CaMad2 Promotes Multiple Aspects of Genome Stability Beyond Its Direct Function in Chromosome Segregation
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain Construction
2.2. Growth Analysis
2.3. Serial Dilution and Plate Spotting
2.4. Minimum Inhibitory Concentration Assay
2.5. Fluctuation Analyses of Chromosome Loss Rates
2.6. SNP-RFLP
2.7. Reverse Transcriptase qPCR
2.8. Microscopy
3. Results
3.1. Deletion of CaMAD2 Reduces Growth
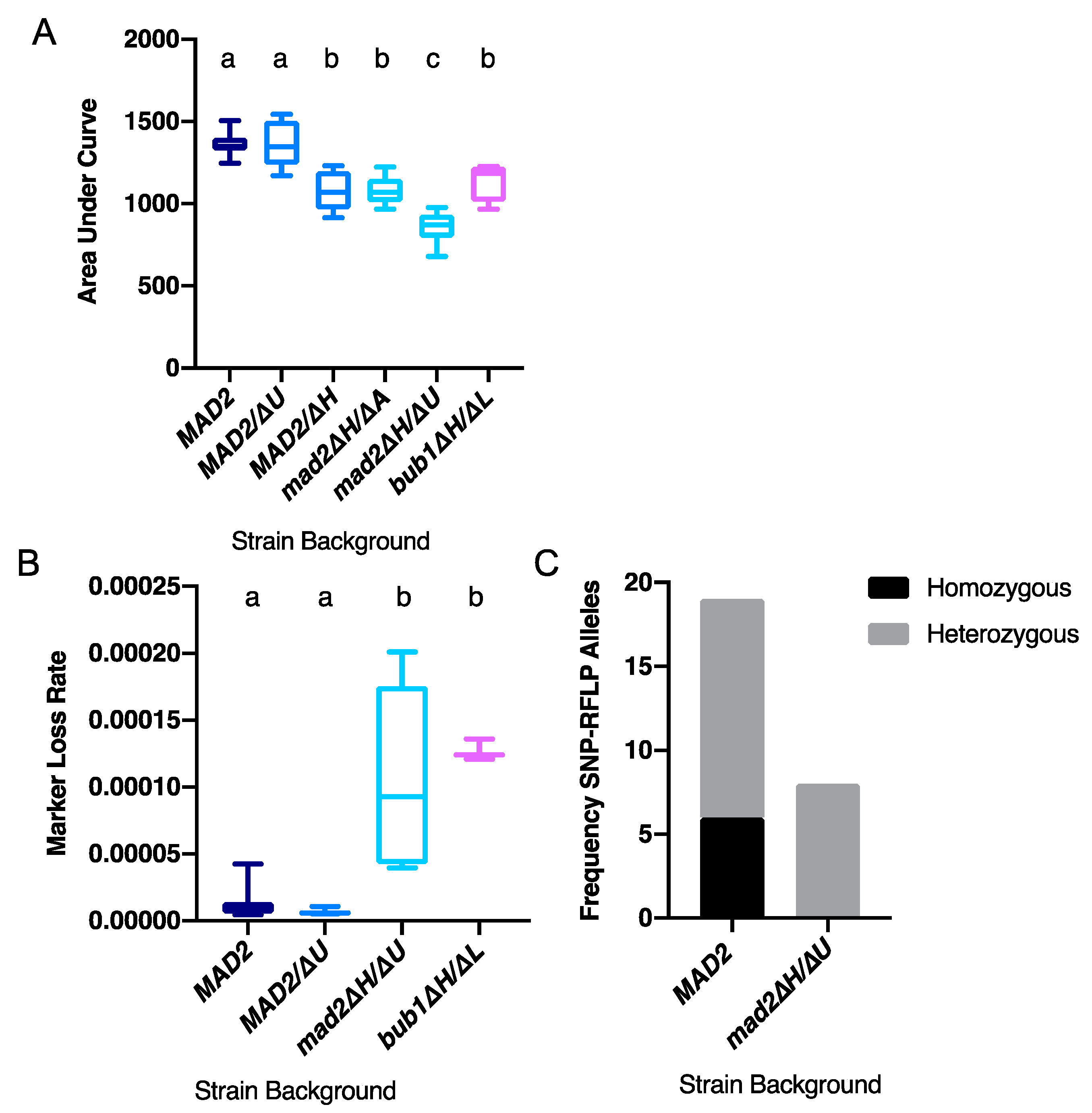
3.2. Deletion of CaMAD2 Increases Genome Instability
3.3. More Kinetochore-Microtubule Attachments Per Centromere Does Not Affect the Requirement for MAD2
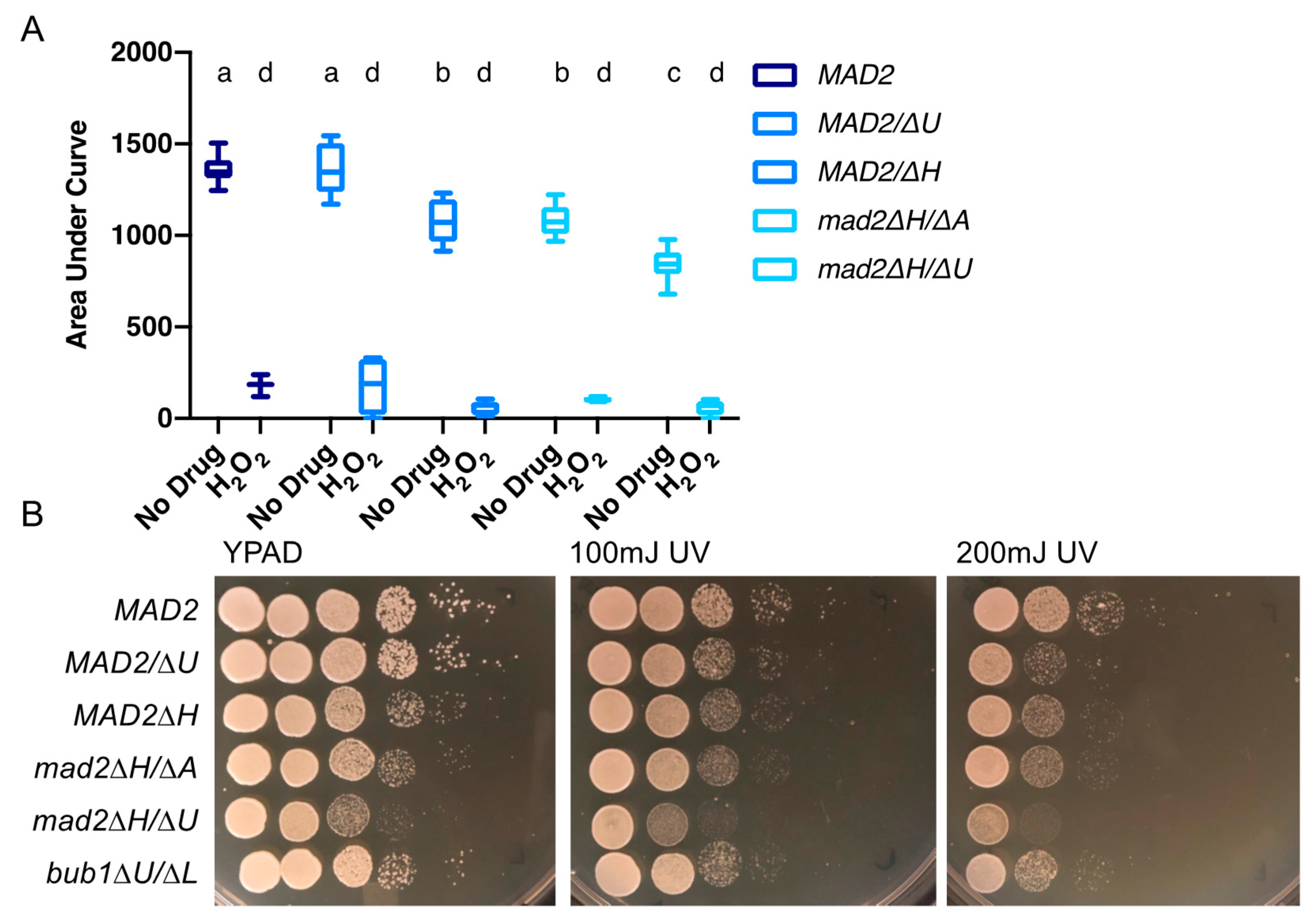
3.4. Loss of MAD2 Increases Sensitivity to Microtubule-Destabilizing Compounds
3.5. Deletion of MAD2 Increases Sensitivity to DNA-Damage Inducing Treatments
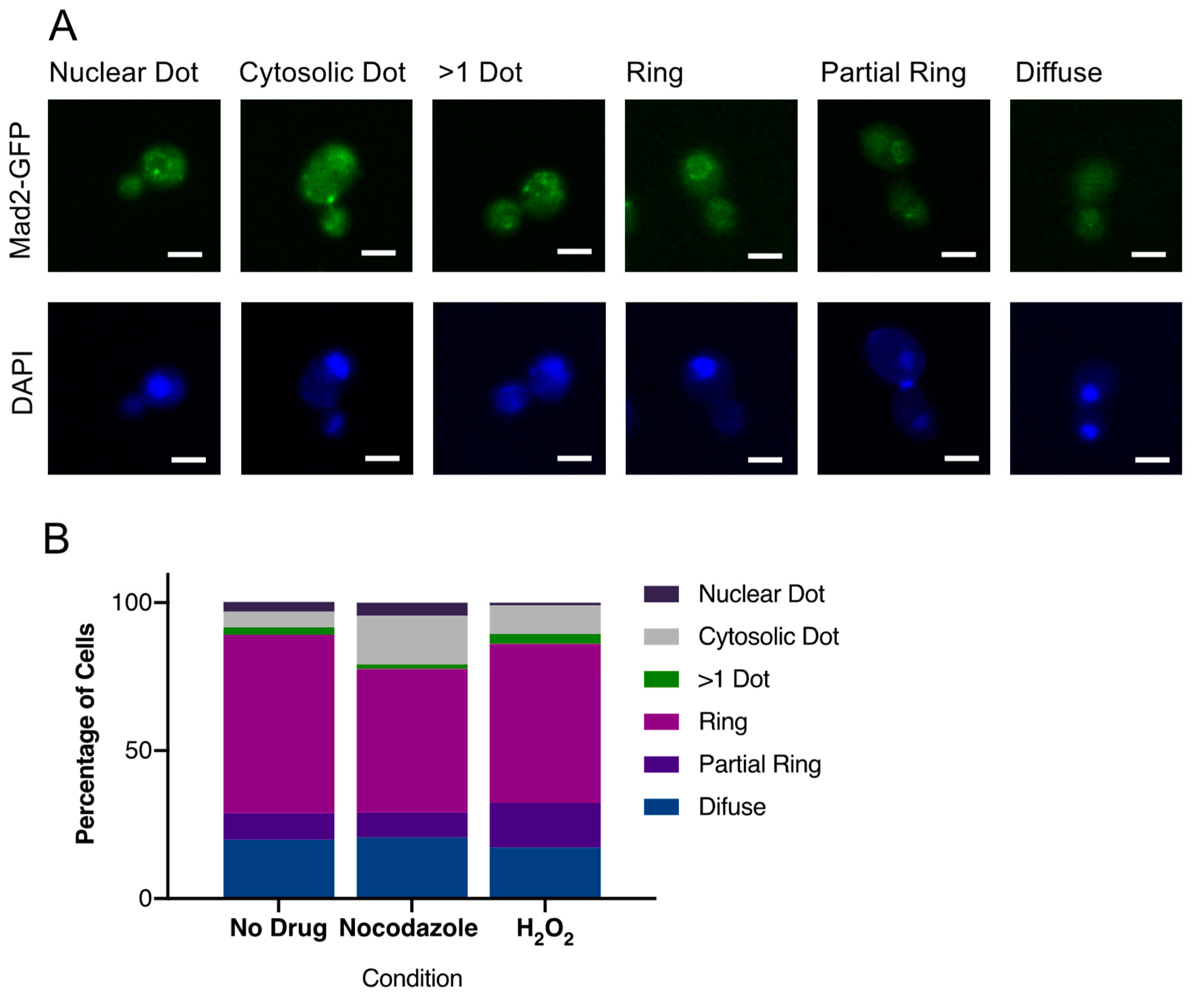
3.6. CaMad2 Localizes to the Nuclear Periphery in C. albicans
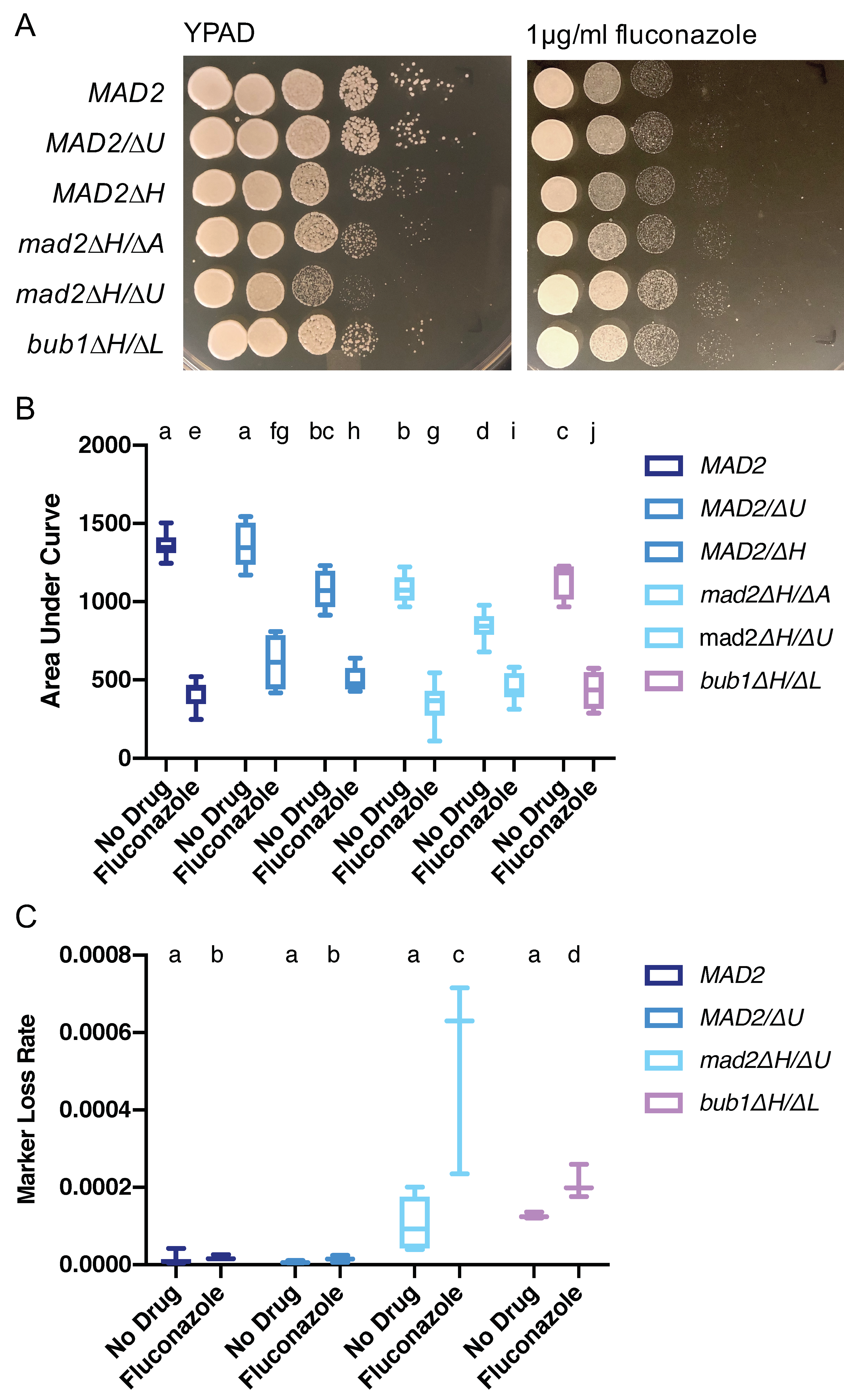
3.7. Increased Growth on Fluconazole Is a Shared Feature of Mutants with Altered Chromosome Segregation Checkpoint Function in C. albicans
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Forche, A.; Cromie, G.; Gerstein, A.C.; Solis, N.V.; Pisithkul, T.; Srifa, W.; Jeffery, E.; Abbey, D.; Filler, S.G.; Dudley, A.M.; et al. Rapid Phenotypic and Genotypic Diversification After Exposure to the Oral Host Niche in Candida albicans. Genetics 2018, 209, 725–741. [Google Scholar] [PubMed]
