Sp1 Mediates the Constitutive Expression and Repression of the PDSS2 Gene in Lung Cancer Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture, Transient Transfection, and Mithramycin A Treatment
2.2. Reporter Plasmid Constructions
2.3. Site-Directed Mutagenesis
2.4. Luciferase Reporter Assay
2.5. DNA Sequence Alignment and Database Analysis
2.6. Chromatin Immunoprecipitation
2.7. RT-PCR
2.8. Western Blotting
2.9. Expression Analysis of Sp1 and PDSS2 in Lung Cancer
2.10. Statistical Analysis
3. Results
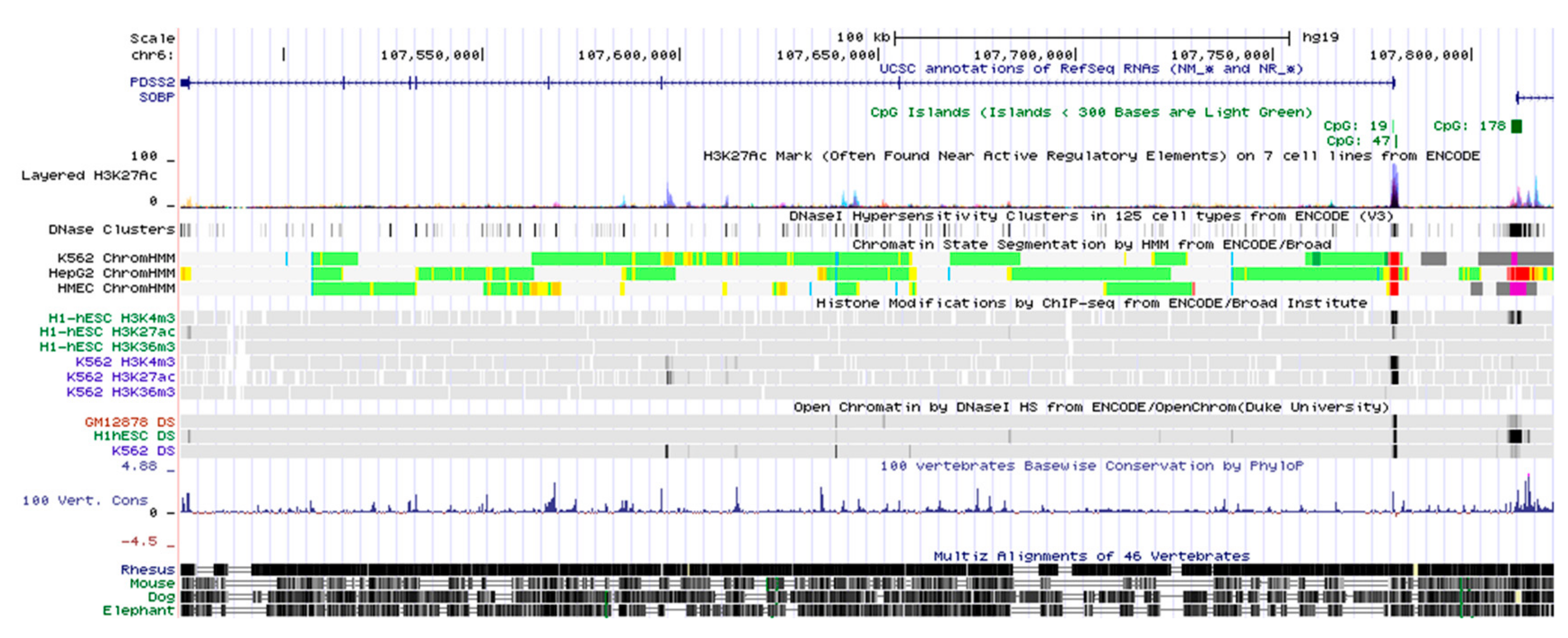
3.1. Genomic Structure of the PDSS2 Gene Locus
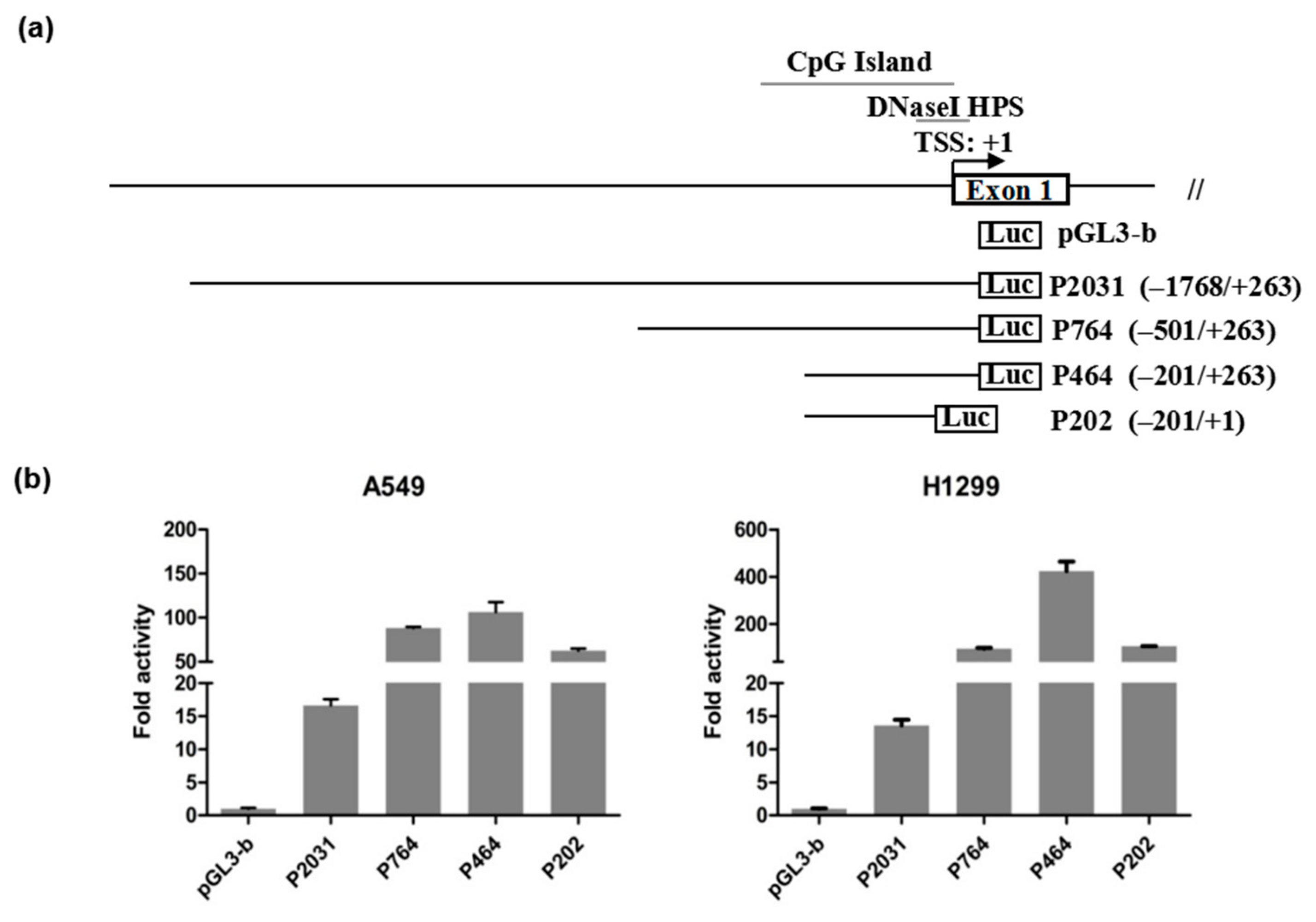
3.2. Identification of the PDSS2 Promoter Region
3.3. Sequence and Homology Analysis of the PDSS2 Promoter Region
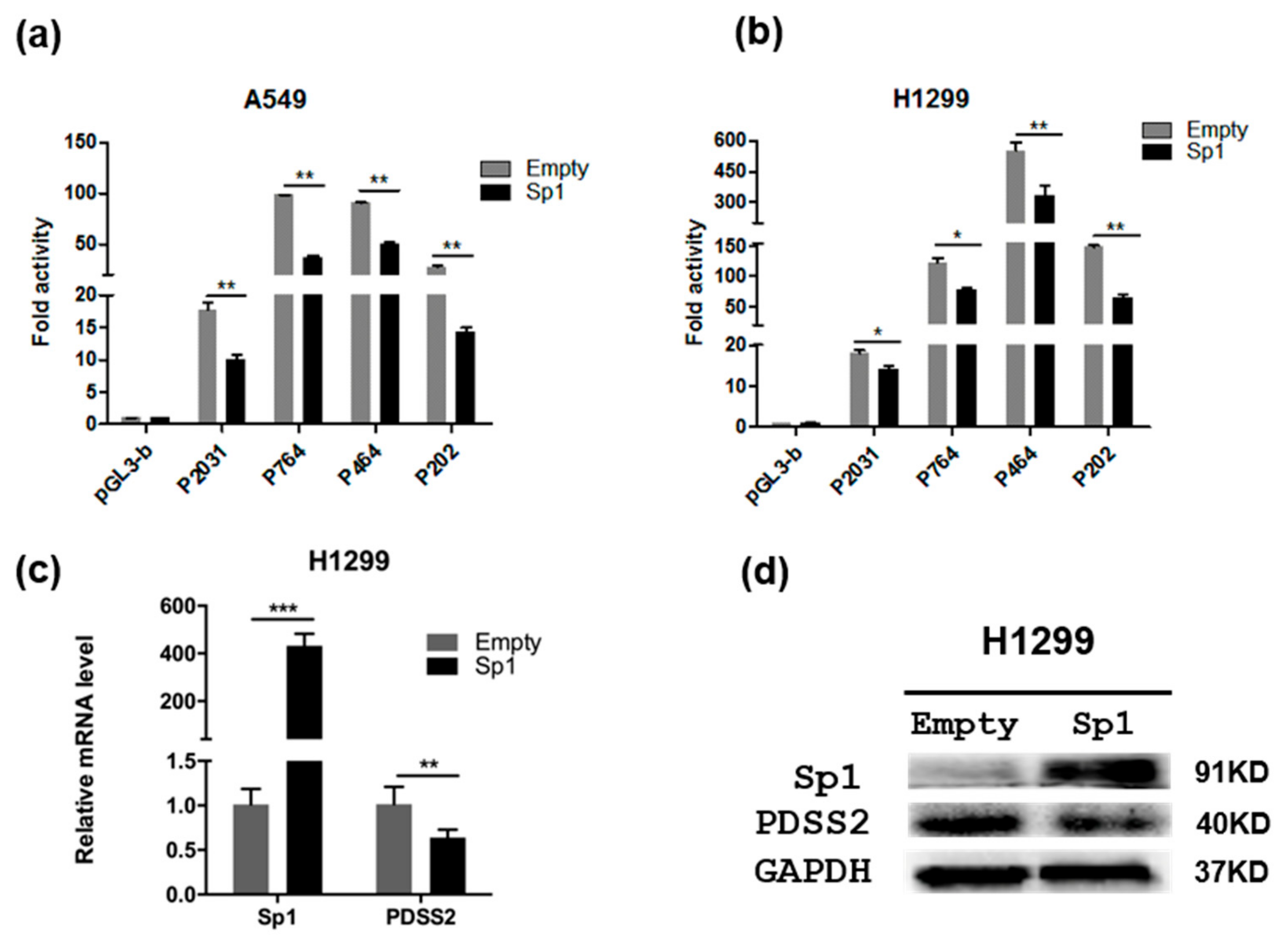
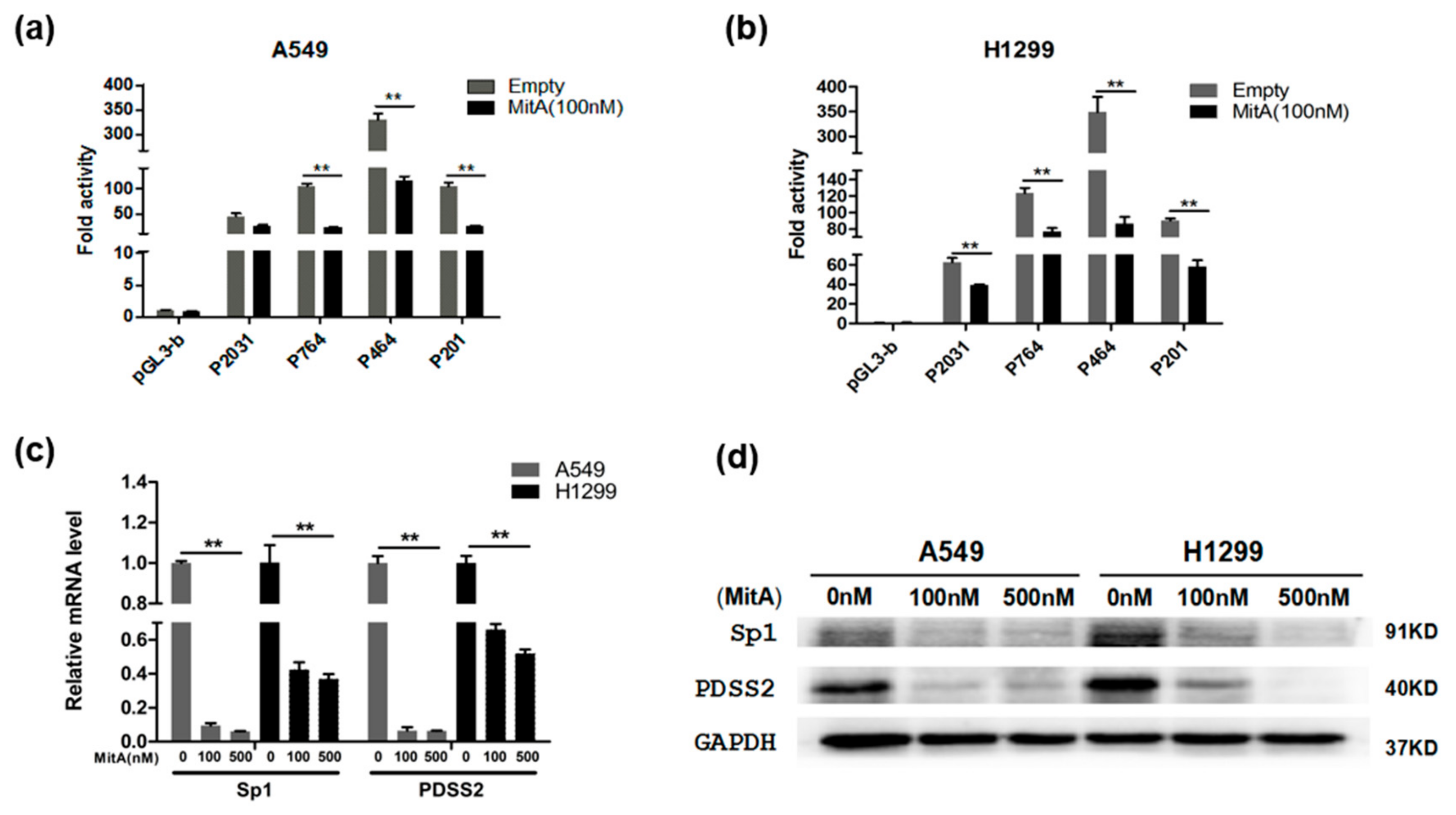
3.4. Sp1 Inhibits PDSS2 Expression
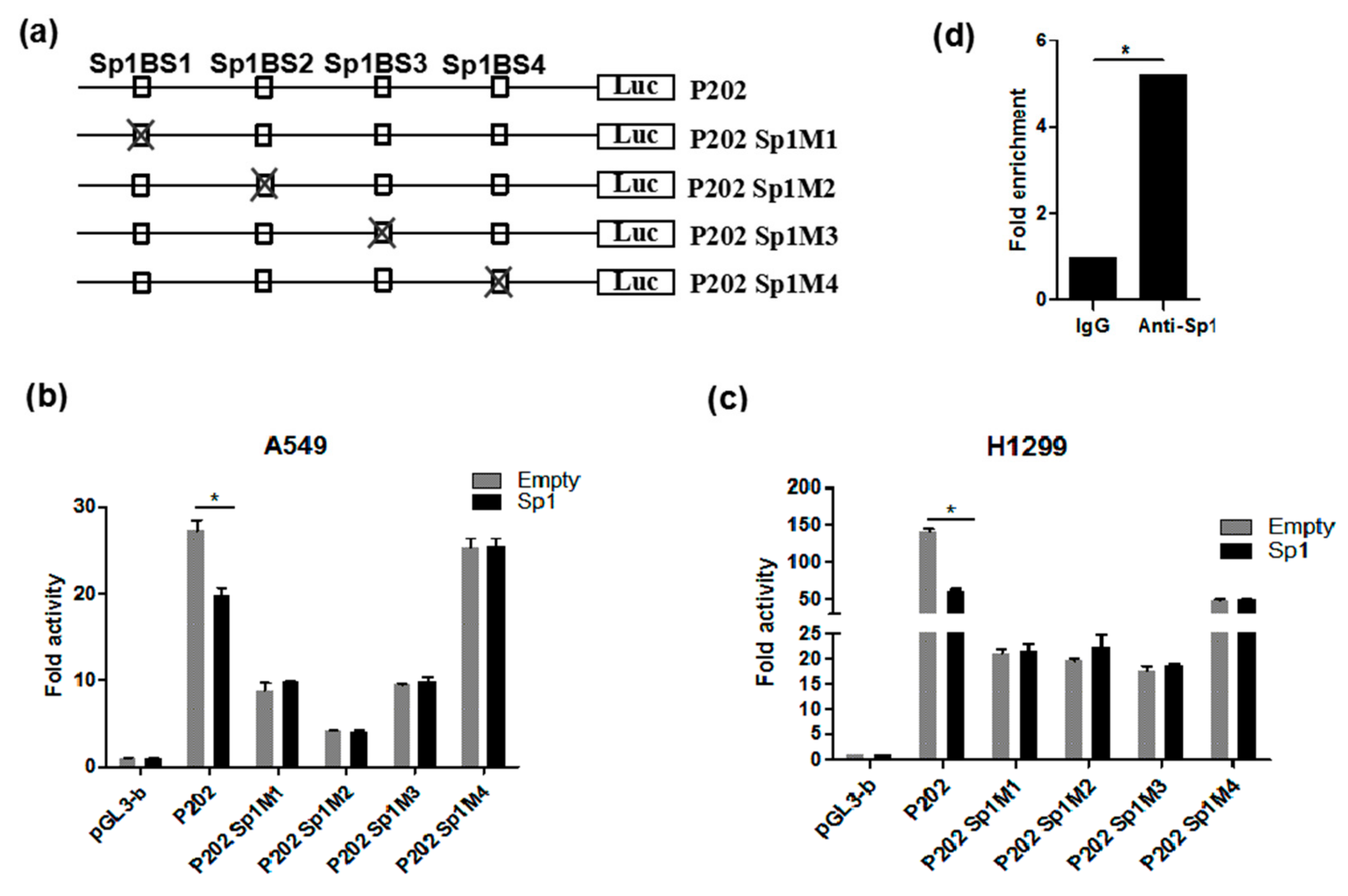
3.5. Functional Analysis of Sp1 Binding Sites in the PDSS2 Core Promoter
3.6. Sp1 Is Essential for the Constitutive Expression of PDSS2 Gene
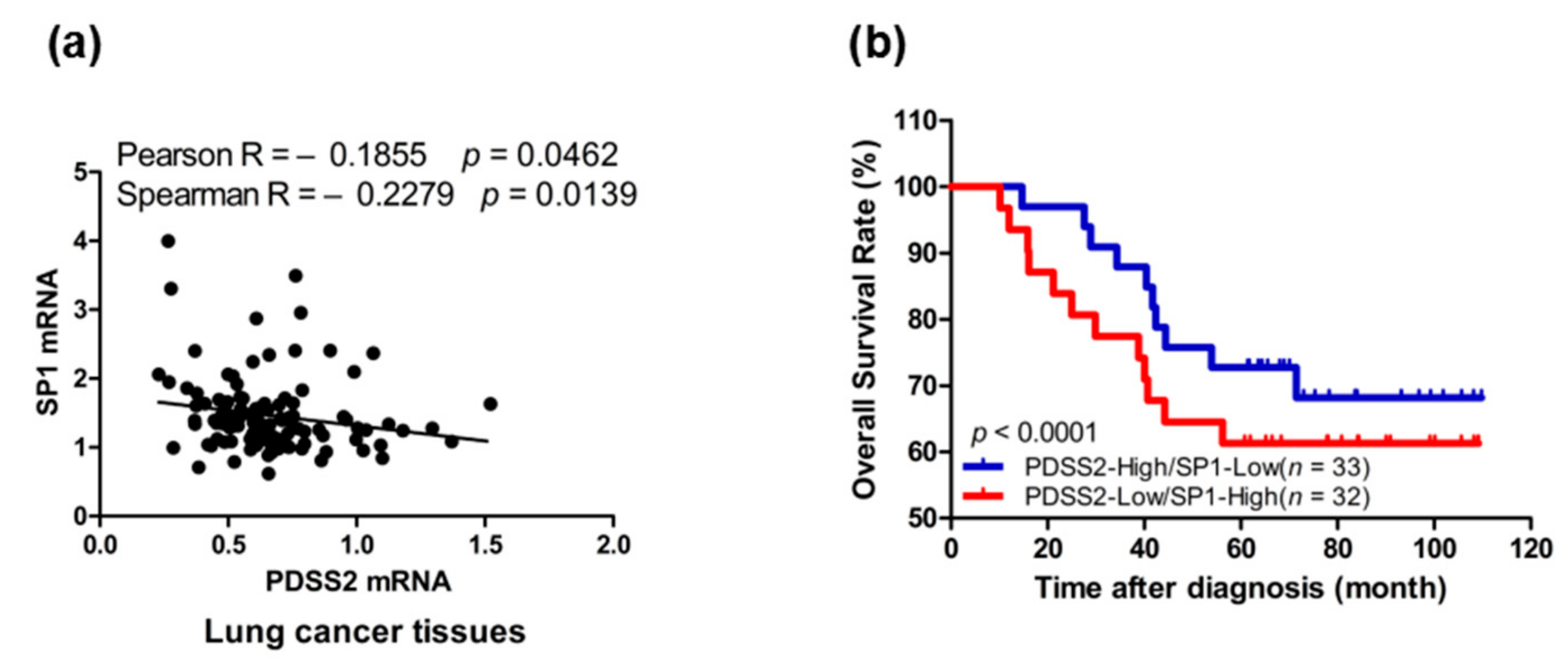
3.7. Sp1-Mediated PDSS2 Expression Is Associated with Poor Prognosis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cm, Q.; López, L.C.; Von-Moltke, J.; Naini, A.; Krishna, S.; Schuelke, M.; Salviati, L.; Navas, P.; DiMauro, S.; Hirano, M. Respiratory chain dysfunction and oxidative stress correlate with severity of primary CoQ10 deficiency. FASEB J. 2008, 22, 1874–1885. [Google Scholar]
