DNA Helicases as Safekeepers of Genome Stability in Plants
Abstract
:1. Introduction
2. RecQ Helicases
2.1. RECQ4 Homologs and the RTR Complex
2.2. Other Classical RecQ Helicases
2.3. HRQ1
3. Fe-S Cluster Helicases
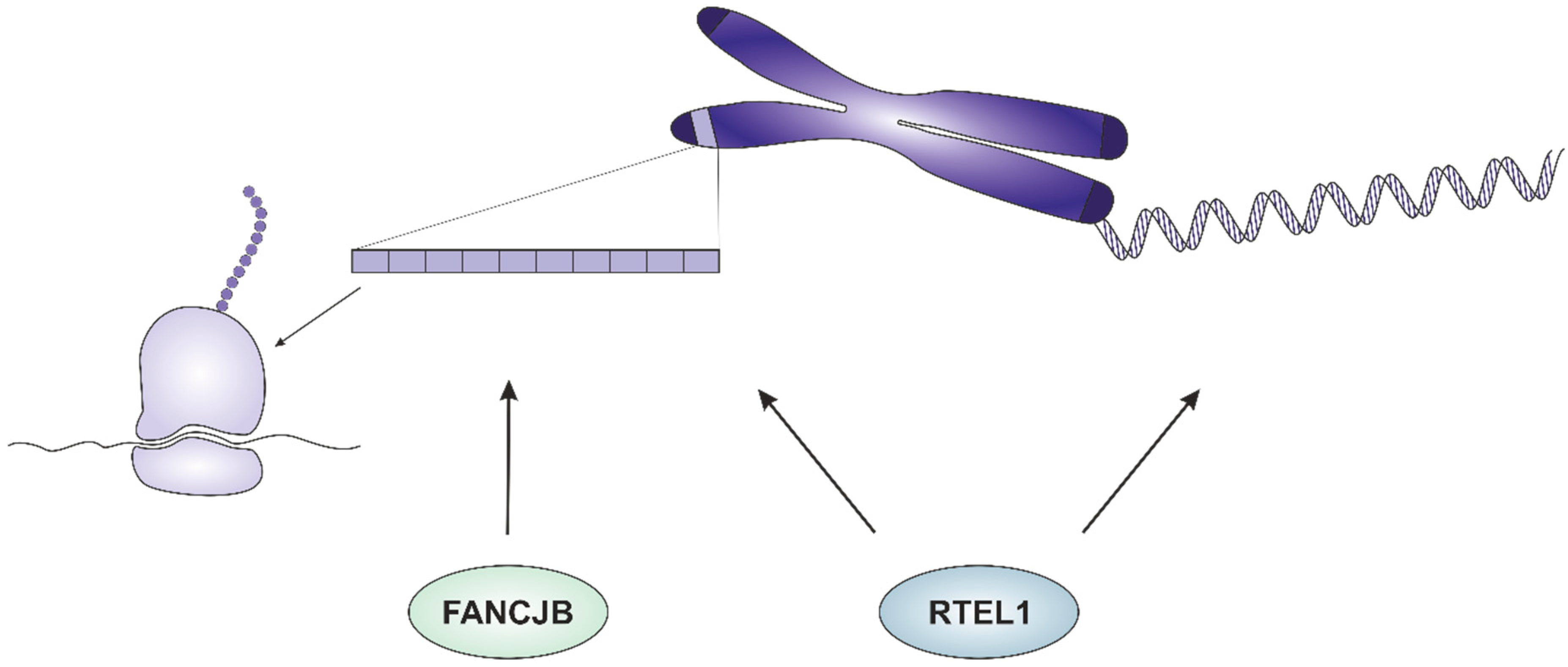
3.1. RTEL1
3.2. FANCJ
4. Further Helicases Involved in Genome Maintenance
4.1. SRS2
4.2. FANCM
4.3. MCM Helicases
4.4. POLQ/Θ (TEBICHI)
5. Helicases are Relevant for Multiple Aspects of Plant Breeding
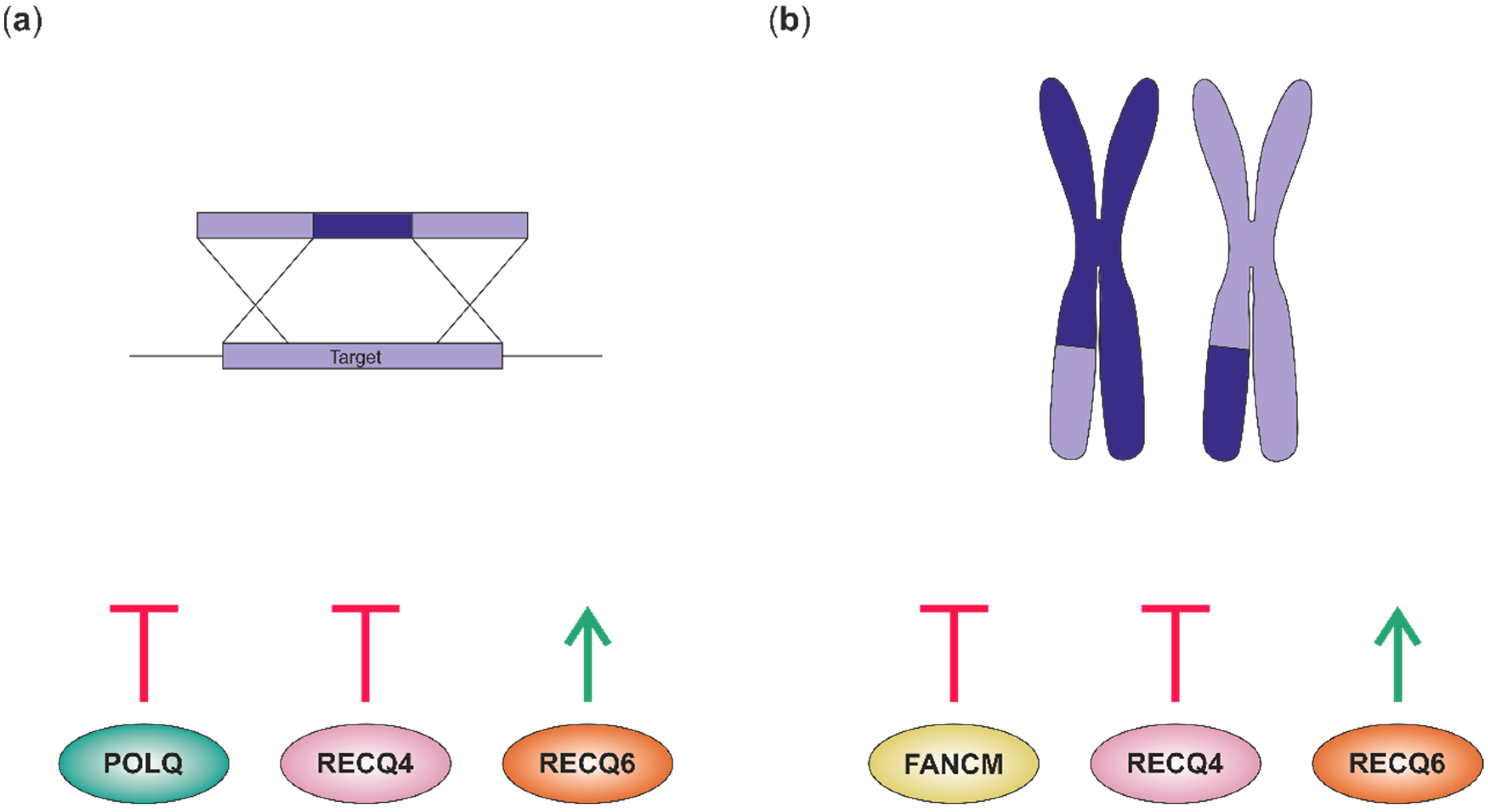
5.1. Gene Targeting
5.2. Manipulation of CO Frequency
6. Conclusion and Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Hartung, F.; Puchta, H. The RecQ gene family in plants. J. Plant Physiol. 2006, 163, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Wiedemann, G.; van Gessel, N.; Köchl, F.; Hunn, L.; Schulze, K.; Maloukh, L.; Nogué, F.; Decker, E.L.; Hartung, F.; Reski, R. RecQ Helicases Function in Development, DNA Repair, and Gene Targeting in Physcomitrella patens. Plant Cell 2018, 30, 717–736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellis, N.A.; Groden, J.; Ye, T.Z.; Straughen, J.; Lennon, D.J.; Ciocci, S.; Proytcheva, M.; German, J. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell 1995, 83, 655–666. [Google Scholar] [CrossRef] [Green Version]
- Chaganti, R.S.; Schonberg, S.; German, J. A manyfold increase in sister chromatid exchanges in Bloom’s syndrome lymphocytes. Proc. Natl. Acad. Sci. USA 1974, 71, 4508–4512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartung, F.; Plchová, H.; Puchta, H. Molecular characterisation of RecQ homologues in Arabidopsis thaliana. Nucleic Acids Res. 2000, 28, 4275–4282. [Google Scholar] [CrossRef] [Green Version]
- Hartung, F.; Suer, S.; Puchta, H. Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2007, 104, 18836–18841. [Google Scholar] [CrossRef] [Green Version]
- Bagherieh-Najjar, M.B.; de Vries, O.M.H.; Hille, J.; Dijkwel, P.P. Arabidopsis RecQI4A suppresses homologous recombination and modulates DNA damage responses. Plant J. 2005, 43, 789–798. [Google Scholar] [CrossRef]
- Schröpfer, S.; Kobbe, D.; Hartung, F.; Knoll, A.; Puchta, H. Defining the roles of the N-terminal region and the helicase activity of RECQ4A in DNA repair and homologous recombination in Arabidopsis. Nucleic Acids Res. 2014, 42, 1684–1697. [Google Scholar] [CrossRef] [Green Version]
- Hartung, F.; Suer, S.; Bergmann, T.; Puchta, H. The role of AtMUS81 in DNA repair and its genetic interaction with the helicase AtRecQ4A. Nucleic Acids Res. 2006, 34, 4438–4448. [Google Scholar] [CrossRef]
- Mannuss, A.; Dukowic-Schulze, S.; Suer, S.; Hartung, F.; Pacher, M.; Puchta, H. RAD5A, RECQ4A, and MUS81 have specific functions in homologous recombination and define different pathways of DNA repair in Arabidopsis thaliana. Plant Cell 2010, 22, 3318–3330. [Google Scholar] [CrossRef] [Green Version]
- Dangel, N.J.; Knoll, A.; Puchta, H. MHF1 plays Fanconi anaemia complementation group M protein (FANCM)-dependent and FANCM-independent roles in DNA repair and homologous recombination in plants. Plant J. 2014, 78, 822–833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herrmann, N.J.; Knoll, A.; Puchta, H. The nuclease FAN1 is involved in DNA crosslink repair in Arabidopsis thaliana independently of the nuclease MUS81. Nucleic Acids Res. 2015, 43, 3653–3666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dorn, A.; Feller, L.; Castri, D.; Röhrig, S.; Enderle, J.; Herrmann, N.J.; Block-Schmidt, A.; Trapp, O.; Köhler, L.; Puchta, H. An Arabidopsis FANCJ helicase homologue is required for DNA crosslink repair and rDNA repeat stability. PLoS Genet. 