- Selmecki, A.M.; Dulmage, K.; Cowen, L.E.; Anderson, J.B.; Berman, J. Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLoS Genet. 2009, 5, e1000705. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Teoh, F.; Tan, A.S.M.; Cao, Y.; Pavelka, N.; Berman, J. Aneuploidy Enables Cross-Adaptation to Unrelated Drugs. Mol. Biol. Evol. 2019, 36, 1768–1782. [Google Scholar] [CrossRef]
- Stone, N.R.; Rhodes, J.; Fisher, M.C.; Mfinanga, S.; Kivuyo, S.; Rugemalila, J.; Segal, E.S.; Needleman, L.; Molloy, S.F.; Kwon-Chung, J.; et al. Dynamic ploidy changes drive fluconazole resistance in human cryptococcal meningitis. J. Clin. Investig. 2019, 129, 999–1014. [Google Scholar] [CrossRef] [PubMed]
- Lara-Gonzalez, P.; Westhorpe, F.G.; Taylor, S.S. The spindle assembly checkpoint. Curr. Biol. 2012, 22, R966–R980. [Google Scholar] [CrossRef]
- Lawrence, K.S.; Chau, T.; Engebrecht, J. DNA damage response and spindle assembly checkpoint function throughout the cell cycle to ensure genomic integrity. PLoS Genet. 2015, 11, e1005150. [Google Scholar] [CrossRef]
- Gay, S.; Piccini, D.; Bruhn, C.; Ricciardi, S.; Soffientini, P.; Carotenuto, W.; Biffo, S.; Foiani, M. A Mad2-Mediated Translational Regulatory Mechanism Promoting S-Phase Cyclin Synthesis Controls Origin Firing and Survival to Replication Stress. Mol. Cell 2018, 70, 628–638. [Google Scholar] [CrossRef]
- Campbell, M.S.; Chan, G.K.; Yen, T.J. Mitotic checkpoint proteins HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. J. Cell Sci. 2001, 114, 953–963. [Google Scholar]
- Iouk, T.; Kerscher, O.; Scott, R.J.; Basrai, M.A.; Wozniak, R.W. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J. Cell Biol. 2002, 159, 807–819. [Google Scholar] [CrossRef]