- Crane, F.L. Biochemical Functions of Coenzyme Q10. J. Am. Coll. Nutr. 2001, 20, 591–598. [Google Scholar] [CrossRef]
- Lopez-Martin, J.M.; Salviati, L.; Trevisson, E.; Montini, G.; DiMauro, S.; Quinzii, C.; Hirano, M.; Rodriguez-Hernandez, A.; Cordero, M.D.; Sánchez-Alcázar, J.A.; et al. Missense mutation of the COQ2 gene causes defects of bioenergetics and de novo pyrimidine synthesis. Hum. Mol. Genet. 2007, 16, 1091–1097. [Google Scholar] [CrossRef] [PubMed]
- Rusciani, L.; Proietti, I.; Rusciani, A.; Paradisi, A.; Sbordoni, G.; Alfano, C.; Panunzi, S.; de Gaetano, A.; Lippa, S. Low plasma coenzyme Q10 levels as an independent prognostic factor for melanoma progression. J. Am. Acad. Dermatol. 2006, 54, 234–241. [Google Scholar] [CrossRef]
- Jolliet, P.; Simon, N.; Barré, J.; Pons, J.Y.; Boukef, M.; Paniel, B.J.; Tillement, J.P. Plasma coenzyme Q10 concentrations in breast cancer: Prognosis and therapeutic consequences. Int. J. Clin. Pharmacol. Ther. 1998, 36, 506–509. [Google Scholar]
- Palan, P.R.; Mikhail, M.S.; Shaban, D.W.; Romney, S.L. Plasma concentrations of coenzyme Q10 and tocopherols in cervical intraepithelial neoplasia and cervical cancer. Eur. J. Cancer Prev. 2003, 12, 321–326. [Google Scholar] [CrossRef]
