Abstract
Listeria monocytogenes is a highly fatal foodborne causative agent that has been implicated in numerous outbreaks and related deaths of listeriosis in the world. In this study, six L. monocytogenes isolated from ready-to-eat (RTE) meat products were analysed using Whole Genome Sequencing (WGS) to identify virulence and resistance genes, prophage sequences, PCR-serogroups, and sequence types (STs). The WGS identified four different STs (ST1, ST121, ST204, and ST876) that belonged to serogroup 4b (lineage I) and 1/2a (lineage II). Core genome, and average nucleotide identity (ANI) phylogenetic analyses showed that the majority of strains from serogroup 4b (lineage I) clustered together. However, two isolates that belong to serogroup 1/2a (lineage II) grouped far from each other and the other strains. Examination of reference-guided scaffolds for the presence of prophages using the PHAge Search Tool Enhanced Release (PHASTER) software identified 24 diverse prophages, which were either intact or incomplete/questionable. The National Center for Biotechnology Information- Nucleotide Basic Local Alignment Search Tool (NCBI-BLASTn) revealed that Listeria monocytogenes strains in this study shared some known major virulence genes that are encoded in Listeria pathogenicity islands 1 and 3. In general, the resistance profiles for all the isolates were similar and encoded for multidrug, heavy metal, antibiotic, and sanitizer resistance genes. All the isolates in this study possessed genes that code for resistance to common food processing antiseptics such as Benzalkonium chloride.
1. Introduction
Listeria species are ubiquitous bacteria widely distributed in the environment of which Listeria monocytogenes is the most important zoonotic species of global public health and economic importance in the genus [1]. The general approach to prevent listeriosis in the human population is to restrict the exposure of the human and animal populations to foods contaminated with L. monocytogenes; however, in the case of a listeriosis outbreak, timeous removal of suspected foods during ongoing epidemiological investigations limits human exposure and spread of the disease [2,3]. Identification of suspected contaminated food during a listeriosis outbreak is primarily performed by patient interview, a process that is long and is usually hampered by several factors such as low incidence of L. monocytogenes, the ubiquitous presence of L. monocytogenes in the environment, and the wide variation of the incubation time which usually range from 3–90 days [4,5]. These drawbacks are exacerbated by limitations in patient memory during interviews and, in some instances, the inability to conduct effective interviews [6].
Recently, molecular-based subtyping comparisons to match human isolates to food or environmental isolates have become critical for tracking and source identification of the cause of outbreak [7]. Traditionally, pulsed-field gel electrophoresis (PFGE) has been used as the “gold standard” for subtyping of L. monocytogenes isolates involved in outbreaks and sporadic cases; however, Whole Genome Sequencing (WGS) has emerged as a powerful tool for subtyping and investigation of L. monocytogenes outbreak cases [8]. Typing in WGS is performed at higher resolution than that of traditional molecular typing methods as it uses the entire genome of a bacterium and, consequently, WGS can reveal the genetic differences between the sequence types, the acquisition, and evolution of virulence as well as the pathogenic traits and antimicrobial resistance profiles of L. monocytogenes [9]. In 2013, the United States employed WGS as a primary method for subtyping of L. monocytogenes, which led to the identification of more outbreaks than could have not been detected by PFGE. The use of WGS further led to the differentiation between strains with indistinguishable PFGE profiles which enhanced resolution in the outbreak investigations [10]. Moreover, WGS is important in helping to understand the biology, phylogeny, and ecology of L. monocytogenes contamination in the food value chain [11].
The cost reduction of WGS has allowed it to become the preferred method for molecular subtyping of L. monocytogenes outbreaks and a viable alternative tool for the source attribution of listeriosis cases [12,13]. Apart from two studies that reported on the use of WGS for typing of L. monocytogenes, which was associated with human listeriosis outbreaks in 2015 and 2018 [14,15], there is no published genomic information on this pathogen in South Africa. Considering that listeriosis is a notifiable human disease in many countries including South Africa and considering its association with ready-to-eat (RTE) food products, a need exists to generate more genomic data on L. monocytogenes obtained from RTE products for epidemiological purposes such as source identification and tracking. Polony and biltong are the most popular RTE meat products in South Africa, accounting for approximately up to 50% of the country RTE meat product production [16,17]. Therefore, the aim of this study was to characterise the strains of L. monocytogenes isolated from RTE meat products in South Africa. The WGS information of the L. monocytogenes strains was analysed in order to identify virulence and resistance genes, prophage sequences, phylogeny, PCR-serogroup, and sequence type (ST).
2. Materials and Methods
2.1. Sample Information
The samples used in this study were collected from supermarkets and butcheries located in four provinces of South Africa, namely Gauteng, Limpopo, Mpumalanga, and Western Cape, as indicated in Figure 1 as part of the routine national survey for L. monocytogenes in meat and meat products in South Africa [18]. Isolates of L. monocytogenes from biltong (n = 5) and Polony (n = 1) samples were sequenced in this study. Samples were collected aseptically between 2015 and 2016 using sterile plastic bags and transported on ice immediately to the Onderstepoort Veterinary Research (OVR): Feed and Food laboratory, SA for microbiological analysis.
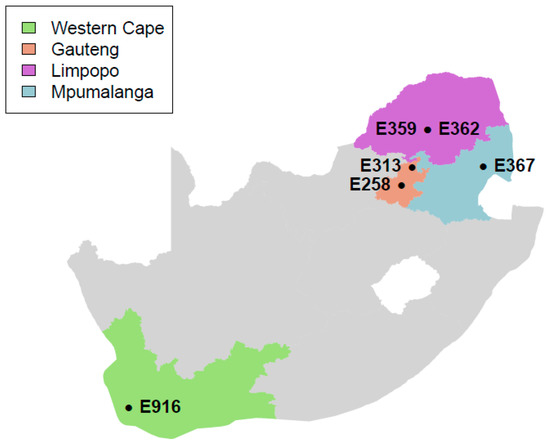
Figure 1.
Location of supermarkets and butcheries from which samples were collected in South Africa.
2.2. Microbiological Analysis
Microbiological analysis of the samples was performed according to procedure described by Matle et al. [18]. Briefly, samples weighing 25 g each were aseptically transferred into 225 mL of ONE broth-Listeria (Oxoid, Basingstoke, UK), followed by homogenization for 2 min using a Stomacher (Stomacher Lab Blender 400, Seward Ltd., West Sussex, UK). After homogenization, the broth sample was incubation at 35 °C for 24 hours. The broth samples (10 µL per sample) were inoculated onto Brilliance-Listeria plates (Oxoid, Basingstoke, UK) and incubated at 35 °C for 24 hours. Presumptive Listeria colonies were subjected to Oxoid Biochemical Identification System (Oxoid, Basingstoke, UK) for identification. The isolates that were confirmed as L. monocytogenes were preserved in brain–heart infusion (Oxoid, Basingstoke, UK) broth supplemented with 35% glycerol and stored at −80 °C at OVR: Feed and Food laboratory.