- De Souza, C.P.; Hashmi, S.B.; Nayak, T.; Oakley, B.; Osmani, S.A. Mlp1 acts as a mitotic scaffold to spatially regulate spindle assembly checkpoint proteins in Aspergillus nidulans. Mol. Biol. Cell 2009, 20, 2146–2159. [Google Scholar] [CrossRef]
- Saitoh, S.; Kobayashi, Y.; Ogiyama, Y.; Takahashi, K. Dual Regulation of Mad2 Localization on Kinetochores by Bub1 and Dam1/DASH that Ensure Proper Spindle Interaction. Mol. Biol. Cell 2008, 19, 3885–3897. [Google Scholar] [CrossRef] [PubMed]
- Scott, R.J.; Lusk, C.P.; Dilworth, D.J.; Aitchison, J.D.; Wozniak, R.W. Interactions between Mad1p and the Nuclear Transport Machinery in the Yeast Saccharomyces cerevisiae. Mol. Biol. Cell 2005, 16, 4362–4374. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Kruse, T.; Guasch Boldu, C.; Garvanska, D.H.; Coscia, F.; Mann, M.; Barisic, M.; Nilsson, J. Efficient mitotic checkpoint signaling depends on integrated activities of Bub1 and the RZZ complex. EMBO J. 2019, 38, e100977. [Google Scholar] [CrossRef] [PubMed]
- Leontiou, I.; London, N.; May, K.M.; Ma, Y.; Grzesiak, L.; Medina-Pritchard, B.; Amin, P.; Jeyaprakash, A.A.; Biggins, S.; Hardwick, K.G. The Bub1-TPR Domain Interacts Directly with Mad3 to Generate Robust Spindle Checkpoint Arrest. Curr. Biol. CB 2019, 29, 2407–2414. [Google Scholar] [CrossRef] [PubMed]
- Steenwyk, J.L.; Opulente, D.A.; Kominek, J.; Shen, X.-X.; Zhou, X.; Labella, A.L.; Bradley, N.P.; Eichman, B.F.; Čadež, N.; Libkind, D.; et al. Extensive loss of cell-cycle and DNA repair genes in an ancient lineage of bipolar budding yeasts. PLoS Biol. 2019, 17, e3000255. [Google Scholar] [CrossRef]
