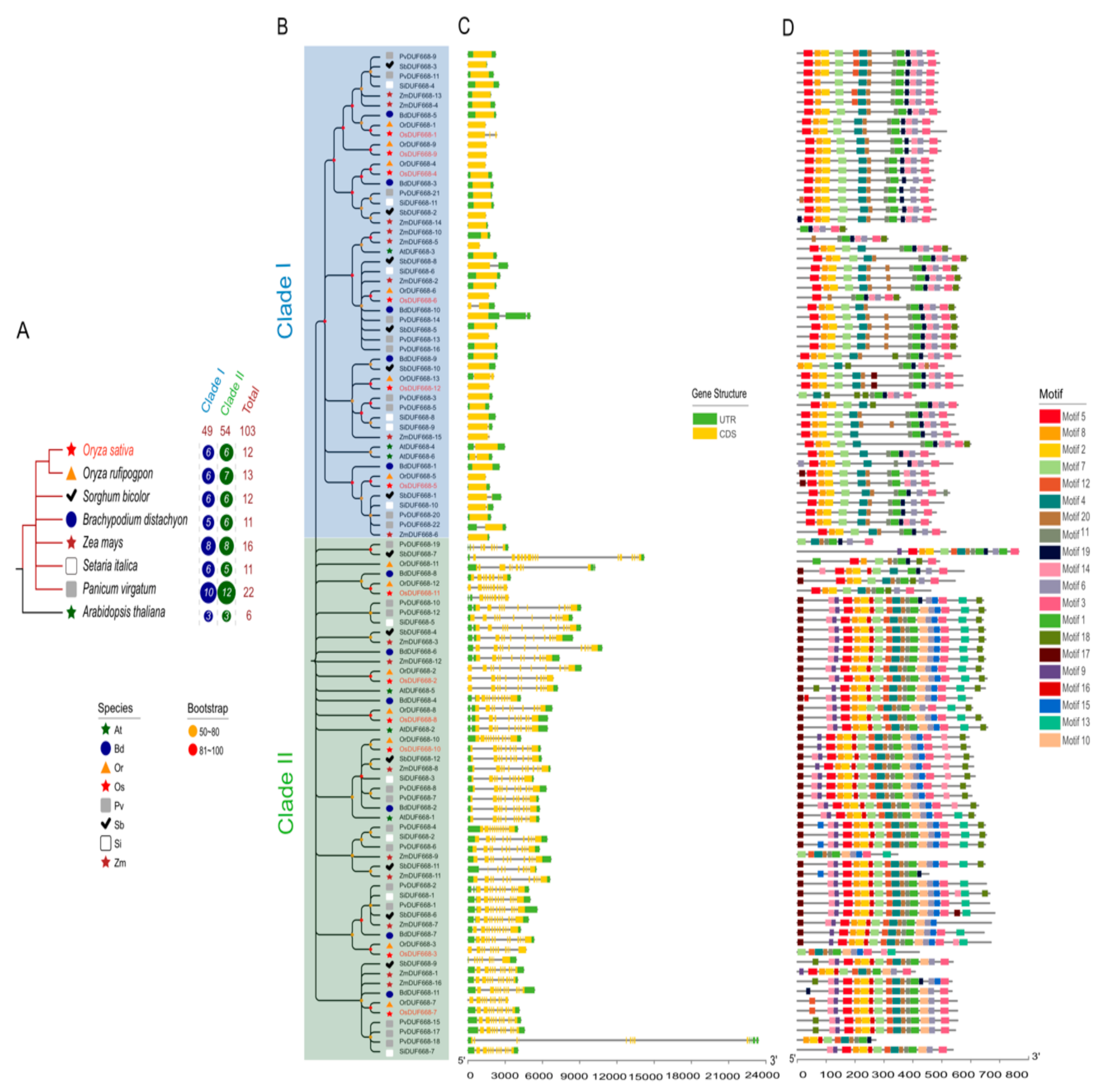
3.2. Phylogenetic Analysis, Gene Structure, and Motif Composition of the DUF668s in Eight Plants
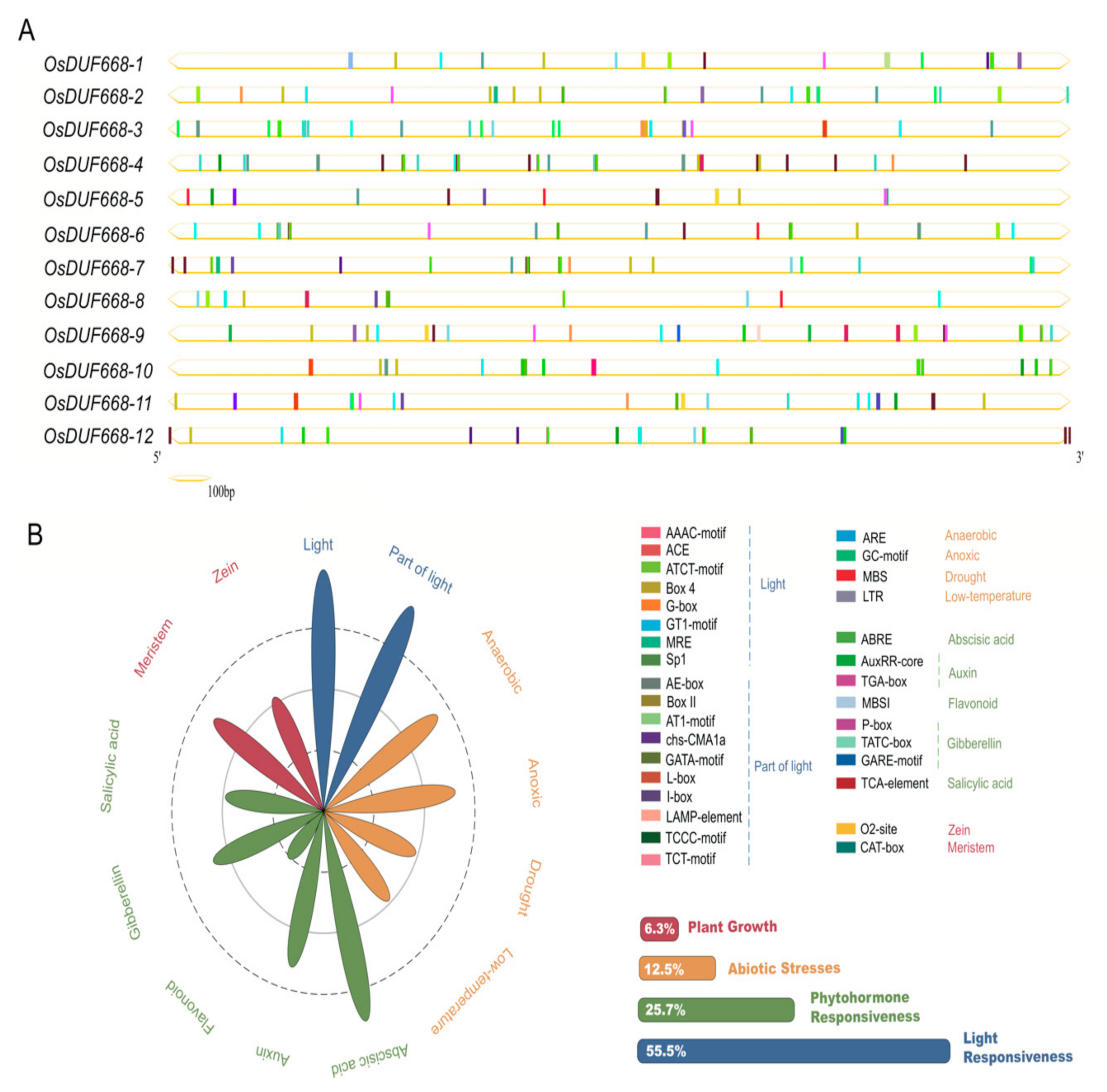
3.3. Cis-acting Elements Prediction of DUF668 Genes in Rice
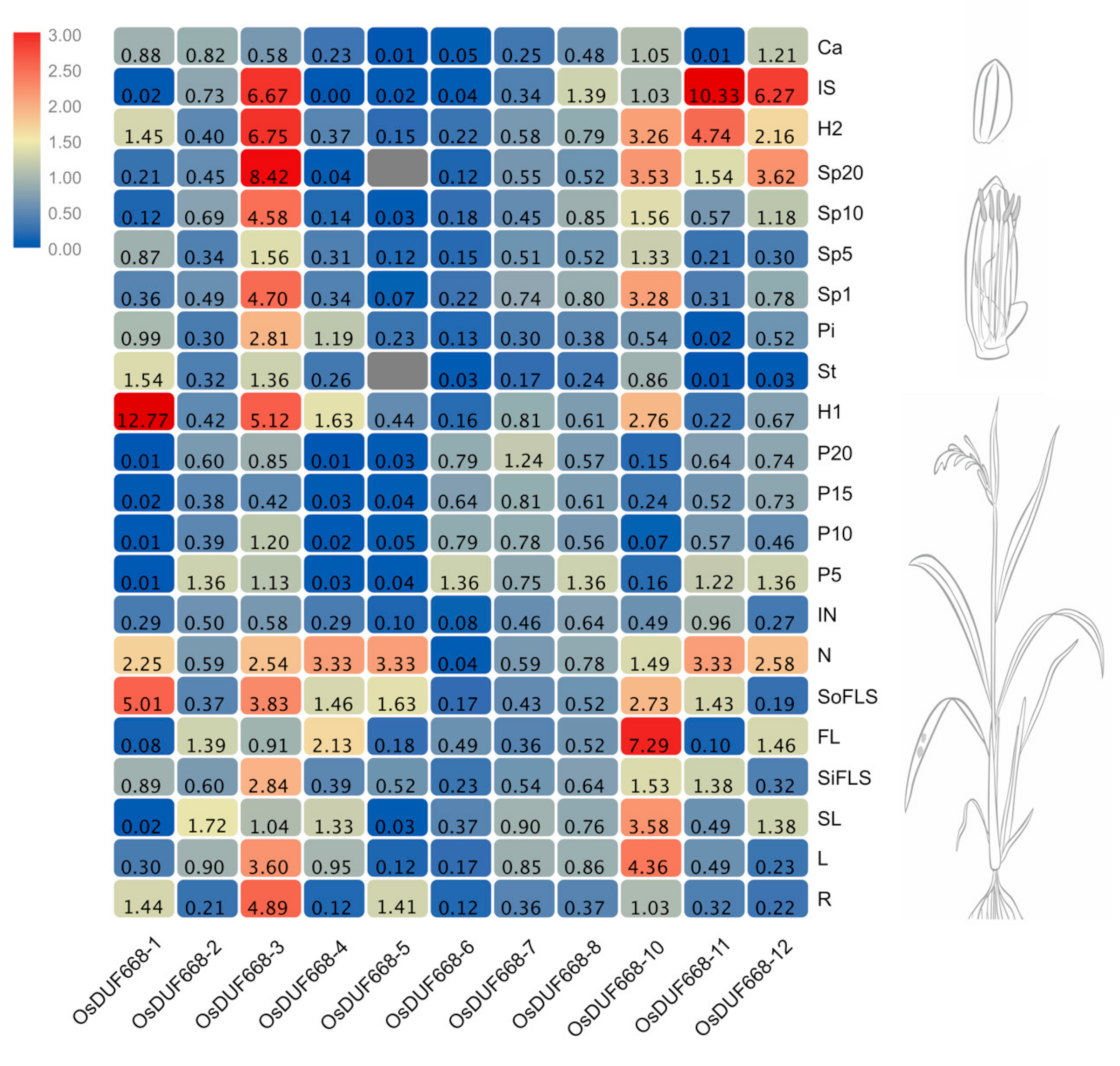
3.4. Tissue-Specific Expression Patterns of OsDUF668s
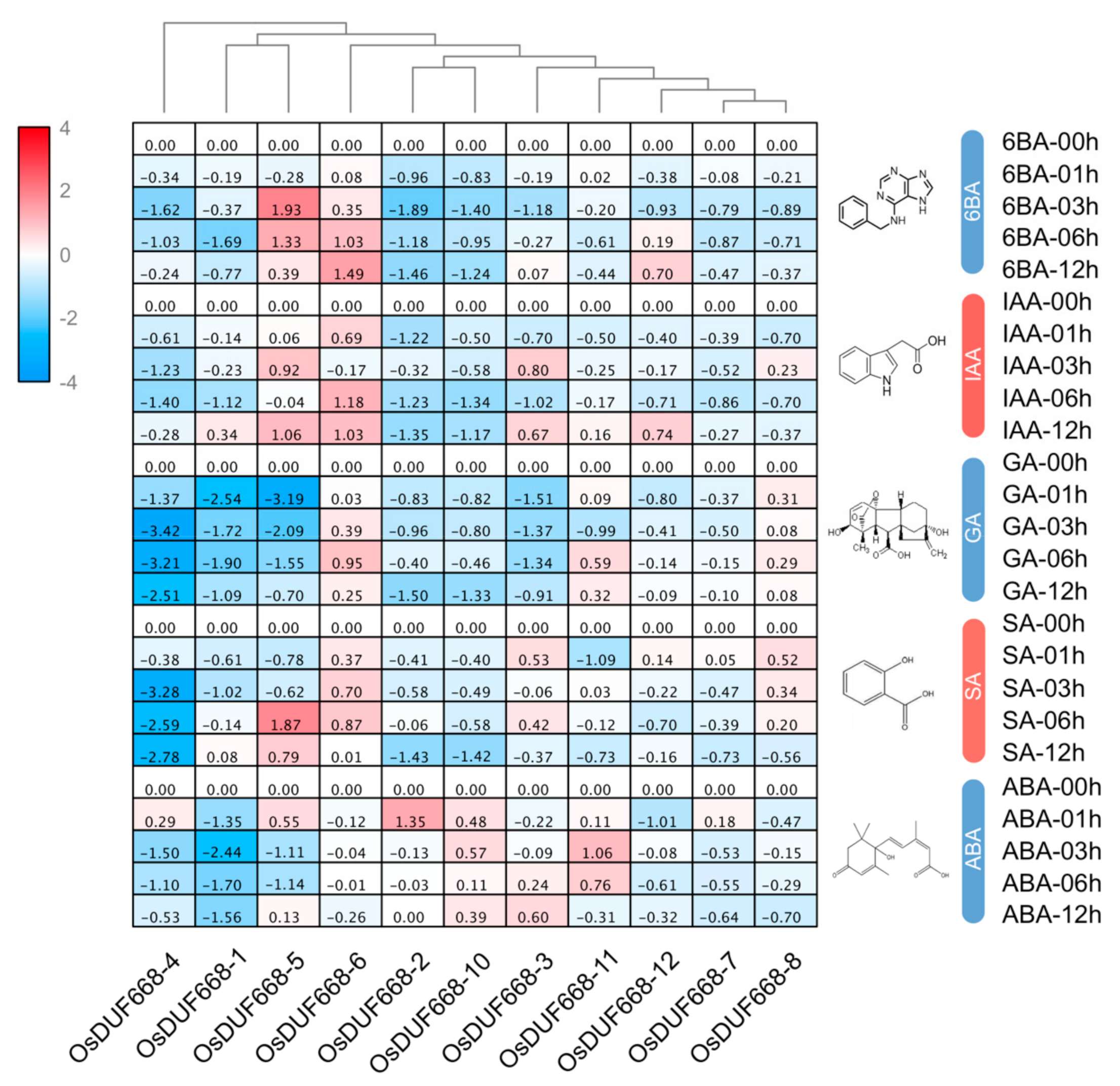
3.5. Expression Profiles of OsDUF668s in Response to Plant Hormone
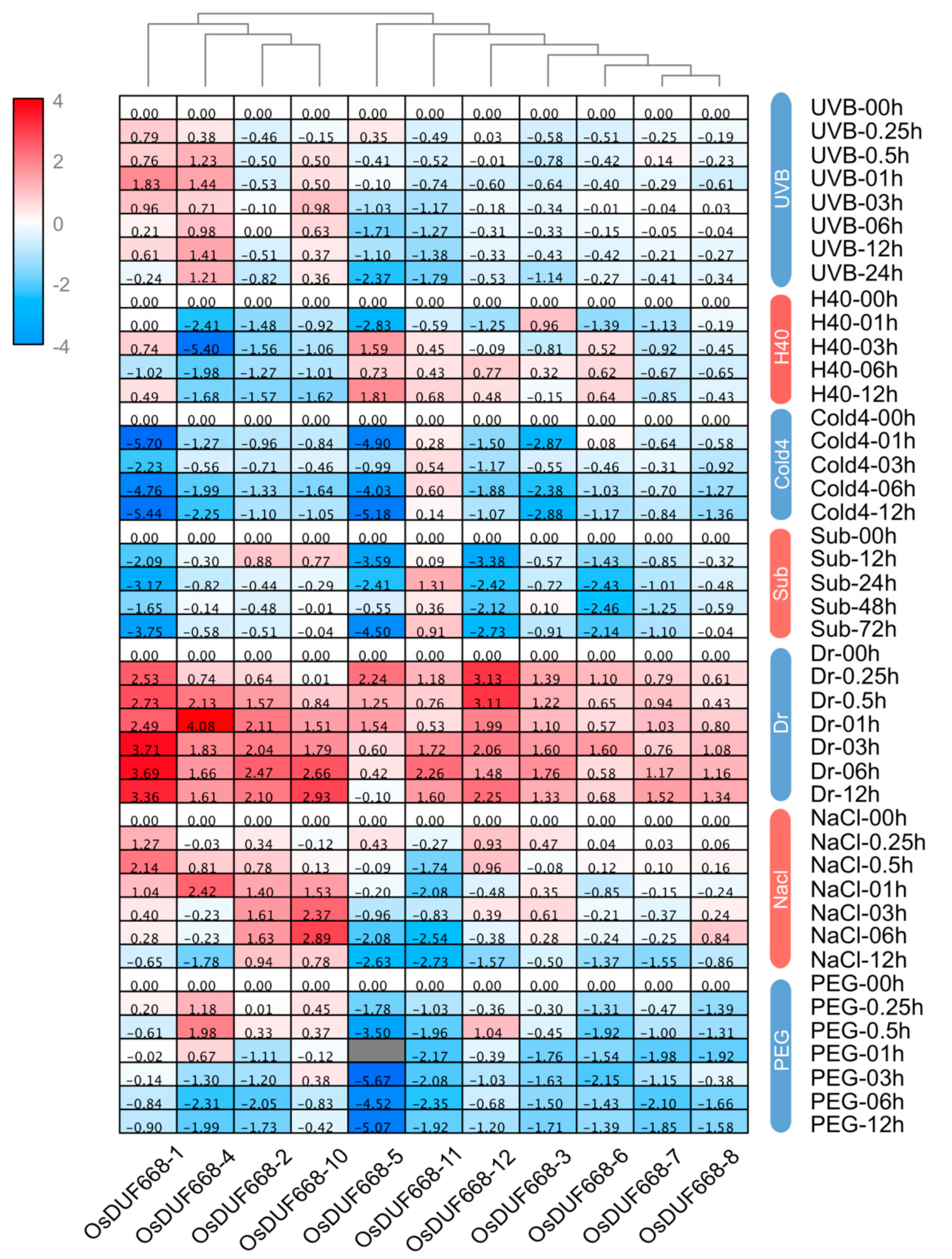
3.6. Transcriptional Responses of OsDUF668s Facing Environmental Stresses
3.7. Transcriptional Responses of OsDUF668s Facing Pathogen-Defense Related Stresses
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ohta, T. Evolution of gene families. Gene 2000, 259, 45–52. [Google Scholar] [CrossRef]
- Bateman, A.; Coggill, P.; Finn, R.D. DUFs: Families in search of function. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 1148–1152. [Google Scholar] [CrossRef]
- Simm, R.; Morr, M.; Kader, A.; Nimtz, M.; Römling, U. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessibility to motility. Mol. Microbiol. 2004, 53, 1123–1134. [Google Scholar] [CrossRef] [PubMed]
- Karras, G.I.; Kustatscher, G.; Buhecha, H.R.; Allen, M.D.; Pugieux, C.; Sait, F.; Bycroft, M.; Ladurner, A.G. The macro domain is an ADP-ribose binding module. EMBO J. 2005, 24, 1911–1920. [Google Scholar] [CrossRef] [PubMed]