2.3. Genomic Deoxyribonucleic Acid (DNA) Extraction
DNA was extracted using the High Pure Polymerase Chain Reaction (PCR) Template preparation kit (Roche, Potsdam, Germany) as per manufacturer’s protocol. Briefly, pure colonies of L. monocytogenes on blood agar (Oxoid, Basingstoke, UK) were inoculated into 50 µL of DNA-free water followed by adding 200 µL of binding buffer and 40 µL of Proteinase K. The mixture was then incubated at 70 °C using heating block for 10 min. After incubation, 100 µL of isopropanol was added and the mixture was applied to a High Pure Filter tube followed by centrifugation at 13,000 rpm for a minute. The flow-through and collection tube were discarded. Then, 500 µL of inhibitor removal buffer was added on the High Pure Filter tube followed by centrifuging at 13,000 rpm for a minute and discarding of the flow-through and collection tube. The filter tube was washed two times with 500 µL of wash buffer and centrifuged at 13,000 rpm for a minute. Then, 200 µL of elution buffer (70 °C) and new collection tubes were added, followed by centrifuging at 13,000 rpm for a minute. The extracted DNA was stored at −80 °C for further analyses. The DNA quantity and purity were assessed by using Qubit fluorimetric quantitation (Thermo Fisher Scientific, Waltham, MA, USA).
2.4. Genome Sequencing and De Novo Assembly
Whole genome sequencing of the samples was performed at the Biotechnology Platform, Agricultural Research Council, Onderstepoort, South Africa. The DNA libraries were prepared using the Nextera XT DNA library preparation kit (Illumina, San Diego, CA, USA), followed by 2 × 300 paired-end sequencing on a MiSeq instrument (Illumina, San Diego, CA, USA). Quality control, including adapter removal of the raw data, was done using Trimmomatic [19]. SPAdes “careful” mode was used to create a de novo assembly of each isolate [20]. All de novo assembled contigs were compared to the reference L. monocytogenes EGD-e chromosome, complete genome (NC_003210.1) using BLAST Ring Image Generator (BRIG) [21].
2.5. Core Genome Determination
Gene prediction in protein format was done for all contigs using PROkaryotic DYnamic programming Gene-finding Algorithm (Prodigal) [22], with the “closed ends” parameter specified to ensure the prediction of complete proteins. A total of 37 other Listeria complete genomes (Table 1) were obtained from the NCBI RefSeq database [23] to aid in the construction of a representative core genome. The PCR-serogroup, lineage, and Multilocus sequence typing (MLST) information for these strains were obtained from the Listeria database hosted by the Pasteur Institute, France (http://bigsdb.pasteur.fr/listeria/) [24]. All proteins from the 37 complete genomes and those predicted in our strains were compared by means of an all-against-all Protein BLAST (BLASTp) [25] with an e-value cut-off set at 1 × 10−30. The Markov Clustering (MCL) algorithm was used to cluster proteins in nonoverlapping groups [26]. For the MCL algorithm, the pairwise BLASTp bit-scores were used as edge weights and the inflation parameter was set at 1.8. Clusters containing single-copy orthologs from all strains as well as strains in this study were used for further phylogenetic analysis.

Table 1.
List of Listeria monocytogenes strains used for the construction of a core genome.
2.6. Core Genome Phylogenetic Analysis
Core genome clusters containing single-copy orthologs from all strains, including the controls, were used for core genome phylogenetic analysis. For each cluster, the protein sequences from all genomes were aligned using Multiple Alignment using Fast Fourier Transform (MAFFT) [27] with the default parameter “auto”. All multiple sequence alignments were concatenated into a single file, which was used for phylogenetic inference by the program IQ-TREE [28] with the “ModelFinder Plus” parameter to calculate the best-fit substitution model, and 1000 bootstrap replicates were specified. The HIVb+F+R2 was chosen as the best-fit model according to BIC. The resulting consensus tree was visualized and edited using the Interactive Tree of Life (iTOL) [29].
2.7. Prophage Identification and Analysis
Reference-guided scaffolding was done by means of an algorithm for genome scaffolding called Multi-Draft based Scaffolder (MeDuSa) [30] using L. monocytogenes EGD-e chromosome, complete genome (NC_003210.1) as a reference. The largest scaffold for each strain was used to predict putative prophages with PHAge Search Tool—Enhanced Release (PHASTER) [31].
2.8. Determination of Plasmids
In order to determine the plasmids, the contig files of the de novo assembled genomes from this study were analysed with Plasmid Finder 2.0 for the specified Gram-positive scheme [32].
2.9. Average Nucleotide Identity Calculation
Average nucleotide identity (ANI) was determined using ANI calculator, using both best hits (one-way ANI) and reciprocal best hits (two-way ANI) between genomic datasets as described [33]. In order to obtain normalized ANI values, a mean of pairwise ANI values were calculated from those obtained for all pairs between strains taking cognisance that the ANI values between genomes of the same species must be above 95%. Statistics on the distribution of ANI values were analysed using the R statistical software (http://www.r-project.org).
2.10. In Silico PCR-Serogroup and ST Prediction
PCR-serogroup and Multi Locus Sequence Type (MLST) profiles were obtained from the Listeria database hosted by the Pasteur Institute, France (http://bigsdb.pasteur.fr/listeria/) [24]. The PCR-serogroup database contains 5 loci with 142 different alleles, and the MLST database contains 7 loci with a total of 1799 different alleles. The presence and combination of these different alleles allow for the prediction of Listeria PCR-serogroups and MLST. A k-mer based, mapping tool, stringMLST [34] was used to align reads against these profiles to determine the PCR-serogroup and MLST for each sample using k-mers of length 35. To validate the k-mer-based predictions, all contigs were further compared against the PCR-serogroup and MLST profiles by BLASTn [25] with an e-value cut-off set to 1 × 10−30 and with allele presence and combination determined.
2.11. Virulence Factors
All predicted proteins for the samples were compared with the Virulence Factor Database (VFDB) [35] by means of BLASTp with an e-value cut-off set at 1 × 10−5 and a minimum identity of 75%. Some of the main virulence factors were examined using the well-annotated L. monocytogenes EDGe as a reference genome. Virulence factor intersections between samples were visualized with the R package UpSetR [36].