2019, 15, e1008174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobbe, S.; Trapp, O.; Knoll, A.; Manuss, A.; Puchta, H. The Translesion Polymerase ζ Has Roles Dependent on and Independent of the Nuclease MUS81 and the Helicase RECQ4A in DNA Damage Repair in Arabidopsis. Plant Physiol. 2015, 169, 2718–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enderle, J.; Dorn, A.; Puchta, H. DNA- and DNA-Protein-Crosslink Repair in Plants. Int. J. Mol. Sci. 2019, 20, 4304. [Google Scholar] [CrossRef] [Green Version]
- Kwon, Y.-I.; Abe, K.; Osakabe, K.; Endo, M.; Nishizawa-Yokoi, A.; Saika, H.; Shimada, H.; Toki, S. Overexpression of OsRecQl4 and/or OsExo1 enhances DSB-induced homologous recombination in rice. Plant Cell Physiol. 2012, 53, 2142–2152. [Google Scholar] [CrossRef] [Green Version]
- Kwon, Y.-I.; Abe, K.; Endo, M.; Osakabe, K.; Ohtsuki, N.; Nishizawa-Yokoi, A.; Tagiri, A.; Saika, H.; Toki, S. DNA replication arrest leads to enhanced homologous recombination and cell death in meristems of rice OsRecQl4 mutants. BMC Plant Biol. 2013, 13, 62. [Google Scholar] [CrossRef] [Green Version]
- Higgins, J.D.; Ferdous, M.; Osman, K.; Franklin, F.C.H. The RecQ helicase AtRECQ4A is required to remove inter-chromosomal telomeric connections that arise during meiotic recombination in Arabidopsis. Plant J. 2011, 65, 492–502. [Google Scholar] [CrossRef]
- Séguéla-Arnaud, M.; Crismani, W.; Larchevêque, C.; Mazel, J.; Froger, N.; Choinard, S.; Lemhemdi, A.; Macaisne, N.; van Leene, J.; Gevaert, K.; et al. Multiple mechanisms limit meiotic crossovers: TOP3α and two BLM homologs antagonize crossovers in parallel to FANCM. Proc. Natl. Acad. Sci. USA 2015, 112, 4713–4718. [Google Scholar] [CrossRef] [Green Version]
- de Maagd, R.A.; Loonen, A.; Chouaref, J.; Pele, A.; Meijer-Dekens, F.; Fransz, P.; Bai, Y. CRISPR/Cas inactivation of RECQ4 increases homeologous crossovers in an interspecific tomato hybrid. Plant Biotechnol. J. 2019. [Google Scholar] [CrossRef]
- Mieulet, D.; Aubert, G.; Bres, C.; Klein, A.; Droc, G.; Vieille, E.; Rond-Coissieux, C.; Sanchez, M.; Dalmais, M.; Mauxion, J.-P.; et al. Unleashing meiotic crossovers in crops. Nat. Plants 2018, 4, 1010–1016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knoll, A.; Schröpfer, S.; Puchta, H. The RTR complex as caretaker of genome stability and its unique meiotic function in plants. Front. Plant Sci. 2014, 5, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chelysheva, L.; Vezon, D.; Belcram, K.; Gendrot, G.; Grelon, M. The Arabidopsis BLAP75/Rmi1 homologue plays crucial roles in meiotic double-strand break repair. PLoS Genet. 2008, 4, e1000309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartung, F.; Suer, S.; Knoll, A.; Wurz-Wildersinn, R.; Puchta, H. Topoisomerase 3alpha and RMI1 suppress somatic crossovers and are essential for resolution of meiotic recombination intermediates in Arabidopsis thaliana. PLoS Genet. 2008, 4, e1000285. [Google Scholar] [CrossRef] [Green Version]
- Röhrig, S.; Schröpfer, S.; Knoll, A.; Puchta, H. The RTR Complex Partner RMI2 and the DNA Helicase RTEL1 Are Both Independently Involved in Preserving the Stability of 45S rDNA Repeats in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006394. [Google Scholar] [CrossRef]