- Dlakić, M. DUF283 domain of dicer proteins has a double-stranded RNA-binding fold. Bioinformatics 2006, 22, 2711–2714. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Chen, F.; Huan, X.; Machida, S.; Song, J.; Yuan, Y.A. Structure of the Arabidopsis thaliana DCL4 DUF283 domain reveals a noncanonical double-stranded RNA-binding fold for protein—Protein interaction. Rna 2010, 16, 474–481. [Google Scholar] [CrossRef] [PubMed]
- Gholizadeh, A. DUF538 protein superfamily is predicted to be chlorophyll hydrolyzing enzymes in plants. Physiol. Mol. Biol. Plants 2016, 22, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Gholizadeh, A. Chlorophyll binding ability of non-chloroplastic DUF538 protein superfamily in plants. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2018, 88, 967–976. [Google Scholar] [CrossRef]
- Palmeros-Suárez, P.A.; Massange-Sánchez, J.A.; Sánchez-Segura, L.; Martínez-Gallardo, N.A.; Espitia Rangel, E.; Gómez-Leyva, J.F.; Délano-Frier, J.P. AhDGR2, an amaranth abiotic stress-induced DUF642 protein gene, modifies cell wall structure and composition and causes salt and ABA hyper-sensibility in transgenic Arabidopsis. Planta 2017, 245, 623–640. [Google Scholar] [CrossRef]
- Harada, K.; Yamashita, E.; Inoue, K.; Yamaguchi, K.; Fujiwara, T.; Nakagawa, A.; Kawasaki, T.; Kojima, C. Plant-specific DUF1110 protein from Oryza sativa: Expression, purification and crystallization. Acta Crystallogr. Sect. Struct. Biol. Commun. 2016, 72, 480–484. [Google Scholar] [CrossRef]