- Buric, S.S.; Podolski-Renic, A.; Dinic, J.; Stankovic, T.; Jovanovic, M.; Hadzic, S.; Ayuso, J.M.; Virumbrales-Munoz, M.; Fernandez, L.J.; Ochoa, I.; et al. Modulation of Antioxidant Potential with Coenzyme Q10 Suppressed Invasion of Temozolomide-Resistant Rat Glioma In Vitro and In Vivo. Oxid. Med. Cell. Longev. 2019, 2019, 3061607. [Google Scholar] [CrossRef]
- Saiki, R.; Nagata, A.; Kainou, T.; Matsuda, H.; Kawamukai, M. Characterization of solanesyl and decaprenyl diphosphate synthases in mice and humans. FEBS J. 2005, 272, 5606–5622. [Google Scholar] [CrossRef]
- López, L.C.; Schuelke, M.; Quinzii, C.M.; Kanki, T.; Rodenburg, R.J.; Naini, A.; Dimauro, S.; Hirano, M. Leigh syndrome with nephropathy and CoQ10 deficiency due to decaprenyl diphosphate synthase subunit 2 (PDSS2) mutations. Am. J. Hum. Genet. 2006, 79, 1125–1129. [Google Scholar] [CrossRef]
- Zhang, Z.; Gasser, D.L.; Rappaport, E.F.; Falk, M.J. Cross-platform expression microarray performance in a mouse model of mitochondrial disease therapy. Mol. Genet. Metab. 2010, 99, 309–318. [Google Scholar] [CrossRef][Green Version]
- Fung, J.M.; Smith, R.; Brown, M.A.; Lau, S.H.; Xie, D.; Lau, G.K.; Guan, X.Y. Identification and characterization of a novel melanoma tumor suppressor gene on human chromosome 6q21. Clin. Cancer Res. 2009, 15, 797–803. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kanda, M.; Nomoto, S.; Oya, H.; Hashimoto, R.; Takami, H.; Shimizu, D.; Sonohara, F.; Kobayashi, D.; Tanaka, C.; Yamada, S.; et al. Decreased expression of prenyl diphosphate synthase subunit 2 correlates with reduced survival of patients with gastric cancer. J. Exp. Clin. Cancer Res. 2014, 33, 88. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Gao, F.; Li, K.; Wang, W.; Lai, Y.R.; Tang, S.H.; Yang, D.H. Decaprenyl diphosphate synthase subunit 2 as a prognosis factor in hepatocellular carcinoma. World J. Gastroenterol. 2015, 21, 3055–3065. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Zhang, Y.; Polireddy, K.; Chen, Q. The tumor-suppressing activity of the prenyl diphosphate synthase subunit 2 gene in lung cancer cells. Anti-Cancer Drugs 2014, 25, 790–798. [Google Scholar] [CrossRef]
- Bu, Y.; Suenaga, Y.; Ono, S.; Koda, T.; Song, F.; Nakagawara, A.; Ozak, T. Sp1-mediated transcriptional regulation of NFBD1/MDC1 plays a critical role in DNA damage response pathway. Genes Cells 2008, 13, 53–66. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Zhang, C.; Weng, H.; Li, Y.; Cai, W.; Xie, M.; Long, Y.; Ai, Q.; Liu, Z.; et al. The gene pair PRR11 and SKA2 shares a NF-Y-regulated bidirectional promoter and contributes to lung cancer development. Biochim. Biophys. Acta 2015, 1849, 1133–1144. [Google Scholar] [CrossRef]