- Plank, J.L.; Wu, J.; Hsieh, T.-S. Topoisomerase IIIalpha and Bloom’s helicase can resolve a mobile double Holliday junction substrate through convergent branch migration. Proc. Natl. Acad. Sci. USA 2006, 103, 11118–11123. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Hickson, I.D. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature 2003, 426, 870–874. [Google Scholar] [CrossRef]
- Raynard, S.; Bussen, W.; Sung, P. A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIalpha, and BLAP75. J. Biol. Chem. 2006, 281, 13861–13864. [Google Scholar] [CrossRef] [Green Version]
- Dorn, A.; Röhrig, S.; Papp, K.; Schröpfer, S.; Hartung, F.; Knoll, A.; Puchta, H. The topoisomerase 3α zinc-finger domain T1 of Arabidopsis thaliana is required for targeting the enzyme activity to Holliday junction-like DNA repair intermediates. PLoS Genet. 2018, 14, e1007674. [Google Scholar] [CrossRef]
- Séguéla-Arnaud, M.; Choinard, S.; Larchevêque, C.; Girard, C.; Froger, N.; Crismani, W.; Mercier, R. RMI1 and TOP3α limit meiotic CO formation through their C-terminal domains. Nucleic Acids Res. 2017, 45, 1860–1871. [Google Scholar] [CrossRef] [Green Version]
- Bonnet, S.; Knoll, A.; Hartung, F.; Puchta, H. Different functions for the domains of the Arabidopsis thaliana RMI1 protein in DNA cross-link repair, somatic and meiotic recombination. Nucleic Acids Res. 2013, 41, 9349–9360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobbe, D.; Blanck, S.; Focke, M.; Puchta, H. Biochemical characterization of AtRECQ3 reveals significant differences relative to other RecQ helicases. Plant Physiol. 2009, 151, 1658–1666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobbe, D.; Blanck, S.; Demand, K.; Focke, M.; Puchta, H. AtRECQ2, a RecQ helicase homologue from Arabidopsis thaliana, is able to disrupt various recombinogenic DNA structures in vitro. Plant J. 2008, 55, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Klaue, D.; Kobbe, D.; Kemmerich, F.; Kozikowska, A.; Puchta, H.; Seidel, R. Fork sensing and strand switching control antagonistic activities of RecQ helicases. Nat. Commun. 2013, 4, 2024. [Google Scholar] [CrossRef] [Green Version]
- Plchova, H.; Hartung, F.; Puchta, H. Biochemical characterization of an exonuclease from Arabidopsis thaliana reveals similarities to the DNA exonuclease of the human Werner syndrome protein. J. Biol. Chem. 2003, 278, 44128–44138. [Google Scholar] [CrossRef] [Green Version]
- Röhrig, S.; Dorn, A.; Enderle, J.; Schindele, A.; Herrmann, N.J.; Knoll, A.; Puchta, H. The RecQ-like helicase HRQ1 is involved in DNA crosslink repair in Arabidopsis in a common pathway with the Fanconi anemia-associated nuclease FAN1 and the postreplicative repair ATPase RAD5A. New Phytol. 2018, 218, 1478–1490. [Google Scholar] [CrossRef] [Green Version]
- Saotome, A.; Kimura, S.; Mori, Y.; Uchiyama, Y.; Morohashi, K.; Sakaguchi, K. Characterization of four RecQ homologues from rice (Oryza sativa L. cv. Nipponbare). Biochem. Biophys. Res. Commun. 2006, 345, 1283–1291. [Google Scholar] [CrossRef]
- Chen, H.; Samadder, P.P.; Tanaka, Y.; Ohira, T.; Okuizumi, H.; Yamaoka, N.; Miyao, A.; Hirochika, H.; Ohira, T.; Tsuchimoto, S.; et al. OsRecQ1, a QDE-3 homologue in rice, is required for RNA silencing induced by particle bombardment for inverted repeat DNA, but not for double-stranded RNA. Plant J. 2008, 56, 274–286. [Google Scholar] [CrossRef]
