Physical Map of FISH 5S rDNA and (AG3T3)3 Signals Displays Chimonanthus campanulatus R.H. Chang & C.S. Ding Chromosomes, Reproduces its Metaphase Dynamics and Distinguishes Its Chromosomes
Abstract
1. Introduction
2. Materials and Methods
2.1. Chromosomes and Probe Preparation
2.2. Hybridization and Image Capture
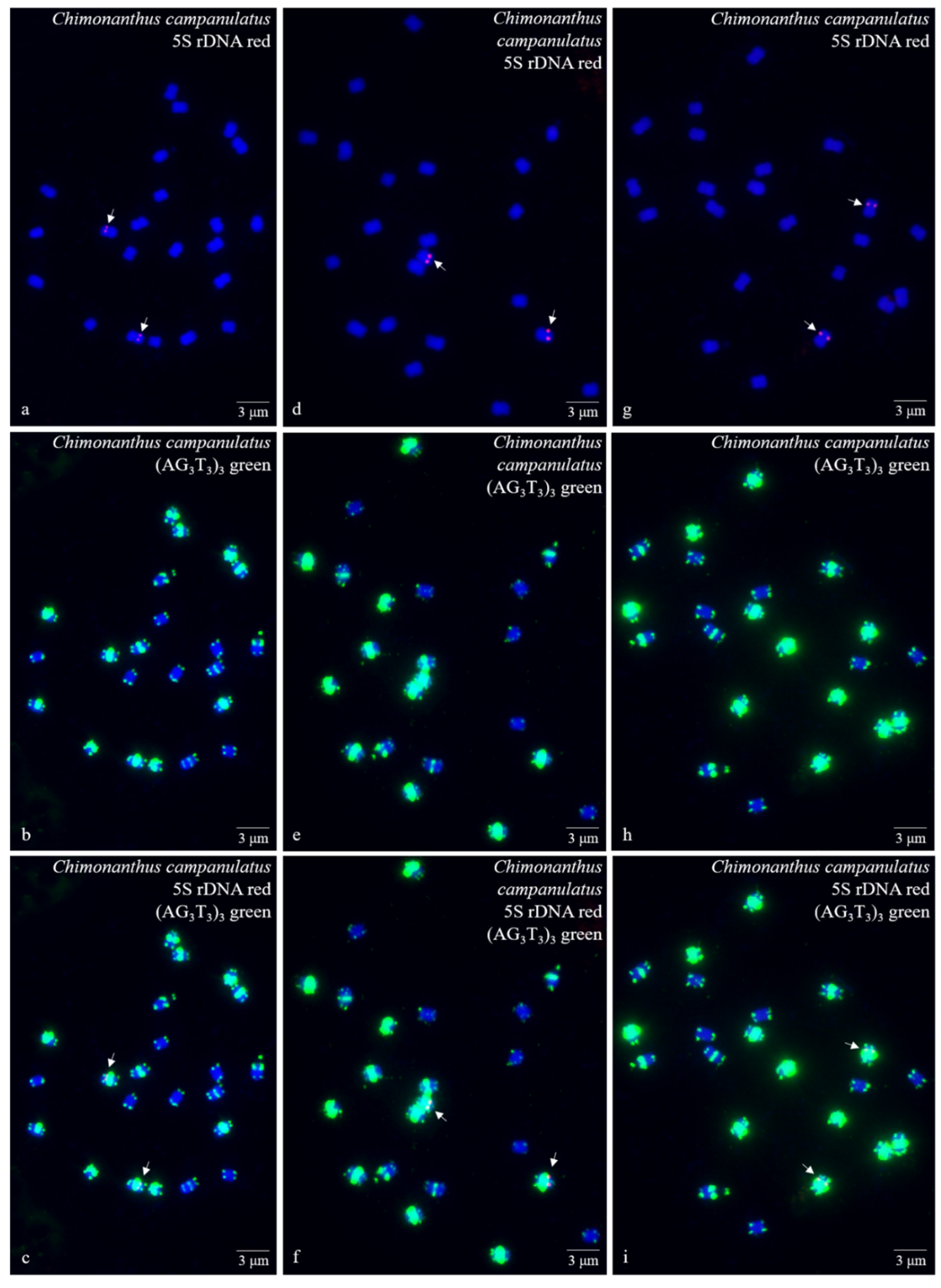
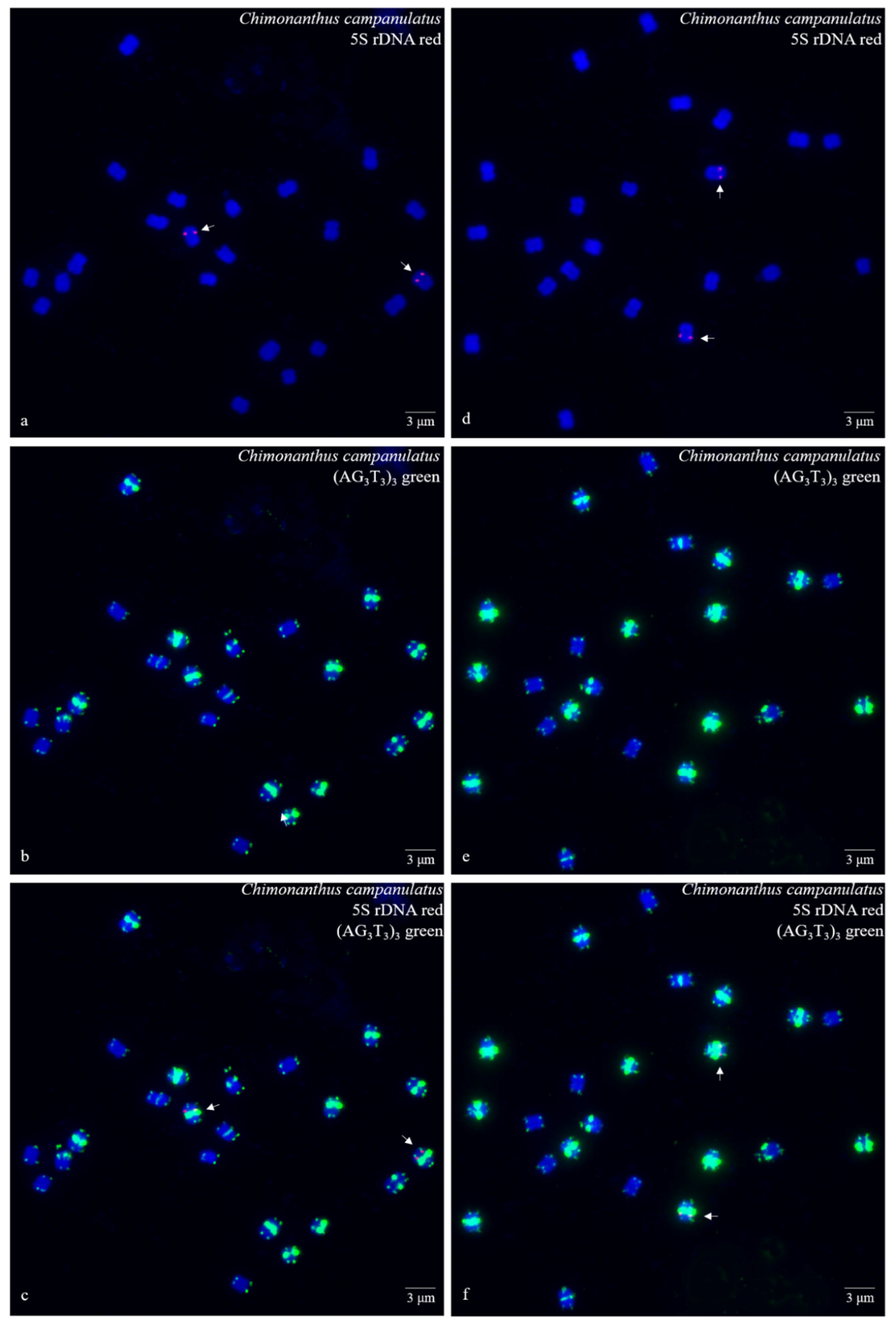
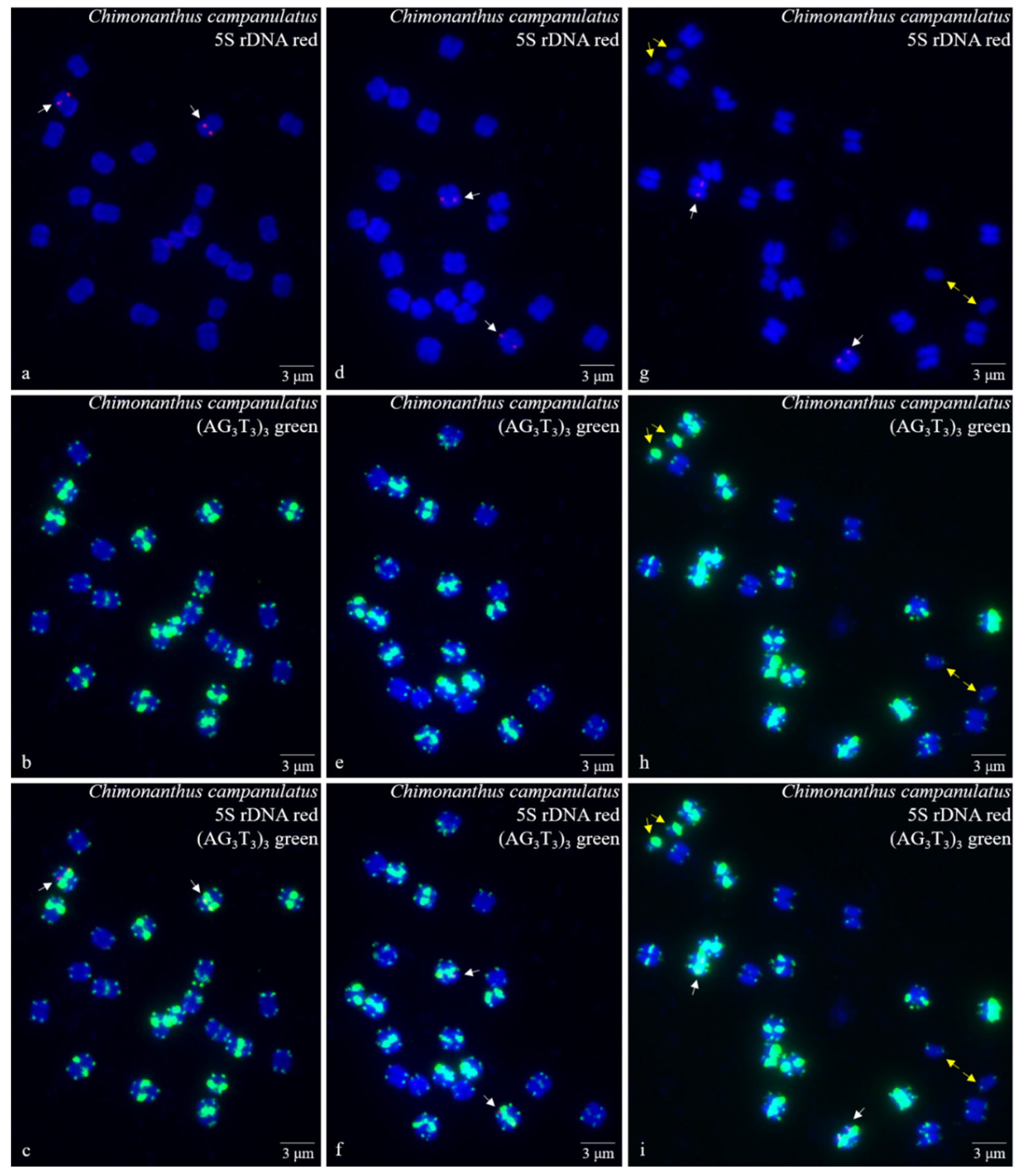
3. Results
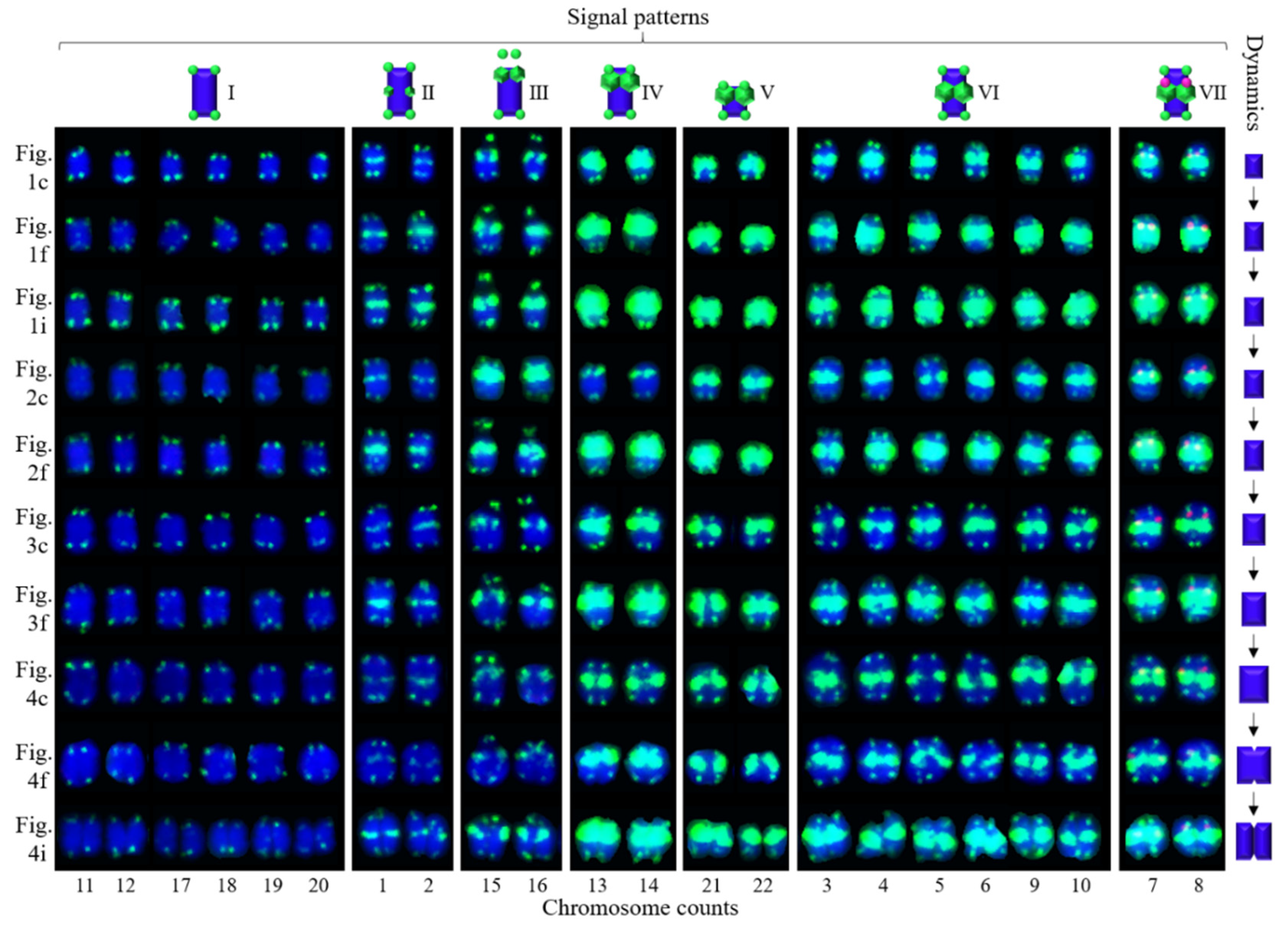
3.1. S rDNA and (AG3T3)3 Enabled Visualization of Ch. Campanulatus Chromosomes