- Ji, Y.; Xie, M.; Lan, H.; Zhan, Y.; Long, Y.; Weng, H.; Li, D.; Cai, W.; Zhu, H.; Yang, Z.; et al. PRR11 is a novel gene implicated in cell cycle progression and lung cancer. Int. J. Biochem. Cell Biol. 2013, 45, 645–656. [Google Scholar] [CrossRef]
- Kanda, M.; Sugimoto, H.; Nomoto, S.; Oya, H.; Shimizu, D.; Takami, H.; Hashimoto, R.; Sonohara, F.; Okamura, Y.; Yamada, S.; et al. Clinical utility of PDSS2 expression to stratify patients at risk for recurrence of hepatocellular carcinoma. Int. J. Oncol. 2014, 45, 2005–2012. [Google Scholar] [CrossRef]
- Li, Y.; Lin, S.; Li, L.; Tang, Z.; Hu, Y.; Ban, X.; Zeng, T.; Zhou, Y.; Zhu, Y.; Gao, S.; et al. Deficiency Induces Hepatocarcinogenesis by Decreasing Mitochondrial Respiration and Reprogramming Glucose Metabolism. Cancer Res. 2018, 78, 4471–4481. [Google Scholar] [CrossRef]
- Wang, F.; Ma, Y.L.; Zhang, P.; Shen, T.Y.; Shi, C.Z.; Yang, Y.Z.; Moyer, M.P.; Zhang, H.Z.; Chen, H.Q.; Liang, Y.; et al. SP1 mediates the link between methylation of the tumour suppressor miR-149 and outcome in colorectal cancer. J. Pathol. 2013, 229, 12–24. [Google Scholar] [CrossRef]
- Zhang, H.W.; Wang, E.W.; Li, L.X.; Yi, S.H.; Li, L.C.; Xu, F.L.; Wang, D.L.; Wu, Y.Z.; Nian, W.Q. A regulatory loop involving miR-29c and Sp1 elevates the TGF-β1 mediated epithelial-to-mesenchymal transition in lung cancer. Oncotarget 2016, 7, 85905–85916. [Google Scholar] [PubMed]