- Chen, H.; Kobayashi, K.; Miyao, A.; Hirochika, H.; Yamaoka, N.; Nishiguchi, M. Both OsRecQ1 and OsRDR1 are required for the production of small RNA in response to DNA-damage in rice. PLoS ONE 2013, 8, e55252. [Google Scholar] [CrossRef] [Green Version]
- Gardiner, L.-J.; Wingen, L.U.; Bailey, P.; Joynson, R.; Brabbs, T.; Wright, J.; Higgins, J.D.; Hall, N.; Griffiths, S.; Clavijo, B.J.; et al. Analysis of the recombination landscape of hexaploid bread wheat reveals genes controlling recombination and gene conversion frequency. Genome Biol. 2019, 20, 69. [Google Scholar] [CrossRef] [Green Version]
- Bagherieh-Najjar, M.B.; de Vries, O.M.H.; Kroon, J.T.M.; Wright, E.L.; Elborough, K.M.; Hille, J.; Dijkwel, P.P. Arabidopsis RecQsim, a plant-specific member of the RecQ helicase family, can suppress the MMS hypersensitivity of the yeast sgs1 mutant. Plant Mol. Biol. 2003, 52, 273–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barea, F.; Tessaro, S.; Bonatto, D. In silico analyses of a new group of fungal and plant RecQ4-homologous proteins. Comput. Biol. Chem. 2008, 32, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Groocock, L.M.; Prudden, J.; Perry, J.J.P.; Boddy, M.N. The RecQ4 orthologue Hrq1 is critical for DNA interstrand cross-link repair and genome stability in fission yeast. Mol. Cell. Biol. 2012, 32, 276–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, S.-H.; Choi, D.-H.; Lee, R.; Bae, S.-H. Saccharomyces cerevisiae Hrq1 requires a long 3’-tailed DNA substrate for helicase activity. Biochem. Biophys. Res. Commun. 2012, 427, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.-H.; Min, M.-H.; Kim, M.-J.; Lee, R.; Kwon, S.-H.; Bae, S.-H. Hrq1 facilitates nucleotide excision repair of DNA damage induced by 4-nitroquinoline-1-oxide and cisplatin in Saccharomyces cerevisiae. J. Microbiol. 2014, 52, 292–298. [Google Scholar] [CrossRef] [PubMed]
- Bochman, M.L.; Paeschke, K.; Chan, A.; Zakian, V.A. Hrq1, a homolog of the human RecQ4 helicase, acts catalytically and structurally to promote genome integrity. Cell Rep. 2014, 6, 346–356. [Google Scholar] [CrossRef] [Green Version]
- Rudolf, J.; Makrantoni, V.; Ingledew, W.J.; Stark, M.J.R.; White, M.F. The DNA repair helicases XPD and FancJ have essential iron-sulfur domains. Mol. Cell 2006, 23, 801–808. [Google Scholar] [CrossRef]
- Suhasini, A.N.; Brosh, R.M. DNA helicases associated with genetic instability, cancer, and aging. Adv. Exp. Med. Biol. 2013, 767, 123–144. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Hong, S.-W.; Escobar, M.; Vierling, E.; Mitchell, D.L.; Mount, D.W.; Hall, J.D. Arabidopsis UVH6, a Homolog of Human XPD and Yeast RAD3 DNA Repair Genes, Functions in DNA Repair and Is Essential for Plant Growth1. Plant Physiol. 2003, 132, 1405–1414. [Google Scholar] [CrossRef] [Green Version]
- Recker, J.; Knoll, A.; Puchta, H. The Arabidopsis thaliana homolog of the helicase RTEL1 plays multiple roles in preserving genome stability. Plant Cell 2014, 26, 4889–4902. [Google Scholar] [CrossRef] [Green Version]