2.12. Resistance Profiles
Antibiotic and antibacterial biocide and metal-resistance profiles for the samples from this study were determined by comparing all the predicted proteins to the BacMet [37], MEGARes [38] and nonredundant antibiotic resistance database (noradab) (noradab.bi.up.ac.za) databases, respectively. This was done by means of BLASTp and tBLASTn for the MEGARes database which is in nucleotide format with an e-value cut-off set at 1 × 10−5 and a minimum identity of 75%.
2.13. Data Availability
The six genome sequences of L. monocytogenes isolates were deposited at the National Centre for Biotechnology Information (NCBI)/GenBank under the accession numbers from SAMN12360665 to SAMN12360670 (BioProject No. PRJNA556582).
3. Results
3.1. Genome Sequencing, Assembly, and Annotation
All de novo assembled contigs from genomes from this study were aligned and compared with L. monocytogenes EGD-e chromosome, complete genome (NC_003210.1) as a reference by means of BRIG (Figure 2). Visualization with BRIG indicates that the genomes displayed typical attributes of L. monocytogenes such as assembly sizes ranging from 2.9 to 3.1 base pairs and low genomic G+C content of 37.8%. The difference in assembly sizes was due to variation in the length of prophages from the sequenced isolates (Figure 3). De novo assembly ranged from 38 to 47 contigs with N50 between 411,134 bp and 637,980 bp (Table 2).
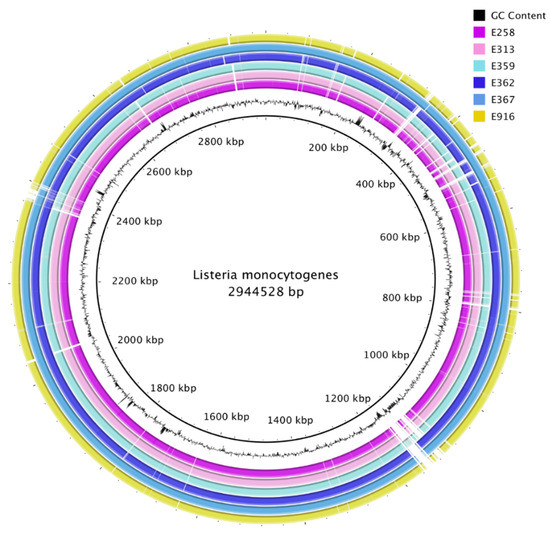
Figure 2.
Comparison of ready-to-eat (RTE) sample contigs against the Listeria monocytogenes EGD-e chromosome, complete genome (NC_003210.1), displayed in the inner black circle.
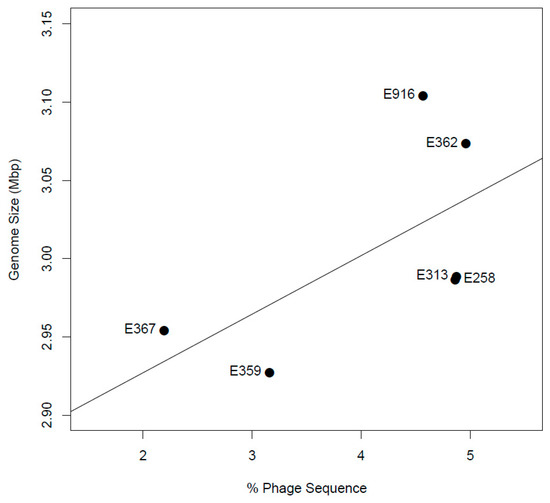
Figure 3.
The correlation of genome size with percentage of phage sequence.

Table 2.
General features of Listeria monocytogenes strains.
3.2. Plasmid Identification
Plasmid identification was performed to determine if the sequenced isolates in this study harboured plasmids that confer resistance of L. monocytogenes to antibiotics. The analysis of the plasmids revealed that none of the isolates possessed known plasmids associated with L. monocytogenes (Table 2).
3.3. Stress Survival Islet
The stress survival islets (SSI-1 and SSI2) are known to be responsible for the proliferation of L. monocytogenes under stressful conditions in food processing facilities [39,40]. In the current study, SSI-1 was detected in a E313 sample belonging to ST1 of lineage I. SSI-2 was present in two samples (E367 and E362) which belong to ST121 and ST204 of lineage II, respectively (Table 2).
3.4. Listeria Pathogenicity Islands
In this study, each of the 6 sequenced samples encoded Listeria pathogenicity islands 1 (LIPI1), which houses a cluster of six virulence genes of L. monocytogenes (Table 2). LIPI3, which is a gene cluster that encodes a potential haemolytic factor (Streptolysin S), was present in samples E258, E313, E359, and E916. Samples E258, E313, E359, and E916 belonged to serogroup 4b and lineage I. The LIPI3 was absent in samples E362 and E367 of serogroup 1/2a and lineage II. LIPI-4, which has been recently described as a gene cluster involved in neural and placental infection, was not detected in this study.
3.5. Multi-Locus Sequence Typing (MLST) and PCR-Serogroups
The MLST analysis, which is used to identify sequence types, lineages, and clone complexes of closely related L. monocytogenes strains, identified four different STs (Table 3). ST1 represented 50% (n = 3/6) of the samples while the other 50% was represented by ST121, ST204, and ST876. When silico PCR-serogroup analysis was used to differentiate among major serotypes of L. monocytogenes, it revealed that the strains from this study belonged to 4b (lineage I) and 1/2a (lineage II).

Table 3.
PCR-serogroup and MLST.
3.6. Core-Genome Phylogenetic Analysis
A total of 124,729 proteins were used for the generation of orthologous clusters which resulted in a core genome size of 1753. Core genome phylogenetic analysis reveals that the majority of samples (E252, E313, E359, and E916) in this study belonged to serogroup 4b (lineage I) clustered together (Figure 4). Two samples (E362 and E367) that were assigned to serogroup 1/2a (lineage II) grouped far from each other compared to the rest of the samples. The core genome phylogenetic analysis was in correlation with the predicted PCR-serogroups and MLST.