- Hu, J.; Hu, H.; Hang, J.J.; Yang, H.Y.; Wang, Z.Y.; Wang, L.; Chen, D.H.; Wang, L.W. Simultaneous high expression of PLD1 and Sp1 predicts a poor prognosis for pancreatic ductal adenocarcinoma patients. Oncotarget 2016, 7, 78557–78565. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Cai, N.; Tao, T.; Zhang, R.; Yan, W.; Li, R.; Zhang, J.; Luo, H.; Shi, Y.; Luan, W.; et al. An axis involving SNAI1, microRNA-128 and SP1 modulates glioma progression. PLoS ONE 2014, 9, e98651. [Google Scholar] [CrossRef] [PubMed]
- Safe, S.; Abbruzzese, J.; Abdelrahim, M.; Hedrick, E. Specificity Protein Transcription Factors and Cancer: Opportunities for Drug Development. Cancer Prev. Res. 2018, 11, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Obinata, D.; Takayama, K.; Urano, T.; Murata, T.; Kumagai, J.; Fujimura, T.; Ikeda, K.; Horie-Inoue, K.; Homma, Y.; Ouchi, Y.; et al. Oct1 regulates cell growth of LNCaP cells and is a prognostic factor for prostate cancer. Int. J. Cancer 2012, 130, 1021–1028. [Google Scholar] [CrossRef]
- Qian, J.; Kong, X.; Deng, N.; Tan, P.; Chen, H.; Wang, J.; Li, Z.; Hu, Y.; Zou, W.; Xu, J.; et al. OCT1 is a determinant of synbindin-related ERK signalling with independent prognostic significance in gastric cancer. Gut 2015, 64, 37–48. [Google Scholar] [CrossRef][Green Version]