- Forche, A.; Alby, K.; Schaefer, D.; Johnson, A.D.; Berman, J.; Bennett, R.J. The Parasexual Cycle in Candida albicans Provides an Alternative Pathway to Meiosis for the Formation of Recombinant Strains. PLoS Biol. 2008, 6, e110. [Google Scholar] [CrossRef]
- Brimacombe, C.A.; Burke, J.E.; Parsa, J.-Y.; Catania, S.; O’Meara, T.R.; Witchley, J.N.; Burrack, L.S.; Madhani, H.D.; Noble, S.M. A natural histone H2A variant lacking the Bub1 phosphorylation site and regulated depletion of centromeric histone CENP-A foster evolvability in Candida albicans. PLoS Biol. 2019, 17, e3000331. [Google Scholar] [CrossRef] [PubMed]
- Kamthan, M.; Nalla, V.K.; Ruhela, D.; Kamthan, A.; Maiti, P.; Datta, A. Characterization of a Putative Spindle Assembly Checkpoint Kinase Mps1, Suggests Its Involvement in Cell Division, Morphogenesis and Oxidative Stress Tolerance in Candida albicans. PLoS ONE 2014, 9, e101517. [Google Scholar] [CrossRef]
- Bai, C.; Ramanan, N.; Wang, Y.M.; Wang, Y. Spindle assembly checkpoint component CaMad2p is indispensable for Candida albicans survival and virulence in mice. Mol. Microbiol. 2002, 45, 31–44. [Google Scholar] [CrossRef]
- Wilson, R.B.; Davis, D.; Mitchell, A.P. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 1999, 181, 1868–1874. [Google Scholar]
- Gerami-Nejad, M.; Berman, J.; Gale, C.A. Cassettes for PCR-mediated construction of green, yellow, and cyan fluorescent protein fusions in Candida albicans. Yeast 2001, 18, 859–864. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, K.; Carbon, J. The CENP-A homolog CaCse4p in the pathogenic yeast Candida albicans is a centromere protein essential for chromosome transmission. Proc. Natl. Acad. Sci. USA 2002, 99, 12969–12974. [Google Scholar] [CrossRef] [PubMed]