- Bakolitsa, C.; Bateman, A.; Jin, K.K.; McMullan, D.; Krishna, S.S.; Miller, M.D.; Abdubek, P.; Acosta, C.; Astakhova, T.; Axelrod, H.L.; et al. The structure of Jann-2411 (DUF1470) from Jannaschia sp. at 1.45 Å resolution reveals a new fold (the ABATE domain) and suggests its possible role as a transcription regulator. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 1198–1204. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Shen, R.; Chen, L.T.; Liu, Y.G. Characterization of a novel DUF1618 gene family in rice. J. Integr. Plant Biol. 2014, 56, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.B.; Bogdanove, A.J.; Sessa, G. Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 2003, 54, 23–61. [Google Scholar] [CrossRef] [PubMed]
- Heese, A.; Ludwig, A.A.; Jones, J.D.G. Rapid phosphorylation of a syntaxin during the Avr9/Cf-9-race-specific signaling pathway. Plant Physiol. 2005, 138, 2406–2416. [Google Scholar] [CrossRef] [PubMed]
- Durrant, W.E.; Rowland, O.; Piedras, P.; Hammond-Kosack, K.E.; Jones, J.D.G. cDNA-AFLP reveals a striking overlap in race-specific resistance and wound response gene expression profiles. Plant Cell 2000, 12, 963–977. [Google Scholar] [CrossRef]
- Van Den Burg, H.A.; Tsitsigiannis, D.I.; Rowland, O.; Lo, J.; Rallapalli, G.; MacLean, D.; Takken, F.L.W.; Jones, J.D.G. The F-box protein ACRE189/ACIF1 regulates cell death and defense responses activated during pathogen recognition in tobacco and tomato. Plant Cell 2008, 20, 697–719. [Google Scholar] [CrossRef]