- Ballew, B.J.; Yeager, M.; Jacobs, K.; Giri, N.; Boland, J.; Burdett, L.; Alter, B.P.; Savage, S.A. Germline mutations of regulator of telomere elongation helicase 1, RTEL1, in Dyskeratosis congenita. Hum. Genet. 2013, 132, 473–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, Z.; Glousker, G.; Molczan, A.; Fox, A.J.; Lamm, N.; Dheekollu, J.; Weizman, O.-E.; Schertzer, M.; Wang, Z.; Vladimirova, O.; et al. Inherited mutations in the helicase RTEL1 cause telomere dysfunction and Hoyeraal-Hreidarsson syndrome. Proc. Natl. Acad. Sci. USA 2013, 110, E3408–E3416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Guen, T.; Jullien, L.; Touzot, F.; Schertzer, M.; Gaillard, L.; Perderiset, M.; Carpentier, W.; Nitschke, P.; Picard, C.; Couillault, G.; et al. Human RTEL1 deficiency causes Hoyeraal-Hreidarsson syndrome with short telomeres and genome instability. Hum. Mol. Genet. 2013, 22, 3239–3249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barber, L.J.; Youds, J.L.; Ward, J.D.; McIlwraith, M.J.; O’Neil, N.J.; Petalcorin, M.I.R.; Martin, J.S.; Collis, S.J.; Cantor, S.B.; Auclair, M.; et al. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell 2008, 135, 261–271. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Hathcock, K.S.; Hande, P.; Lansdorp, P.M.; Seldin, M.F.; Hodes, R.J. Telomere length regulation in mice is linked to a novel chromosome locus. Proc. Natl. Acad. Sci. USA 1998, 95, 8648–8653. [Google Scholar] [CrossRef] [Green Version]
- Uringa, E.-J.; Youds, J.L.; Lisaingo, K.; Lansdorp, P.M.; Boulton, S.J. RTEL1: an essential helicase for telomere maintenance and the regulation of homologous recombination. Nucleic Acids Res. 2011, 39, 1647–1655. [Google Scholar] [CrossRef] [Green Version]
- Vannier, J.-B.; Pavicic-Kaltenbrunner, V.; Petalcorin, M.I.R.; Ding, H.; Boulton, S.J. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell 2012, 149, 795–806. [Google Scholar] [CrossRef] [Green Version]
- Hu, Z.; Cools, T.; Kalhorzadeh, P.; Heyman, J.; de Veylder, L. Deficiency of the Arabidopsis helicase RTEL1 triggers a SOG1-dependent replication checkpoint in response to DNA cross-links. Plant Cell 2015, 27, 149–161. [Google Scholar] [CrossRef] [Green Version]
- Kamisugi, Y.; Whitaker, J.W.; Cuming, A.C. The Transcriptional Response to DNA-Double-Strand Breaks in Physcomitrella patens. PLoS ONE 2016, 11, e0161204. [Google Scholar] [CrossRef] [Green Version]
- Goffová, I.; Vágnerová, R.; Peška, V.; Franek, M.; Havlová, K.; Holá, M.; Zachová, D.; Fojtová, M.; Cuming, A.; Kamisugi, Y.; et al. Roles of RAD51 and RTEL1 in telomere and rDNA stability in Physcomitrella patens. Plant J. 2019, 98, 1090–1105. [Google Scholar] [CrossRef]
- Olivier, M.; Charbonnel, C.; Amiard, S.; White, C.I.; Gallego, M.E. RAD51 and RTEL1 compensate telomere loss in the absence of telomerase. Nucleic Acids Res. 2018, 46, 2432–2445. [Google Scholar] [CrossRef] [PubMed]
- Procházková Schrumpfová, P.; Fojtová, M.; Fajkus, J. Telomeres in Plants and Humans: Not So Different, Not So Similar. Cells 2019, 8, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.; D’Andrea, A.D. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 2012, 26, 1393–1408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cantor, S.B.; Bell, D.W.; Ganesan, S.; Kass, E.M.; Drapkin, R.; Grossman, S.; Wahrer, D.C.; Sgroi, D.C.; Lane, W.S.; Haber, D.A.; et al. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell 2001, 105, 149–160. [Google Scholar] [CrossRef] [Green Version]
- Dohrn, L.; Salles, D.; Siehler, S.Y.; Kaufmann, J.; Wiesmüller, L. BRCA1-mediated repression of mutagenic end-joining of DNA double-strand breaks requires complex formation with BACH1. Biochem. J. 2012, 441, 919–926. [Google Scholar] [CrossRef]