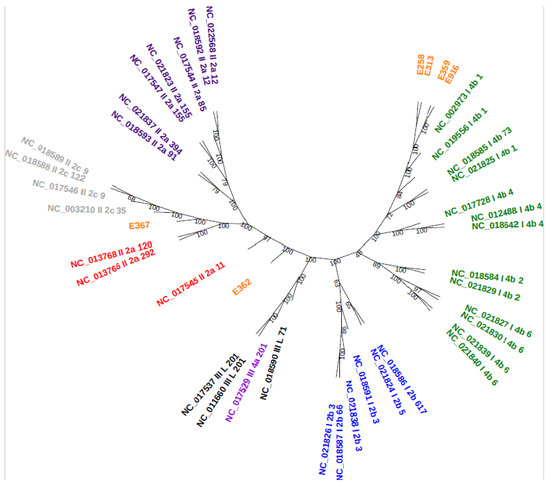
Figure 4.
Core genome phylogenetic tree: RefSeq accessions are followed by lineage, PCR-serogroup, and MLST. The RTE samples are indicated in orange.
3.7. Average Nucleotide Identities
Average Nucleotide Identity (ANI) is a measure of nucleotide-level genomic similarity between the coding regions of two genomes. The ANI phylogenetic analysis corroborated the core genome phylogeny analysis and MLST prediction results. The analysis indicated that samples E258 (1544), E313 (1545), E359 (1545), and E916 (1549) clustered together, while samples E362 (1547) and E367 (1548) were far from all the other samples (Figure 5). Sample E359 was sequence typed as ST876 from lineage I, which belong to serogroup IVb on MLST; however, on ANI (Figure 5), it clustered with most of the ST1s that belongs to the same lineage and serogroup.
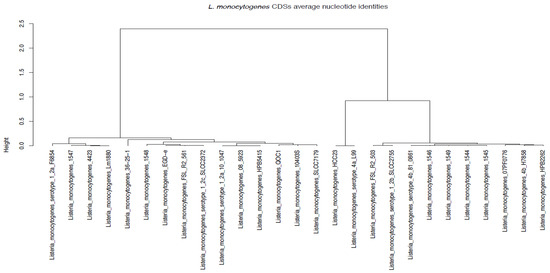
Figure 5.
Listeria monocytogenes CDS average nucleotide identities using Gower distance metric.
3.8. Virulence Factors
The distribution of main virulence genes of L. monocytogenes was surveyed in this study. A total of 142 virulence genes were identified across all six sequenced L. monocytogenes samples. Each sample contained between 80 and 86 virulence genes. Forty-six similar virulence genes were present in all the six sequenced strains. The NCBI-BLASTn revealed that samples from this study share some of the known major virulence genes. Those similar virulence genes include prfA, plcA, plcB, hly, iap/cwha, iapB, actA, and mpl as well as the internalin AB operon, which is encoded in LIPI1. However, other internalin family member genes (inlF, InlC, inlK, and InlJ) as well the genes vip, ami, gtcA, igt, oat, pdgA, agrA, agrC, prsA2, oppA, hbp2, srtA, and srtB were identified in all the samples.
Interestingly, samples E362 and E367 belonging to lineage II and serogroup 1/2a harboured the largest number of unique virulence genes and contained 21 similar genes, which were not present in the other samples. The unique virulence genes were shared by samples including the inlGHE cluster among others. The virulence genes associated with LIPI3 were not identified in samples E362 and E367. The LIPI3-associated genes were mostly identified in samples belonging to lineage I and included haemolysin known as listeriolysin S (LLS), llsB, llsG, llsY, llsX, and llsD genes. The truncated internalin A (inlA) gene and deletion in the ActA protein were not observed in all the samples.
3.9. Prophage Identification and Analysis
Reference-guided scaffolding resulted in near full-length assemblies for each sample, which allowed for the prediction of putative prophages (Table 4). A total of 24 prophages (either intact or incomplete/questionable) were identified in all genomes by PHASTER software including one intact prophage identified in sample E258 and two intact prophages identified in samples E313 and E362 (Table 4). Samples E258 and E916 harboured three intact prophages each, while no intact prophage was identified in E367. Furthermore, incomplete and questionable prophages were identified among various samples in this study. Sample E362 carried the highest number of incomplete prophages, followed by E258, E313, and E367 with single incomplete prophage each. Questionable prophages were observed in all the samples except E362. The Phage Classification Tool Set (PHACTS) [41] also gives a possible phage source for the intact prophages with PHAGE_Lister_ vB_LmoS_188 and PHAGE_Lister_vB_LmoS_293 phages being predicted in the majority of the samples. Other intact prophages that were identified included PHAGE_Lister LP030-2, PHAGE_Lister B054, PHAGE_Lister A006, and PHAGE_Lister LP101.

Table 4.
Scaffold length and predicted prophages of Listeria monocytogenes strains.
3.10. Resistance Genes
In general, the resistance profiles for all the samples sequenced in this study were similar. The examination of heavy metal, multidrug, and antibiotic resistance genes revealed the presence of resistance genes that coded for multidrug resistance genes (EmrB/QacA and Bcr/CflA family) quaternary ammonium compound resistance (SugE, Tn6188, and bcrABC), fosfomycin resistance (fosX), lead/cadmium/zinc resistance, copper resistance (CopC), cobalt/zinc/cadmium resistance, and aluminium resistance (CorA and CzcD) in all samples. Other resistance genes included tetA, tetM, mecC, mrB, msrA, lde, and mdrL.
4. Discussion
The current study characterised the genomic diversity of six L. monocytogenes isolates that were isolated from biltong (E258, E313, E359, E362, and E367) and polony (E916) samples collected in four provinces of South Africa between 2015 and 2016. The alignment of samples from this study to L. monocytogenes EGD-e reference strain displayed features that are in line with those found in most Listeria genomes as reported in previous studies by Den Bakker et al. [4] in the US, by Kuenne et al. [42] in Germany, and by Schmitz-Esser et al. [43] in Austria. These features include assembly size, N50, and genomic G+C content. However, the difference in assembly size (2.9 to 3.1 base pairs) among the genomes might be influenced by the presence of prophages as their length varied and did not occur in the same places in the genomes in this study.
The application of MLST to subtype L. monocytogenes has provided important information into the population structure of this pathogen. Listeria monocytogenes sequence types reported in this study have been shown to have global distribution [44,45,46]. Listeria monocytogenes ST1 comprised 50% of all samples in this study and is known to be overrepresented in clinical and food isolates in the world as previously reported [43]. Listeria monocytogenes ST121 and ST204 are regarded as the most common persistent strains in food-processing environments [47]. These sequence types have the ability to survive and persist for months and even years in food-processing environments and to keep contaminating food products [46]. Importantly, the STs reported in the current study were also identified in human listeriosis cases associated with the 2017–2018 outbreak in SA [15].