- Cuenca-Estrella, M.; Moore, C.B.; Barchiesi, F.; Bille, J.; Chryssanthou, E.; Denning, D.W.; Donnelly, J.P.; Dromer, F.; Dupont, B.; Rex, J.H.; et al. Multicenter evaluation of the reproducibility of the proposed antifungal susceptibility testing method for fermentative yeasts of the Antifungal Susceptibility Testing Subcommittee of the European Committee on Antimicrobial Susceptibility Testing (AFST-EUCAST). Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2003, 9, 467–474. [Google Scholar]
- Spell, R.M.; Jinks-Robertson, S. Examination of the roles of Sgs1 and Srs2 helicases in the enforcement of recombination fidelity in Saccharomyces cerevisiae. Genetics 2004, 168, 1855–1865. [Google Scholar] [CrossRef]
- Lea, D.E.; Coulson, C.A. The distribution of the numbers of mutants in bacterial populations. J. Genet. 1949, 49, 264–285. [Google Scholar] [CrossRef]
- Forche, A.; Abbey, D.; Pisithkul, T.; Weinzierl, M.A.; Ringstrom, T.; Bruck, D.; Petersen, K.; Berman, J. Stress alters rates and types of loss of heterozygosity in Candida albicans. MBio 2011, 2, e00129-11. [Google Scholar] [CrossRef]
- Michel, L.; Benezra, R.; Diaz-Rodriguez, E. MAD2 dependent mitotic checkpoint defects in tumorigenesis and tumor cell death: A double edged sword. Cell Cycle 2004, 3, 990–992. [Google Scholar] [CrossRef]
- Wilmes, A.; Hanna, R.; Heathcott, R.W.; Northcote, P.T.; Atkinson, P.H.; Bellows, D.S.; Miller, J.H. Chemical genetic profiling of the microtubule-targeting agent peloruside A in budding yeast Saccharomyces cerevisiae. Gene 2012, 497, 140–146. [Google Scholar] [CrossRef]