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef] [Green Version]
- Lansdorp, P.; van Wietmarschen, N. Helicases FANCJ, RTEL1 and BLM Act on Guanine Quadruplex DNA in Vivo. Genes 2019, 10, 870. [Google Scholar] [CrossRef] [Green Version]
- Knoll, A.; Puchta, H. The role of DNA helicases and their interaction partners in genome stability and meiotic recombination in plants. J. Exp. Bot. 2011, 62, 1565–1579. [Google Scholar] [CrossRef] [Green Version]
- Niu, H.; Klein, H.L. Multifunctional roles of Saccharomyces cerevisiae Srs2 protein in replication, recombination and repair. FEMS Yeast Res. 2017, 17. [Google Scholar] [CrossRef] [Green Version]
- Blanck, S.; Kobbe, D.; Hartung, F.; Fengler, K.; Focke, M.; Puchta, H. A SRS2 homolog from Arabidopsis thaliana disrupts recombinogenic DNA intermediates and facilitates single strand annealing. Nucleic Acids Res. 2009, 37, 7163–7176. [Google Scholar] [CrossRef] [Green Version]
- Charlot, F.; Chelysheva, L.; Kamisugi, Y.; Vrielynck, N.; Guyon, A.; Epert, A.; Le Guin, S.; Schaefer, D.G.; Cuming, A.C.; Grelon, M.; et al. RAD51B plays an essential role during somatic and meiotic recombination in Physcomitrella. Nucleic Acids Res. 2014, 42, 11965–11978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knoll, A.; Higgins, J.D.; Seeliger, K.; Reha, S.J.; Dangel, N.J.; Bauknecht, M.; Schröpfer, S.; Franklin, F.C.H.; Puchta, H. The Fanconi anemia ortholog FANCM ensures ordered homologous recombination in both somatic and meiotic cells in Arabidopsis. Plant Cell 2012, 24, 1448–1464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roth, N.; Klimesch, J.; Dukowic-Schulze, S.; Pacher, M.; Mannuss, A.; Puchta, H. The requirement for recombination factors differs considerably between different pathways of homologous double-strand break repair in somatic plant cells. Plant J. 2012, 72, 781–790. [Google Scholar] [CrossRef] [PubMed]
- Crismani, W.; Girard, C.; Froger, N.; Pradillo, M.; Santos, J.L.; Chelysheva, L.; Copenhaver, G.P.; Horlow, C.; Mercier, R. FANCM limits meiotic crossovers. Science 2012, 336, 1588–1590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Girard, C.; Crismani, W.; Froger, N.; Mazel, J.; Lemhemdi, A.; Horlow, C.; Mercier, R. FANCM-associated proteins MHF1 and MHF2, but not the other Fanconi anemia factors, limit meiotic crossovers. Nucleic Acids Res. 2014, 42, 9087–9095. [Google Scholar] [CrossRef] [PubMed]
- Blary, A.; Gonzalo, A.; Eber, F.; Bérard, A.; Bergès, H.; Bessoltane, N.; Charif, D.; Charpentier, C.; Cromer, L.; Fourment, J.; et al. FANCM Limits Meiotic Crossovers in Brassica Crops. Front. Plant Sci. 2018, 9, 368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Girard, C.; Chelysheva, L.; Choinard, S.; Froger, N.; Macaisne, N.; Lemhemdi, A.; Lehmemdi, A.; Mazel, J.; Crismani, W.; Mercier, R. AAA-ATPase FIDGETIN-LIKE 1 and Helicase FANCM Antagonize Meiotic Crossovers by Distinct Mechanisms. PLoS Genet. 2015, 11, e1005369. [Google Scholar] [CrossRef] [Green Version]
- Ziolkowski, P.A.; Berchowitz, L.E.; Lambing, C.; Yelina, N.E.; Zhao, X.; Kelly, K.A.; Choi, K.; Ziolkowska, L.; June, V.; Sanchez-Moran, E.; et al. Juxtaposition of heterozygous and homozygous regions causes reciprocal crossover remodelling via interference during Arabidopsis meiosis. Elife 2015, 4. [Google Scholar] [CrossRef]