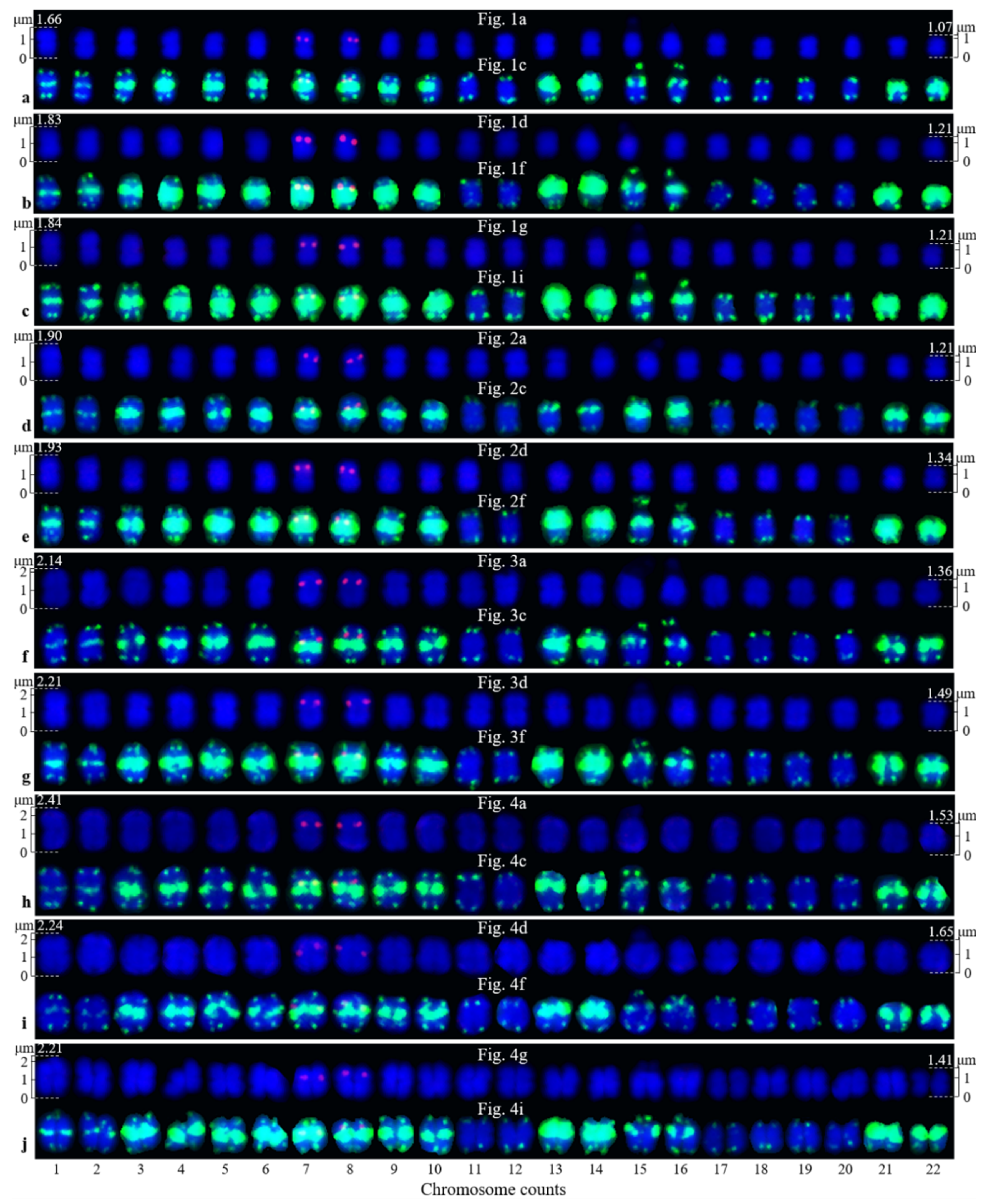
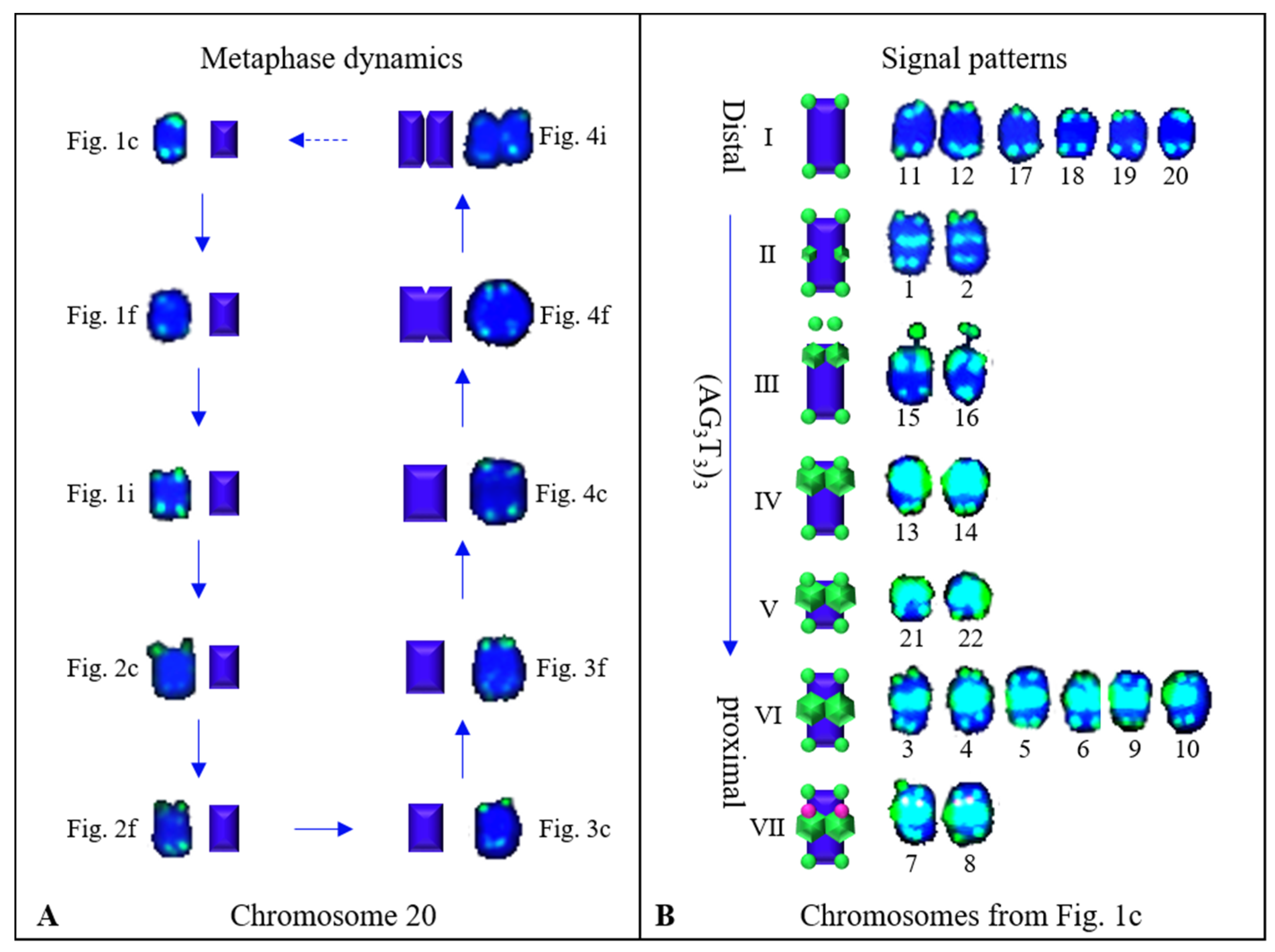
3.2. Physical Map Reproduced Metaphase Dynamics and Distinguished Chromosomes
4. Discussion
4.1. Karyotype Analysis and Dynamic Metaphase Progression
4.2. Distinguishing Ch. campanulatus chromosomes
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chang, R.H.; Ding, C.C. The seedling characters of Chinese Calycanthaceae with a new species of Chimonanthus Lindl. J. Univ. Chin. Acad. Sci. 1980, 18, 328–332. Available online: http://journal.ucas.ac.cn/EN/Y1980/V18/I3/328 (accessed on 15 October 2019).