- Rowland, O.; Ludwig, A.A.; Merrick, C.J.; Baillieul, F.; Tracy, F.E.; Durrant, W.E.; Fritz-Laylin, L.; Nekrasov, V.; Sjölander, K.; Yoshioka, H.; et al. Functional analysis of Avr9/Cf-9 rapidly elicited genes identifies a protein kinase, ACIK1, that is essential for full Cf-9-dependent disease resistance in tomato. Plant Cell 2005, 17, 295–310. [Google Scholar] [CrossRef]
- Savatin, D.V.; Gramegna, G.; Modesti, V.; Cervone, F. Wounding in the plant tissue: The defense of a dangerous passage. Front. Plant Sci. 2014, 5, 1–11. [Google Scholar] [CrossRef]
- Walley, J.W.; Coughlan, S.; Hudson, M.E.; Covington, M.F.; Kaspi, R.; Banu, G.; Harmer, S.L.; Dehesh, K. Mechanical stress induces biotic and abiotic stress responses via a novel cis-element. PLoS Genet. 2007, 3, 1800–1812. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Chou, K.C.; Shen, H. Bin Cell-PLoc: A package of web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Xia, R.; Chen, H.; He, Y. TBtools, a toolkit for biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv 2018. [Google Scholar] [CrossRef]
- He, Z.; Zhang, H.; Gao, S.; Lercher, M.J.; Chen, W.H.; Hu, S. Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016, 44, W236–W241. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, 1–14. [Google Scholar] [CrossRef]