- Tuteja, N.; Tran, N.Q.; Dang, H.Q.; Tuteja, R. Plant MCM proteins: role in DNA replication and beyond. Plant Mol. Biol. 2011, 77, 537–545. [Google Scholar] [CrossRef]
- Di Ni, A.; Sozzani, R.; Blanchet, S.; Domenichini, S.; Reuzeau, C.; Cella, R.; Bergounioux, C.; Raynaud, C. The Arabidopsis MCM2 gene is essential to embryo development and its over-expression alters root meristem function. New Phytol. 2009, 184, 311–322. [Google Scholar] [CrossRef]
- Herridge, R.P.; Day, R.C.; Macknight, R.C. The role of the MCM2–7 helicase complex during Arabidopsis seed development. Plant Mol. Biol. 2014, 86, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, I.; Choudhury, N.R.; Tuteja, N. Arabidopsis thaliana MCM3 single subunit of MCM2–7 complex functions as 3’ to 5’ DNA helicase. Protoplasma 2016, 253, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Tran, N.Q.; Dang, H.Q.; Tuteja, R.; Tuteja, N. A single subunit MCM6 from pea forms homohexamer and functions as DNA helicase. Plant Mol. Biol. 2010, 74, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Dang, H.Q.; Tran, N.Q.; Gill, S.S.; Tuteja, R.; Tuteja, N. A single subunit MCM6 from pea promotes salinity stress tolerance without affecting yield. Plant Mol. Biol. 2011, 76, 19–34. [Google Scholar] [CrossRef]
- Tuteja, R. Helicases from all domains of life; Academic Press: London, UK, 2019; pp. 39–52. [Google Scholar]
- Crismani, W.; Portemer, V.; Froger, N.; Chelysheva, L.; Horlow, C.; Vrielynck, N.; Mercier, R. MCM8 is required for a pathway of meiotic double-strand break repair independent of DMC1 in Arabidopsis thaliana. PLoS Genet. 2013, 9, e1003165. [Google Scholar] [CrossRef] [Green Version]
- Black, S.J.; Kashkina, E.; Kent, T.; Pomerantz, R.T. DNA Polymerase θ: A Unique Multifunctional End-Joining Machine. Genes 2016, 7, 67. [Google Scholar] [CrossRef] [Green Version]
- Chan, S.H.; Yu, A.M.; McVey, M. Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet. 2010, 6, e1001005. [Google Scholar] [CrossRef] [Green Version]
- Roerink, S.F.; van Schendel, R.; Tijsterman, M. Polymerase theta-mediated end joining of replication-associated DNA breaks in C. elegans. Genome Res. 2014, 24, 954–962. [Google Scholar] [CrossRef] [Green Version]
- Ozdemir, A.Y.; Rusanov, T.; Kent, T.; Siddique, L.A.; Pomerantz, R.T. Polymerase θ-helicase efficiently unwinds DNA and RNA-DNA hybrids. J. Biol. Chem. 2018, 293, 5259–5269. [Google Scholar] [CrossRef] [Green Version]
- Seki, M.; Marini, F.; Wood, R.D. POLQ (Pol theta), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res. 2003, 31, 6117–6126. [Google Scholar] [CrossRef] [Green Version]
- Newman, J.A.; Cooper, C.D.O.; Aitkenhead, H.; Gileadi, O. Structure of the Helicase Domain of DNA Polymerase Theta Reveals a Possible Role in the Microhomology-Mediated End-Joining Pathway. Structure 2015, 23, 2319–2330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inagaki, S.; Suzuki, T.; Ohto, M.-A.; Urawa, H.; Horiuchi, T.; Nakamura, K.; Morikami, A. Arabidopsis TEBICHI, with helicase and DNA polymerase domains, is required for regulated cell division and differentiation in meristems. Plant Cell 2006, 18, 879–892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klemm, T.; Mannuß, A.; Kobbe, D.; Knoll, A.; Trapp, O.; Dorn, A.; Puchta, H. The DNA translocase RAD5A acts independently of the other main DNA repair pathways, and requires both its ATPase and RING domain for activity in Arabidopsis thaliana. Plant J. 2017, 91, 725–740. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, S.; Nakamura, K.; Morikami, A. A link among DNA replication, recombination, and gene expression revealed by genetic and genomic analysis of TEBICHI gene of Arabidopsis thaliana. PLoS Genet. 2009, 5, e1000613. [Google Scholar] [CrossRef] [Green Version]