- Skrzypek, M.S.; Binkley, J.; Binkley, G.; Miyasato, S.R.; Simison, M.; Sherlock, G. The Candida Genome Database (CGD): Incorporation of Assembly 22, systematic identifiers and visualization of high throughput sequencing data. Nucleic Acids Res. 2017, 45, D592–D596. [Google Scholar] [CrossRef]
- Burrack, L.S.; Applen, S.E.; Berman, J. The requirement for the Dam1 complex is dependent upon the number of kinetochore proteins and microtubules. Curr. Biol. CB 2011, 21, 889–896. [Google Scholar] [CrossRef]
- Li, Y.; Benezra, R. Identification of a human mitotic checkpoint gene: HsMAD2. Science 1996, 274, 246–248. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Murray, A.W. Feedback control of mitosis in budding yeast. Cell 1991, 66, 519–531. [Google Scholar] [CrossRef]
- Sawin, K.E.; Nurse, P. Regulation of Cell Polarity by Microtubules in Fission Yeast. J. Cell Biol. 1998, 142, 457–471. [Google Scholar] [CrossRef] [PubMed]
- Lyon, G.M.; Karatela, S.; Sunay, S.; Adiri, Y. Candida Surveillance Study Investigators. Antifungal Susceptibility Testing of Candida Isolates from the Candida Surveillance Study. J. Clin. Microbiol. 2010, 48, 1270–1275. [Google Scholar] [CrossRef]
- Anderson, C.A.; Roberts, S.; Zhang, H.; Kelly, C.M.; Kendall, A.; Lee, C.; Gerstenberger, J.; Koenig, A.B.; Kabeche, R.; Gladfelter, A.S. Ploidy variation in multinucleate cells changes under stress. Mol. Biol. Cell 2015, 26, 1129–1140. [Google Scholar] [CrossRef]
- Zhang, L.G.; Zhang, Y.; Li, B.C.; Jia, X.J.; Chen, C.J.; Zhou, M.G. Involvement of FgMad2 and FgBub1 in regulating fungal development and carbendazim resistance in Fusarium graminearum. Plant Pathol. 2015, 64, 1014–1028. [Google Scholar] [CrossRef]