Phylogenetic analysis was used to determine the genetic relationship among the samples from different sources, geographic areas, and time by inferring evolutionary relatedness [46]. Two different phylogenetic analyses were employed to establish relatedness amongst strains in the present study to other international strains. The core genome and ANI phylogenetic analyses revealed similar results, indicating that majority of the samples (E258, E313, E359, and E916) belonged to serogroup 4b, lineage I, and ST1 except E359, which belonged to ST876 clustered together. Based on ANI results, ST876 and ST1 shared an ANI of >95% similarities and all belonged to lineage I. This suggests that the L. monocytogenes isolates might have a common ancestor, hence the clustering. Further phylogenetic analyses indicated that two of the samples (E362 and E367) belonging to serogroup 1/2a and lineage II grouped far from each other and the rest of the strains. The MLST validated this clustering as samples E362 and E367 belong to ST121 and ST204, respectively.
The genomes from the present study were compared with 37 global genomes from different countries. Importantly, it was observed that samples E258, E313, E359, and E916 clustered with L. monocytogenes strains isolated from fatal outbreaks, clinical cases, food, and environment. Furthermore, samples E362 and E367 clustered with known important international strains of L. monocytogenes such as L. monocytogenes EGD-e, L. monocytogenes 36_25-1, and L. monocytogenes_F6654. This observation suggests that a theoretical virulence potential exists and that the samples could cause diseases in humans in South Africa.
Listeria monocytogenes is known to carry several plasmids that often confer resistance to antibiotics and to increase stress tolerance [42]. Those plasmids include pLM4423, pLM6179, and pIP501, which code for common features found in Listeria spp such as the CadAC Cadmium resistance transposon Tn5422 [42]. The analysis of the plasmids identified that none of the strains in this study carried a known plasmid. Plasmids are movable elements, and bacteria can gain or lose them; therefore, their absence in our strains is not surprising. Furthermore, Reference [48] reported L. monocytogenes isolates that possess no plasmids in their study.
Examination of reference-guided scaffolds for the presence of prophages using the PHASTER software identified 24 diverse prophages. However, the distribution of these prophages was uneven among the samples, as some samples contained intact prophages while others had questionable or incomplete prophages. The PHASTER also identified about 11 intact prophages from 6 different Listeria phages (LmoS188, LmoS293, A006, LP101, LP303-2, and B054) and gave the possible source for the phages in which the prophage was detected. The intact prophages detected in this study either belong to siphoviridae (A006, A118, B054, LP 100, and LP 030-3) and myoviridae (LmoS188 and LmoS293) Listeria phages as previously reported in other studies [42,49,50,51]. The identified phages in the present study are commonly found in Listeria strains globally and have been associated with the survival evolution and persistence of L. monocytogenes in food-processing facilities [42,52]. The virulence and pathogenicity of L. monocytogenes is also influenced by the presence of these prophages [53].
The WGS can also be used to identify the presence of pathogenicity islands linked to virulence or particular modes of pathogenesis [47]. In the present study, each of the six L. monocytogenes samples encode LIPI-1, which harbour Prf-A dependent virulence cluster genes that are critical in the infectious cycle of L. monocytogenes [1]. This could explain the reason why all sequenced isolates share about 40% similar genes that are strongly associated with LIPI-1. The genes encoded by LIPI-1 a common feature often found in RTE food and food production environment: L. monocytogenes isolates [54]. The LIPI-3 was identified only in strains that belong to lineage I and serogroup 4b, and it encodes a haemolytic factor (lysteriolysin, LLS), which increases the virulence of L. monocytogenes. LIPI3 is overrepresented among lineage I strains in human isolates and is strongly associated with serotype 4b isolates [55]. The absence of LIPI3 in two samples of lineage II was expected as the sequence types have been reported to lack it LIPI [1]. The LIPI4 is a newly described Listeria pathogenicity island [56] which encodes for the genes that annotated as a cellobiose family PTS system [55], and it was not identified in any of the strains identified in this study. The LIPI4 appears to be strongly associated with ST4 isolates [1]; hence, it was not observed in the current study. Several studies have identified mutations that are important in L. monocytogenes virulence markers [40,56,57,58]. The analysis of all sequenced strains in the present study indicated that they all encode for full-length InlA and ActA proteins.
Examination of resistance genes revealed that all sequenced strains of L. monocytogenes in the current study contained identical genes. The tetracycline resistance genes (tetA, tetL, tetT, and tetM) were found in all the samples, which have been identified in strains isolated from various meat samples [57]. Moreover, all the L. monocytogenes isolates in this study carried the mecC gene that is known to confer resistance to beta-lactam drugs, which is the primary therapeutic option for human listeriosis together with aminoglycosides [16]. The genes encoding for lincomycin resistance protein (lmrB), fosfomycin resistance protein (fosX), and erythromycin resistance ATP-binding protein (msrA) were also identified across all samples. The efflux pump-related genes, lde and mdrL, which confer resistance to quinolone and macrolides, respectively, were also identified. Different multidrug resistance transporter and efflux pump proteins that are known to confer resistance in L. monocytogenes and other bacteria [59] were observed in this study.
Quaternary ammonium compounds (QACs) such as benzalkonium chloride (BC) are widely used for cleaning and disinfection of food-processing environments [60]. The known molecular mechanisms of BC resistance are due to the activity of efflux pump systems encoded through the brcABC [61] and qacH on the Tn6188 transposon [62] genes that can be acquired through horizontal gene transfer leading to BC resistance in L. monocytogenes. In this study, QAC resistance genes (SugE, Tn6188, and BcrA) were identified, which suggests that these isolates are adapted to survival in the food-processing environment where sanitizers such as QACs are commonly used [62]. The BC is recommended in many countries as the most effective disinfectant against L. monocytogenes in food processing facilities [63]; however, the presence of BcrA and Tn6188 genes in strains from this study poses a serious hygiene management concern. Therefore, a need exists to evaluate the use of BC sanitary products in food-processing facilities in South Africa.
Listeria monocytogenes is equipped with mechanisms that allow it to adapt to and survive under stressful conditions [39]. However, there is a high degree of strain divergence in stress response and environmental adaptation, which is mostly associated with the presence of SSI-1 and SSI-2 [40]. According to Reference [39], SSI-1 supported the growth of L. monocytogenes under salt, acidic, bile, and gastric stress conditions and is overrepresented in human clinical isolates, whereas SSI-2 is mostly found in isolates from food and food-processing environments. In the current study, SSI-1 was found in the isolates while SSI-2 was detected from only two isolates that belonged to the sequence types ST121 and ST204, which are known to be persistent in food-processing environments [64]. The presence of SSI-2 in ST121 and ST204 can be suggested to be the factor for the persistence of these strains [63].