- Mara, K.; Charlot, F.; Guyon-Debast, A.; Schaefer, D.G.; Collonnier, C.; Grelon, M.; Nogué, F. POLQ plays a key role in the repair of CRISPR/Cas9-induced double-stranded breaks in the moss Physcomitrella patens. New Phytol. 2019, 222, 1380–1391. [Google Scholar] [CrossRef]
- Gelvin, S.B. Integration of Agrobacterium T-DNA into the Plant Genome. Annu. Rev. Genet. 2017, 51, 195–217. [Google Scholar] [CrossRef]
- van Kregten, M.; de Pater, S.; Romeijn, R.; van Schendel, R.; Hooykaas, P.J.J.; Tijsterman, M. T-DNA integration in plants results from polymerase-θ-mediated DNA repair. Nat. Plants 2016, 2, 16164. [Google Scholar] [CrossRef]
- Huang, T.-K.; Puchta, H. CRISPR/Cas-mediated gene targeting in plants: Finally a turn for the better for homologous recombination. Plant Cell Rep. 2019, 38, 443–453. [Google Scholar] [CrossRef]
- Wolter, F.; Puchta, H. In planta gene targeting can be enhanced by the use of CRISPR/Cas12a. Plant J. 2019. [Google Scholar] [CrossRef] [Green Version]
- Fauser, F.; Roth, N.; Pacher, M.; Ilg, G.; Sánchez-Fernández, R.; Biesgen, C.; Puchta, H. In planta gene targeting. Proc. Natl. Acad. Sci. USA 2012, 109, 7535–7540. [Google Scholar] [CrossRef] [Green Version]
- Blary, A.; Jenczewski, E. Manipulation of crossover frequency and distribution for plant breeding. Theor. Appl. Genet. 2019, 132, 575–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lambing, C.; Heckmann, S. Tackling Plant Meiosis: From Model Research to Crop Improvement. Front. Plant Sci. 2018, 9, 829. [Google Scholar] [CrossRef] [PubMed]
- Fayos, I.; Mieulet, D.; Petit, J.; Meunier, A.C.; Périn, C.; Nicolas, A.; Guiderdoni, E. Engineering meiotic recombination pathways in rice. Plant Biotechnol. J. 2019, 17, 2062–2077. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.B.; Séguéla-Arnaud, M.; Larchevêque, C.; Lloyd, A.H.; Mercier, R. Unleashing meiotic crossovers in hybrid plants. Proc. Natl. Acad. Sci. USA 2018, 115, 2431–2436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
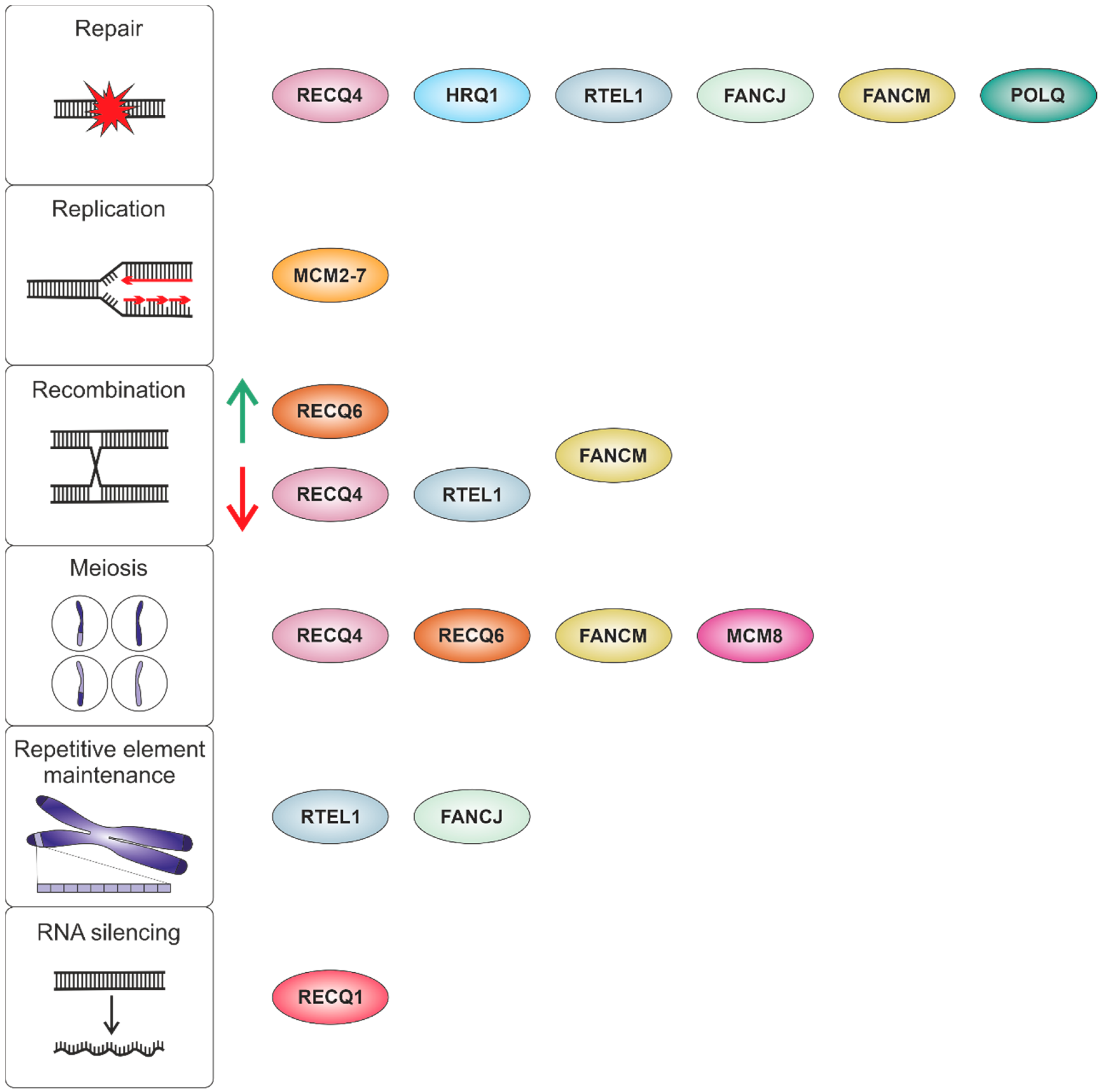
| RecQ Subfamily | Human | Arabidopsis thaliana | Physcomitrella patens |
|---|---|---|---|
| RecQ1 | - | RECQ1 | - |
| RecQ2 | RECQ1 | RECQ2 | RECQ2 |
| RecQ3 | RECQ5 | RECQ3 | - |
| RecQ4 | BLM | RECQ4ARECQ4B | RECQ4 |
| RecQ5 | RECQ4 | RECQ5 | RECQ5 |
| (HRQ1) | |||
| RecQ6/RecQsim | WRN | RECQsim | RECQ6 RECQsim1 RECQsim2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dorn, A.; Puchta, H. DNA Helicases as Safekeepers of Genome Stability in Plants. Genes 2019, 10, 1028. https://doi.org/10.3390/genes10121028
Dorn A, Puchta H. DNA Helicases as Safekeepers of Genome Stability in Plants. Genes. 2019; 10(12):1028. https://doi.org/10.3390/genes10121028
Chicago/Turabian StyleDorn, Annika, and Holger Puchta. 2019. "DNA Helicases as Safekeepers of Genome Stability in Plants" Genes 10, no. 12: 1028. https://doi.org/10.3390/genes10121028
APA StyleDorn, A., & Puchta, H. (2019). DNA Helicases as Safekeepers of Genome Stability in Plants. Genes, 10(12), 1028. https://doi.org/10.3390/genes10121028