- Michel, L.S.; Liberal, V.; Chatterjee, A.; Kirchwegger, R.; Pasche, B.; Gerald, W.; Dobles, M.; Sorger, P.K.; Murty, V.V.V.S.; Benezra, R. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature 2001, 409, 355–359. [Google Scholar] [CrossRef]
- Hisaoka, M.; Matsuyama, A.; Hashimoto, H. Aberrant MAD2 expression in soft-tissue sarcoma. Pathol. Int. 2008, 58, 329–333. [Google Scholar] [CrossRef]
- Wang, L.; Yin, F.; Du, Y.; Du, W.; Chen, B.; Zhang, Y.; Wu, K.; Ding, J.; Liu, J.; Fan, D. MAD2 as a key component of mitotic checkpoint: A probable prognostic factor for gastric cancer. Am. J. Clin. Pathol. 2009, 131, 793–801. [Google Scholar] [CrossRef]
- Nezi, L.; Rancati, G.; De Antoni, A.; Pasqualato, S.; Piatti, S.; Musacchio, A. Accumulation of Mad2–Cdc20 complex during spindle checkpoint activation requires binding of open and closed conformers of Mad2 in Saccharomyces cerevisiae. J. Cell Biol. 2006, 174, 39–51. [Google Scholar] [CrossRef]
- Elow, S. Bub1 and BubR1: At the Interface between Chromosome Attachment and the Spindle Checkpoint. Mol. Cell Biol. 2011, 31, 3085–3093. [Google Scholar] [CrossRef]
- Gillett, E.S.; Espelin, C.W.; Sorger, P.K. Spindle checkpoint proteins and chromosome–microtubule attachment in budding yeast. J. Cell Biol. 2004, 164, 535–546. [Google Scholar] [CrossRef]
- Dotiwala, F.; Harrison, J.C.; Jain, S.; Sugawara, N.; Haber, J.E. Mad2 prolongs DNA damage checkpoint arrest caused by a double-strand break via a centromere-dependent mechanism. Curr. Biol. 2010, 20, 328–332. [Google Scholar] [CrossRef]