- Sugiura, T. A list of chromosome numbers in angiospermous plants. Bot. Mag. Tokyo 1931, 45, 353. [Google Scholar] [CrossRef]
- Liu, H.E.; Zhang, R.H.; Huang, S.F.; Zhao, Z.F. Study on Chromosomes of 8 Species of Calycathaceae. J. Zhejiang For. Coll. 1996, 13, 28–33. Available online: http://europepmc.org/abstract/CBA/288874 (accessed on 15 October 2019).
- Song, X.C.; Liu, Y.X.; Jiang, X.M. Analysis of chromosome karotype of Chimonanthus nitens and Chimonanthus zhejiangensis. Jiangxi For. Sci. Technol. 2013, 2, 10–12. Available online: http://qikan.cqvip.com/Qikan/Article/Detail?id=45635921 (accessed on 15 October 2019).
- Tang, Y.C.; Xiang, Q.Y. Cytological studies on some plants of East China. Acta Phytotaxon. Sin. 1987, 25, 1–8. Available online: http://www.plantsystematics.com/CN/Y1987/V25/I1/145635921 (accessed on 15 October 2019).
- Li, L.C. Karyotype analysis on Calycanthus chinensis. Guihaia 1986, 6, 221–224. Available online: http://kns.cnki.net/KCMS/detail/detail.aspx?dbname=cjfd1986&filename=gxzw198603011 (accessed on 15 October 2019).
- Li, L.C. Cytogeographical study of Calycanthus Linneus. Guihaia 1989, 4, 311–316. Available online: http://qikan.cqvip.com/Qikan/Article/Detail?id=41148 (accessed on 15 October 2019).
- Zhang, L.; Cao, B.; Bai, C. New reports of nuclear DNA content for 66 traditional Chinese medicinal plant taxa in China. Caryologia 2013, 66, 375–383. [Google Scholar] [CrossRef]
- Fridley, J.D.; Craddock, A. Contrasting growth phenology of native and invasive forest shrubs mediated by genome size. New Phytol. 2015, 207, 659–668. [Google Scholar] [CrossRef]
- Chen, L.Q. Biological studies on the genus Chimonanthus Lindley. Beijing For. Univ. 1998. Available online: http://www.wanfangdata.com.cn/details/detail.do?_type=degree&id=Y280512 (accessed on 15 October 2019).
- Dai, P.F. Phylogeography, phylogeny and genetic diversity studies on Chimonanthus Lindley. Northwest Univ. 2012. Available online: http://kns.cnki.net/KCMS/detail/detail.aspx?dbcode=CDFD&dbname=CDFD2013&filename=1013136315.nh (accessed on 15 October 2019).