- Reymann, S.; Borlak, J. Transcription profiling of lung adenocarcinomas of c-myc-transgenic mice: Identification of the c-myc regulatory gene network. BMC Syst. Biol. 2008, 2, 46. [Google Scholar] [CrossRef]
- Xiao, S.; Liao, S.; Zhou, Y.; Jiang, B.; Li, Y.; Xue, M. High expression of octamer transcription factor 1 in cervical cancer. Oncol. Lett. 2014, 7, 1889–1894. [Google Scholar] [CrossRef]
- Shimizu, R.; Engel, J.D.; Yamamoto, M. GATA1-related leukaemias. Nat. Rev. Cancer 2008, 8, 279–287. [Google Scholar] [CrossRef]
- Zheng, R.; Blobel, G.A. GATA Transcription Factors and Cancer. Genes Cancer 2010, 1, 1178–1188. [Google Scholar] [CrossRef]
- Boidot, R.; Végran, F.; Jacob, D.; Chevrier, S.; Cadouot, M.; Feron, O.; Solary, E.; Lizard-Nacol, S. The transcription factor GATA-1 is overexpressed in breast carcinomas and contributes to survivin upregulation via a promoter polymorphism. Oncogene 2010, 29, 2577–2584. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ke, Q.; Shao, Y.; Zhu, G.; Li, Y.; Geng, N.; Jin, F.; Li, F. GATA1 induces epithelial-mesenchymal transition in breast cancer cells through PAK5 oncogenic signaling. Oncotarget 2015, 6, 4345–4356. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, J.; Lin, J.; Zhou, L.; Song, Y.; Wei, B.; Luo, X.; Chen, Z.; Chen, Y.; Xiong, J.; et al. The transcription factor GATA1 and the histone methyltransferase SET7 interact to promote VEGF-mediated angiogenesis and tumor growth and predict clinical outcome of breast cancer. Oncotarget 2016, 7, 9859–9875. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.J.; Liu, M.; Liu, H.; Zhou, L. GATA1 promotes colorectal cancer cell proliferation, migration and invasion via activating AKT signaling pathway. Mol. Cell. Biochem. 2019, 457, 191–199. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, L.; Chen, Q.; Wang, Y.; Zhang, N.; Meng, P.; Liu, T.; Bu, Y. Sp1 Mediates the Constitutive Expression and Repression of the PDSS2 Gene in Lung Cancer Cells. Genes 2019, 10, 977. https://doi.org/10.3390/genes10120977
Hu L, Chen Q, Wang Y, Zhang N, Meng P, Liu T, Bu Y. Sp1 Mediates the Constitutive Expression and Repression of the PDSS2 Gene in Lung Cancer Cells. Genes. 2019; 10(12):977. https://doi.org/10.3390/genes10120977
Chicago/Turabian StyleHu, Lanyue, Quanmei Chen, Yitao Wang, Na Zhang, Peixin Meng, Tong Liu, and Youquan Bu. 2019. "Sp1 Mediates the Constitutive Expression and Repression of the PDSS2 Gene in Lung Cancer Cells" Genes 10, no. 12: 977. https://doi.org/10.3390/genes10120977
APA StyleHu, L., Chen, Q., Wang, Y., Zhang, N., Meng, P., Liu, T., & Bu, Y. (2019). Sp1 Mediates the Constitutive Expression and Repression of the PDSS2 Gene in Lung Cancer Cells. Genes, 10(12), 977. https://doi.org/10.3390/genes10120977