- Lescot, M. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Y.; Yang, J.; Hu, K.; An, B.; Deng, X.; Li, Y. Reliable selection and holistic stability evaluation of reference genes for rice under 22 different experimental conditions. Appl. Biochem. Biotechnol. 2016, 179, 753–775. [Google Scholar] [CrossRef]
- Jain, M.; Nijhawan, A.; Tyagi, A.K.; Khurana, J.P. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2006, 345, 646–651. [Google Scholar] [CrossRef]
- Song, Y.; Wang, L.; Xiong, L. Comprehensive expression profiling analysis of OsIAA gene family in developmental processes and in response to phytohormone and stress treatments. Planta 2009, 229, 577–591. [Google Scholar] [CrossRef] [PubMed]
- Hazman, M.; Sühnel, M.; Schäfer, S.; Zumsteg, J.; Lesot, A.; Beltran, F.; Marquis, V.; Herrgott, L.; Miesch, L.; Riemann, M.; et al. Characterization of jasmonoyl-isoleucine (JA-Ile) hormonal catabolic pathways in rice upon wounding and salt stress. Rice 2019, 12, 45. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Wang, L.; Pan, Q. Identification and characterization of a new blast resistance gene located on rice chromosome 1 through linkage and differential analyses. Phytopathology 2004, 94, 515–519. [Google Scholar] [CrossRef] [PubMed]
- Peterson, P.M. Poaceae (Gramineae). Encycl. Life Sci. 2013. [Google Scholar] [CrossRef]
- Soltani, B.M.; Ehlting, J.; Hamberger, B.; Douglas, C.J. Multiple cis-regulatory elements regulate distinct and complex patterns of developmental and wound-induced expression of Arabidopsis thaliana 4CL gene family members. Planta 2006, 224, 1226–1238. [Google Scholar] [CrossRef]