- Shu, R.G.; Wan, Y.L.; Wang, X.M. Non-Volatile constituents and pharmacology of Chimonanthus: A review. Chin. J. Nat. Med. 2019, 17, 161–186. [Google Scholar] [CrossRef]
- Liu, D.; Sui, S.; Ma, J.; Li, Z.; Guo, Y.; Luo, D.; Yang, J.; Li, M. Transcriptomic analysis of flower development in wintersweet (Chimonanthus praecox). PLoS ONE 2014, 9, e86976. [Google Scholar] [CrossRef]
- Li, Z.; Jiang, Y.; Liu, D.; Ma, J.; Li, J.; Li, M.; Sui, S. Floral scent emission from nectaries in the adaxial side of the innermost and middle petals in Chimonanthus praecox. Int. J. Mol. Sci. 2018, 19, 3278. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.P.; Ma, Z.Y.; Zhao, K.G.; Zhang, J.; Xiang, L.; Chen, L.Q. Transcriptomic and proteomic approaches to explore the differences in monoterpene and benzenoid biosynthesis between scented and unscented genotypes of wintersweet. Physiol. Plant 2019, 166, 478–493. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Zhao, K.; Li, X.; Zhao, R.; Aslam, M.Z.; Yu, L.; Chen, L. Comprehensive analysis of wintersweet flower reveals key structural genes involved in flavonoid biosynthetic pathway. Gene 2018, 676, 279–289. [Google Scholar] [CrossRef]
- Baral, A. Finding the fragrance genes of wintersweet. Physiol. Plant 2019, 166, 475–477. [Google Scholar] [CrossRef]
- Li, L. Morphological comparison flower bud differentiation and development of four species in Calycanthaceae. Huazhong Agric. Univ. 2018. Available online: http://www.wanfangdata.com.cn/details/detail.do?type=degree&id=Y3396525 (accessed on 15 October 2019).
- Deng, C.; Song, G.; Hu, Y. Rapid determination of volatile compounds emitted from Chimonanthus praecox flowers by HS-SPME-GC-MS. Z. Nat. C 2004, 59, 636–640. [Google Scholar] [CrossRef]
- Lv, J.S.; Zhang, L.L.; Chu, X.Z.; Zhou, J.F. Chemical composition, antioxidant and antimicrobial activity of the extracts of the flowers of the Chinese plant Chimonanthus praecox. Nat. Prod. Res. 2012, 26, 1363–1367. [Google Scholar] [CrossRef]
- Li, H.; Zhang, Y.; Liu, Q.; Sun, C.; Li, J.; Yang, P.; Wang, X. Preparative separation of phenolic compounds from Chimonanthus praecox flowers by high-speed counter-current chromatography using a stepwise elution mode. Molecules 2016, 21, 1016. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.W.; Gao, J.M.; Xu, T.; Zhang, X.C.; Ma, Y.T.; Jarussophon, S.; Konishi, Y. Antifungal activity of alkaloids from the seeds of Chimonanthus praecox. Chem. Biodivers. 2009, 6, 838–845. [Google Scholar] [CrossRef] [PubMed]
- Morikawa, T.; Nakanishi, Y.; Ninomiya, K.; Matsuda, H.; Nakashima, S.; Miki, H.; Miyashita, Y.; Yoshikawa, M.; Hayakawa, T.; Muraoka, O. Dimeric pyrrolidinoindoline-type alkaloids with melanogenesis inhibitory activity in flower buds of Chimonanthus praecox. J. Nat. Med. 2014, 68, 539–549. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, N.; Ninomiya, K.; Okugawa, S.; Motaia, C.; Nakanishi, Y.; Yoshikawa, M.; Muraoka, O.; Morikawa, T. Quantitative determination of principal alkaloid and flavonoid constituents in wintersweet, the flower buds of Chimonanthus praecox. Nat. Prod. Commun. 2016, 11, 953–956. [Google Scholar] [CrossRef]
- Lou, H.Y.; Zhang, Y.; Ma, X.P.; Jiang, S.; Wang, X.P.; Yi, P.; Liang, G.Y.; Wu, H.M.; Feng, J.; Jin, F.Y.; et al. Novel sesquiterpenoids isolated from Chimonanthus praecox and their antibacterial activities. Chin. J. Nat. Med. 2018, 16, 621–627. [Google Scholar] [CrossRef]
- Zeng, J. In vitro culture and optimization genetic transformation system of Calycanthus praecox. Southwest Univ. 2007. [Google Scholar] [CrossRef]
- Chen, D.W. Wintersweet in vitro culture and genetic map construct. Huazhong Agric. Univ. 2010. Available online: http://kns.cnki.net/KCMS/detail/detail.aspx?dbcode=CDFD&dbname=CDFDTEMP&filename=2010271821.nh (accessed on 15 October 2019).