5. Conclusions
Listeria monocytogenes is one of the most well-characterized foodborne pathogens in the world; however, in South Africa, there is lack of information on the molecular characterisation of the pathogen in meat and RTE products. The current study is the first to report on the characterisation of L. monocytogenes strains isolated from biltong and polony products using whole genome sequencing in South Africa. The application of WGS in the current study has provided a partial overview of genomic diversity of L. monocytogenes strains that are circulating in South African RTE meat products. It also adds to the global data on genetic diversity of L. monocytogenes from South Africa. This overview has revealed virulence potential associated with presence of cluster LIPI-1 genes in all the isolates. In addition, it has permitted the evaluation of resistance genomic characteristics of the isolates, which indicate the presence of resistance genes to common antimicrobial agents and sanitizer. Prophages, which were observed in South African isolates, are largely found across the global clonal diversity of L. monocytogenes, most probably due to the influx of imported raw materials such as mechanically separated meat (MSM), which commonly is used in the manufacturing of polony. The information provided in this study is important for enhancing the understanding of evolutionary divergence, adaptation, and survival of L. monocytogenes in products of animal origin.
Author Contributions
E.M. conceived and supervised the project. K.R.M. also supervised the project and consolidated the input from all authors. I.M. performed the laboratory experiments and wrote the paper with significant contribution. R.P. carried out the bioinformatic analysis. K.M. obtain funding for the project, designed the study, supervised sample collection and data capture, and reviewed the article.
Funding
The study was funded by the Department of Agriculture, Forestry, and Fisheries (DAFF) under project number 21.1.1/VPH-01/OVI. The human resource capacity for sample collection was provided by the DAFF—Directorate: Veterinary Public Health. Sample testing was conducted by the Food and Feed Analysis and General Bacteriology Laboratories of the Agricultural Research Council—Onderstepoort Veterinary Institute.
Acknowledgments
The following organisations and individuals are acknowledged for their contributions: Department of Agriculture, Forestry, and Fisheries—Directorate: Veterinary Public Health for project funding and the use of data for this study. The officials from the Department of Agriculture, Forestry, and Fisheries—Directorate: Veterinary Public Health (Lizzy Molele, Pauline Modibane, Maphaseka Mosia, Mavis Phaswane, and Maruping Ntsatsi) for the field collection of samples for this study and Mphane Molefe for the approval of the study. The authors are grateful to the Agricultural Research Council: Onderstepoort Veterinary Research for providing all research facilities.
Conflicts of Interest
The authors declare no conflict of interests.
References
- Hilliard, A.; Leong, D.; Callaghan, A.O.; Culligan, E.P.; Morgan, C.A.; Delappe, N.; Hill, C.; Id, K.J.; Cormican, M.; Gahan, C.G.M. Genomic Characterization of Listeria monocytogenes Isolates Associated with Clinical Listeriosis and the Food Production Environment in Ireland. Genes 2018, 9, 171. [Google Scholar] [CrossRef] [PubMed]
- Ryan, S.; Begley, M.; Hill, C.; Gahan, C.G.M. A five-gene stress survival islet (SSI-1) that contributes to the growth of Listeria monocytogenes in suboptimal conditions. J. Appl. Microbiol. 2010, 109, 984–995. [Google Scholar] [CrossRef] [PubMed]
- Burall, L.S.; Grim, C.J.; Mammel, M.K.; Datta, A.R. Whole genome sequence analysis using jspecies tool establishes clonal relationships between Listeria monocytogenes strains from epidemiologically unrelated listeriosis outbreaks. PLoS ONE 2016, 11, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Den Bakker, H.C.; Cummings, C.A.; Ferreira, V.; Vatta, P.; Orsi, R.H.; Degoricija, L.; Barker, M.; Petrauskene, O.; Furtado, M.R.; Wiedmann, M. Comparative genomics of the bacterial genus Listeria: Genome evolution is characterized by limited gene acquisition and limited gene loss. BMC Genomics 2010, 11, 688. [Google Scholar] [CrossRef] [PubMed]
- Haase, J.K.; Didelot, X.; Lecuit, M.; Korkeala, H.; Achtman, M.; Leclercq, A.; Grant, K.; Wiedmann, M.; Apfalter, P. The ubiquitous nature of Listeria monocytogenes clones: A large-scale Multilocus Sequence Typing study. Environ. Microbiol. 2014, 16, 405–416. [Google Scholar] [CrossRef]
- Laksanalamai, P.; Huang, B.; Sabo, J.; Burall, L.S.; Zhao, S.; Bates, J.; Datta, A.R. Genomic characterization of novel Listeria monocytogenes serotype 4b variant strains. PLoS ONE 2014, 9, e89024. [Google Scholar] [CrossRef]
- Hyden, P.; Pietzka, A.; Lennkh, A.; Murer, A.; Springer, B.; Blaschitz, M.; Indra, A.; Huhulescu, S.; Allerberger, F.; Ruppitsch, W.; et al. Whole genome sequence-based serogrouping of Listeria monocytogenes isolates. J. Biotechnol. 2016, 235, 181–186. [Google Scholar] [CrossRef]
- Liu, D. Identification, subtyping and virulence determination of Listeria monocytogenes, an important foodborne pathogen. J. Med. Microbiol. 2006, 55, 645–659. [Google Scholar] [CrossRef]
- Franz, E.; Gras, L.M.; Dallman, T. Significance of whole genome sequencing for surveillance, source attribution and microbial risk assessment of foodborne pathogens. Curr. Opin. Food Sci. 2016, 8, 74–79. [Google Scholar] [CrossRef]