- Walther, D.; Brunnemann, R.; Selbig, J. The regulatory code for transcriptional response diversity and its relation to genome structural properties in A. thaliana. PLoS Genet. 2007, 3, 216–229. [Google Scholar] [CrossRef]
- Abdullah, M.; Cheng, X.; Cao, Y.; Su, X.; Manzoor, M.A.; Gao, J.; Cai, Y.; Lin, Y. Zinc finger-homeodomain transcriptional factors (ZHDs) in upland cotton (Gossypium hirsutum): Genome-wide identification and expression analysis in fiber development. Front. Genet. 2018, 9, 357. [Google Scholar] [CrossRef]
- Marciniak, K.; Przedniczek, K. Comprehensive insight into gibberellin-and jasmonate-mediated stamen development. Genes 2019, 10, 811. [Google Scholar] [CrossRef]
- Deng, X.; An, B.; Zhong, H.; Yang, J.; Kong, W.; Li, Y. A novel insight into functional divergence of the MST gene family in rice based on comprehensive expression patterns. Genes 2019, 10, 239. [Google Scholar] [CrossRef]
- Zhong, H.; Kong, W.; Gong, Z.; Fang, X.; Deng, X.; Liu, C.; Li, Y. Evolutionary analyses reveal diverged patterns of SQUAMOSA promoter binding protein-like (Spl) gene family in Oryza genus. Front. Plant Sci. 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Guo, M.; Chen, K.; Zhang, P. Transcriptome profile analysis of resistance induced by burdock fructooligosaccharide in tobacco. J. Plant Physiol. 2012, 169, 1511–1519. [Google Scholar] [CrossRef] [PubMed]