- Daniel, J.A.; Keyes, B.E.; Ng, Y.P.Y.; Freeman, C.O.; Burke, D.J. Diverse functions of spindle assembly checkpoint genes in Saccharomyces cerevisiae. Genetics 2006, 172, 53–65. [Google Scholar] [CrossRef]
- Selmecki, A.; Forche, A.; Berman, J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 2006, 313, 367–370. [Google Scholar] [CrossRef]
- Harrison, B.D.; Hashemi, J.; Bibi, M.; Pulver, R.; Bavli, D.; Nahmias, Y.; Wellington, M.; Sapiro, G.; Berman, J. A tetraploid intermediate precedes aneuploid formation in yeasts exposed to fluconazole. PLoS Biol. 2014, 12, e1001815. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vossen, M.L.; Alhosawi, H.M.; Aney, K.J.; Burrack, L.S. CaMad2 Promotes Multiple Aspects of Genome Stability Beyond Its Direct Function in Chromosome Segregation. Genes 2019, 10, 1013. https://doi.org/10.3390/genes10121013
Vossen ML, Alhosawi HM, Aney KJ, Burrack LS. CaMad2 Promotes Multiple Aspects of Genome Stability Beyond Its Direct Function in Chromosome Segregation. Genes. 2019; 10(12):1013. https://doi.org/10.3390/genes10121013
Chicago/Turabian StyleVossen, Maicy L., Hanaa M. Alhosawi, Katherine J. Aney, and Laura S. Burrack. 2019. "CaMad2 Promotes Multiple Aspects of Genome Stability Beyond Its Direct Function in Chromosome Segregation" Genes 10, no. 12: 1013. https://doi.org/10.3390/genes10121013
APA StyleVossen, M. L., Alhosawi, H. M., Aney, K. J., & Burrack, L. S. (2019). CaMad2 Promotes Multiple Aspects of Genome Stability Beyond Its Direct Function in Chromosome Segregation. Genes, 10(12), 1013. https://doi.org/10.3390/genes10121013