- Schmid, D.; Allerberger, F.; Huhulescu, S.; Pietzka, A.; Amar, C.; Kleta, S.; Prager, R.; Preußel, K.; Aichinger, E. Whole genome sequencing as a tool to investigate a cluster of seven cases of listeriosis in Austria and Germany, 2011–2013. Clin. Microbiol. Infect. 2014, 20, 431–436. [Google Scholar] [CrossRef]
- Jackson, B.R.; Tarr, C.; Strain, E.; Jackson, K.A.; Conrad, A.; Carleton, H.; Katz, L.S.; Stroika, S.; Gould, L.H.; Mody, R.K.; et al. Implementation of Nationwide Real-time Whole-genome Sequencing to Enhance Listeriosis Outbreak Detection and Investigation. Clin. Infect. Dis. 2016, 63, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Gilmour, M.W.; Graham, M.; Van Domselaar, G.; Tyler, S.; Kent, H.; Trout-Yakel, K.M.; Larios, O.; Allen, V.; Lee, B.; Nadon, C. High-throughput genome sequencing of two Listeria monocytogenes clinical isolates during a large foodborne outbreak. BMC Genomics 2010, 11, 120. [Google Scholar] [CrossRef] [PubMed]
- Jensen, A. Factors Influencing Persistence and Virulence of Listeria monocytogenes. Ph.D. Thesis, Technical University of Denmark, Copenhagen, Denmark, 2007. [Google Scholar]
- Smith, A.M.; Naicker, P.; Bamford, C.; Shuping, L.; McCarthy, K.M.; Sooka, A.; Smouse, S.L.; Tau, N.; Keddy, K.H. Genome Sequences for a Cluster of Human Isolates of Listeria monocytogenes Identified in South Africa in 2015. Genome Announc. 2016, 4. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.M.; Tau, N.P.; Smouse, S.L.; Allam, M.; Ismail, A.; Ramalwa, N.R.; Disenyeng, B.; Ngomane, M.; Thomas, J. Outbreak of Listeria monocytogenes in South Africa, 2017–2018: Laboratory Activities and Experiences Associated with Whole-Genome Sequencing Analysis of Isolates. Foodborne Pathog. Dis. 2019, 16, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Leong, D.; Alvarez-Ordóñez, A.; Jooste, P.; Jordan, K. Listeria monocytogenes in food: Control by monitoring the food processing environment. Afr. J. Microbiol. Res. 2016, 10, 1–14. [Google Scholar]
- Naidoo, K. The Microbial Ecology of Biltong in South Africa during Production and at Point-of-Sale. Ph.D. Thesis, University of the Witwatersrand, Johannesburg, South Afrika, 2014. [Google Scholar]
- Matle, I.; Mbatha, K.R.; Lentsoane, O.; Magwedere, K.; Morey, L.; Madoroba, E. Occurrence, serotypes, and characteristics of Listeria monocytogenes in meat and meat products in South Africa between 2014 and 2016. J. Food Saf. 2019, 39, e12629. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genomics 2011, 12, 402. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef] [PubMed]
- Moura, A.; Tourdjman, M.; Leclercq, A.; Hamelin, E.; Laurent, E.; Fredriksen, N.; Van Cauteren, D.; Bracq-dieye, H.; Thouvenot, P.; Vales, G.; et al. Real-time whole-genome sequencing for surveillance of Listeria monocytogenes, France. Emerg. Infect. Dis. 2017, 23, 1462–1470. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Lipman, D.J. Protein database searches for multiple alignments. Proc. Natl. Acad. Sci. USA 2019, 87, 5509–5513. [Google Scholar] [CrossRef] [PubMed]
- Enright, A.J.; Van Dongen, S.; Ouzounis, C.A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002, 30, 1575–1584. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, 242–245. [Google Scholar] [CrossRef]
- Bosi, E.; Donati, B.; Galardini, M.; Brunetti, S.; Sagot, M.F.; Lió, P.; Crescenzi, P.; Fani, R.; Fondi, M. MeDuSa: A multi-draft based scaffolder. Bioinformatics 2015, 31, 2443–2451. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, 16–21. [Google Scholar] [CrossRef]
- Carattoli, A.; Miriagou, V.; Bertini, A.; Loli, A.; Colinon, C.; Villa, L.; Whichard, J.M.; Rossolini, G.M. Replicon typing of plasmids encoding resistance to newer β-lactams. Emerg. Infect. Dis. 2006, 12, 1145–1148. [Google Scholar] [CrossRef]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Jordan, I.K.; Rishishwar, L. stringMLST: A fast k-mer based tool for multilocus sequence typing. Bioinformatics 2017, 33, 119–121. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB 2016: Hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 2016, 44, D694–D697. [Google Scholar] [CrossRef] [PubMed]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef] [PubMed]
- Pal, C.; Bengtsson-Palme, J.; Rensing, C.; Kristiansson, E.; Larsson, D.G.J. BacMet: Antibacterial biocide and metal resistance genes database. Nucleic Acids Res. 2014, 42, 737–743. [Google Scholar] [CrossRef] [PubMed]
- Lakin, S.M.; Dean, C.; Noyes, N.R.; Dettenwanger, A.; Ross, A.S.; Doster, E.; Rovira, P.; Abdo, Z.; Jones, K.L.; Ruiz, J.; et al. MEGARes: An antimicrobial resistance database for high throughput sequencing. Nucleic Acids Res. 2017, 45, D574–D580. [Google Scholar] [CrossRef]
- Harter, E.; Wagner, E.M.; Zaiser, A.; Halecker, S.; Wagner, M.; Rychli, K. Stress Survival Islet 2, Predominantly Present in Listeria monocytogenes Strains of Sequence Type 121, Is Involved in the Alkaline and Oxidative Stress Responses. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef]
- Muhterem-Uyar, M.; Ciolacu, L.; Wagner, K.; Wagner, M.; Schmitz-Esser, S.; Stessl, B. New Aspects on Listeria monocytogenes ST5-ECVI Predominance in a Heavily Contaminated Cheese Processing Environment. Front. Microbiol. 2018, 9, 1–14. [Google Scholar] [CrossRef]
- McNair, K.; Bailey, B.A.; Edwards, R.A. PHACTS, a computational approach to classifying the lifestyle of phages. Bioinformatics 2012, 28, 614–618. [Google Scholar] [CrossRef]
- Kuenne, C.; Billion, A.; Mraheil, M.A.; Strittmatter, A.; Daniel, R.; Goesmann, A.; Barbuddhe, S.; Hain, T.; Chakraborty, T. Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genomics 2013, 14, 1–19. [Google Scholar] [CrossRef]