| Gene ID | Gene Name | Clade | Chr | mRNA Length (bp) | Protein (aa) | MW (Da) | Theoretical pI | Predicted Location | Annotation |
|---|---|---|---|---|---|---|---|---|---|
| LOC_Os01g62670 | OsDUF668-1 | clade-I | Chr1 | 2317 | 516 | 55952.4 | 8.31 | Chloroplast | avr9/Cf-9 rapidly elicited protein |
| LOC_Os01g65330 | OsDUF668-2 | clade-II | Chr1 | 7233 | 656 | 73065.2 | 9.86 | Nucleus | expressed protein |
| LOC_Os02g34650 | OsDUF668-3 | clade-II | Chr2 | 3899 | 422 | 46119.6 | 10.36 | Chloroplast | expressed protein |
| LOC_Os03g16390 | OsDUF668-4 | clade-I | Chr3 | 1936 | 471 | 52064.3 | 10.07 | Peroxisome | avr9/Cf-9 rapidly elicited protein |
| LOC_Os03g64130 | OsDUF668-5 | clade-I | Chr3 | 1757 | 476 | 52051.3 | 10.31 | Chloroplast | avr9/Cf-9 rapidly elicited protein |
| LOC_Os04g08764 | OsDUF668-6 | clade-I | Chr4 | 2151 | 357 | 38783 | 7.39 | Chloroplast | avr9/Cf-9 rapidly elicited protein |
| LOC_Os04g35410 | OsDUF668-7 | clade-II | Chr4 | 4154 | 554 | 61523.2 | 6.37 | Chloroplast | expressed protein |
| LOC_Os05g35530 | OsDUF668-8 | clade-II | Chr5 | 6418 | 641 | 71108.8 | 9.98 | Cytoplasm | expressed protein |
| LOC_Os05g38320 | OsDUF668-9 | clade-I | Chr5 | 1490 | 497 | 53685 | 10.21 | Chloroplast | avr9/Cf-9 rapidly elicited protein |
| LOC_Os06g50220 | OsDUF668-10 | clade-II | Chr6 | 5930 | 598 | 67106.3 | 9.14 | Nucleus | expressed protein |