- Schmitz-Esser, S.; Müller, A.; Stessl, B.; Wagner, M. Genomes of sequence type 121 Listeria monocytogenes strains harbor highly conserved plasmids and prophages. Front. Microbiol. 2015, 6, 6. [Google Scholar] [CrossRef] [PubMed]
- Ragon, M.; Wirth, T.; Hollandt, F.; Lavenir, R.; Lecuit, M.; Monnier, A.L.; Brisse, S. A New Perspective on Listeria monocytogenes Evolution. PLoS Pathog. 2008, 4, e1000146. [Google Scholar] [CrossRef] [PubMed]
- Chenal-Francisque, V.; Lopez, J.; Cantinelli, T.; Caro, V.; Tran, C.; Leclercq, A.; Lecuit, M.; Brisse, S. World wide distribution of major clones of listeria monocytogenes. Emerg. Infect. Dis. 2011, 17, 1110–1112. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, A.; Zhu, R.; Lan, R.; Jin, D.; Cui, Z.; Wang, Y.; Li, Z.; Wang, Y.; Xu, J.; et al. Genetic diversity and molecular typing of Listeria monocytogenes in China. BMC Microbiol. 2012, 12, 119. [Google Scholar] [CrossRef] [PubMed]
- Fox, E.M.; Allnutt, T.; Bradbury, M.I.; Fanning, S.; Chandry, P.S. Comparative Genomics of the Listeria monocytogenes ST204 Subgroup. Front. Microbiol. 2016, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Suokko, A.; Savijoki, K.; Malinen, E.; Palva, A.; Varmanen, P. Characterization of a mobile clpL gene from Lactobacillus rhamnosus. Appl. Environ. Microbiol. 2005, 71, 2061–2069. [Google Scholar] [CrossRef]
- Klumpp, J.; Loessner, M.J. Listeria phages: Genomes, evolution, and application. J. Bacteriophage 2013, 3, e26861. [Google Scholar] [CrossRef]
- Denes, T.; Vongkamjan, K.; Ackermann, H.; Switt, I.M.; Wiedmann, M. Comparative Genomic and Morphological Analyses of Listeria Phages Isolated from Farm Environments. Appl. Environ. Microbiol. 2014, 80, 4616–4625. [Google Scholar] [CrossRef]
- Casey, A.; Jordan, K.; Neve, H.; Coffey, A.; Mcauliffe, O. A tail of two phages: Genomic and functional analysis of Listeria monocytogenes phages vB_LmoS_188 and vB_LmoS_293 reveal the receptor-binding proteins involved in host specificity. Front. Microbiol. 2015, 6, 1–14. [Google Scholar] [CrossRef]
- Verghese, B.; Lok, M.; Wen, J.; Alessandria, V.; Chen, Y.; Kathariou, S.; Knabel, S. comK Prophage junction fragments as markers for Listeria monocytogenes Genotypes unique to individual meat and poultry processing plants and a model for rapid niche-specific adaptation, biofilm formation, and persistence. Appl. Environ. Microbiol. 2011, 77, 5064. [Google Scholar] [CrossRef]
- Rabinovich, L.; Sigal, N.; Borovok, I.; Nir-Paz, R.; Herskovits, A.A. Prophage excision activates Listeria competence genes that promote phagosomal escape and virulence. Cell 2012, 150, 792–802. [Google Scholar] [CrossRef] [PubMed]
- Nightingale, K.K.; Schukken, Y.H.; Nightingale, C.R.; Fortes, E.D.; Ho, A.J.; Her, Z.; Grohn, Y.T.; Mcdonough, P.L.; Wiedmann, M. Ecology and Transmission of Listeria monocytogenes infecting ruminants and in the farm environment. Appl. Environ. Microbiol. 2004, 70, 4458–4467. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.D.; Draper, L.A.; Lawton, E.M.; Daly, K.M.; Groeger, D.S.; Casey, P.G.; Ross, R.P.; Hill, C. Listeriolysin S, a novel peptide haemolysin associated with a subset of lineage I Listeria monocytogenes. PLoS Pathog. 2008, 4, e1000144. [Google Scholar] [CrossRef] [PubMed]
- Chenal-francisque, V.; Maury, M.M.; Lavina, M.; Touchon, M.; Leclercq, A.; Lecuit, M. Clonogrouping, A Rapid Multiplex PCR Method for Identification of Major Clones of Listeria monocytogenes. J. Clin. Microbiol. 2015, 53, 3355–3358. [Google Scholar] [CrossRef] [PubMed]
- Ducey, T.F.; Page, B.; Usgaard, T.; Borucki, M.K.; Pupedis, K.; Ward, T.J. A Single-Nucleotide-Polymorphism-Based Multilocus Genotyping Assay for Subtyping Lineage I Isolates of Listeria monocytogenes. Appl. Environ. Microbiol. 2006, 73, 133–147. [Google Scholar] [CrossRef] [PubMed]
- Rychli, K.; Wagner, E.M.; Ciolacu, L.; Zaiser, A.; Tasara, T.; Wagner, M.; Schmitz-Esser, S. Comparative genomics of human and non- human Listeria monocytogenes sequence type 121 strains. PLoS ONE 2017, 12, e0176857. [Google Scholar] [CrossRef] [PubMed]
- Mata, M.T.; Baquero, F.; Pe, J.C. A multidrug efflux transporter in Listeria monocytogenes. FEMS Microbiol. Lett. 2000, 187, 185–188. [Google Scholar] [CrossRef]
- Mereghetti, L.; Lanotte, P.; Savoye-Marczuk, V.; Marquet-Van Der Mee, N.; Audurier, A.; Quentin, R. Combined ribotyping and random multiprimer DNA analysis to probe the population structure of Listeria monocytogenes. Appl. Environ. Microbiol. 2002, 68, 2849–2857. [Google Scholar] [CrossRef][Green Version]
- Elhanafi, D.; Utta, V.; Kathariou, S. Genetic characterization of plasmid-associated benzalkonium chloride resistance determinants in a Listeria monocytogenes train from the 1998–1999 outbreak. Appl. Environ. Microbiol. 2010, 76, 8231–8238. [Google Scholar] [CrossRef]
- Müller, A.; Rychli, K.; Muhterem-Uyar, M.; Zaiser, A.; Stessl, B.; Guinane, C.M.; Cotter, P.D.; Wagner, M.; Schmitz-Esser, S. Tn6188—A Novel Transposon in Listeria monocytogenes Responsible for Tolerance to Benzalkonium Chloride. PLoS ONE 2013, 8, e76835. [Google Scholar] [CrossRef]
- Meier, A.B.; Guldimann, C.; Markkula, A.; Pöntinen, A. Comparative Phenotypic and Genotypic Analysis of Swiss and Finnish Listeria monocytogenes Isolates with Respect to Benzalkonium Chloride Resistance. Front. Microbiol. 2017, 8, 57. [Google Scholar] [CrossRef] [PubMed]
- O’Grady, J.; Sedano-Balbás, S.; Maher, M.; Smith, T.; Barry, T. Rapid real-time PCR detection of Listeria monocytogenes in enriched food samples based on the ssrA gene, a novel diagnostic target. Food Microbiol. 2008, 25, 75–84. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).