| LOC_Os11g07840 | OsDUF668-11 | clade-II | Chr11 | 3264 | 462 | 51730.7 | 7.38 | Mitochondrion | expressed protein |
| LOC_Os12g05180 | OsDUF668-12 | clade-I | Chr12 | 1970 | 573 | 62417.4 | 10.5 | Chloroplast | avr9/Cf-9 rapidly elicited protein |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, H.; Zhang, H.; Guo, R.; Wang, Q.; Huang, X.; Liao, J.; Li, Y.; Huang, Y.; Wang, Z. Characterization and Functional Divergence of a Novel DUF668 Gene Family in Rice Based on Comprehensive Expression Patterns. Genes 2019, 10, 980. https://doi.org/10.3390/genes10120980
Zhong H, Zhang H, Guo R, Wang Q, Huang X, Liao J, Li Y, Huang Y, Wang Z. Characterization and Functional Divergence of a Novel DUF668 Gene Family in Rice Based on Comprehensive Expression Patterns. Genes. 2019; 10(12):980. https://doi.org/10.3390/genes10120980
Chicago/Turabian StyleZhong, Hua, Hongyu Zhang, Rong Guo, Qiang Wang, Xiaoping Huang, Jianglin Liao, Yangsheng Li, Yingjin Huang, and Zhaohai Wang. 2019. "Characterization and Functional Divergence of a Novel DUF668 Gene Family in Rice Based on Comprehensive Expression Patterns" Genes 10, no. 12: 980. https://doi.org/10.3390/genes10120980
APA StyleZhong, H., Zhang, H., Guo, R., Wang, Q., Huang, X., Liao, J., Li, Y., Huang, Y., & Wang, Z. (2019). Characterization and Functional Divergence of a Novel DUF668 Gene Family in Rice Based on Comprehensive Expression Patterns. Genes, 10(12), 980. https://doi.org/10.3390/genes10120980