- Chen, Y.X. Wintersweet two moecular marker develop and F1 population segregation evaluation. Huazhong Agric. Univ. 2014. [Google Scholar] [CrossRef]
- Sui, S.; Luo, J.; Ma, J.; Zhu, Q.; Lei, X.; Li, M. Generation and analysis of expressed sequence tags from Chimonanthus praecox (Wintersweet) flowers for discovering stress-responsive and floral development-related genes. Comp. Funct. Genom. 2012, 2012, 134596. [Google Scholar] [CrossRef]
- Zhou, M.Q. Molecular marker analysis germplasm of Calycanthus praecox and genetic diversity of Chimonanthus nitens. Huazhong Agric. Univ. 2007. [Google Scholar] [CrossRef]
- Zhao, S.P. Calycanthus praecox ANL2 cloning and function identification. Huazhong Agric. Univ. 2015. [Google Scholar] [CrossRef]
- Luo, D.P. Chimonanthus praecox (L.) link CpAGL2 cloning and function identification. Southwest Univ. 2014. Available online: http://www.wanfangdata.com.cn/details/detail.do?_type=degree&id=Y2572451 (accessed on 15 October 2019).
- Zhang, Q.; Wang, B.G.; Duan, K.; Wang, L.G.; Wang, M.; Tang, X.M.; Pan., A.H.; Sui, S.Z.; Wang, G.D. The paleoAP3-type gene CpAP3, an ancestral B-class gene from the basal angiosperm Chimonanthus praecox, can affect stamen and petal development in higher eudicots. Dev. Genes Evol. 2011, 221, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.Q. Calycanthus praecox CpCAF1 cloning and function identification. Southwest Univ. 2016. Available online: http://kns.cnki.net/KCMS/detail/detail.aspx?filename=1016767365.nh&dbname=CMFDTEMP (accessed on 15 October 2019).
- Liu, H.; Huang, R.; Ma, J.; Sui, S.; Guo, Y.; Liu, D.; Li, Z.; Lin, Y.; Li, M. Two C3H type zinc finger protein genes, CpCZF1 and CpCZF2, from Chimonanthus praecox affect stamen development in Arabidopsis. Genes 2017, 8, 199. [Google Scholar] [CrossRef]
- Lin, M. Chimonanthus praecox Cpcor413pm1 stress resistance and subcellular localization. Southwest Univ. 2008. [Google Scholar] [CrossRef]
- Li, Z. Calycanthus praecox CpEXP1 expression and function identification. Southwest Univ. 2014. Available online: http://kns.cnki.net/KCMS/detail/detail.aspx?dbcode=CDFD&dbname=CDFDTEMP&filename=1015547733.nh (accessed on 15 October 2019).
- Yang, H.Y. Calycanthus praecox CpH3 cloning and function identification. Southwest Univ. 2013. Available online: http://www.wanfangdata.com.cn/details/detail.do?type=degree&id=Y2309710# (accessed on 15 October 2019).
- Liu, Y.; Xie, L.; Liang, X.; Zhang, S. CpLEA5, the late embryogenesis abundant protein gene from Chimonanthus praecox, possesses low temperature and osmotic resistances in prokaryote and eukaryotes. Int. J. Mol. Sci. 2015, 16, 26978–26990. [Google Scholar] [CrossRef]
- Chen, Y. Wintersweet CpNAC8 cloning and function identification. Southwest Univ. 2017. Available online: http://kns.cnki.net/KCMS/detail/detail.aspx?filename=1017847224.nh&dbname=CMFDTEMP (accessed on 15 October 2019).
- Wang, B. Calycanthus praecox CpRALF cloning and function identification. Southwest Univ. 2012. [Google Scholar] [CrossRef]
- Yang, Y. Calycanthus praecox CpRBL cloning and function identification. Southwest Univ. 2012. Available online: http://www.wan.fangdata.com.cn/details/detail.do?_type=degree&id=Y2309707 (accessed on 15 October 2019).
- Xiang, L.; Zhao, K.; Chen, L. Molecular cloning and expression of Chimonanthus praecox farnesyl pyrophosphate synthase gene and its possible involvement in the biosynthesis of floral volatile sesquiterpenoids. Plant Physiol. Biochem. 2010, 48, 845–850. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.H.; Liu, X.; Gao, B.W.; Zhang, Z.X.; Shi, S.P.; Tu, P.F. Cloning and expression analysis of glucose-6-phosphate dehydrogenase 1 (G6PDH1) gene from Chimonanthus praecox. Zhongguo Zhong Yao Za Zhi 2015, 40, 4160–4164. [Google Scholar] [PubMed]
- Huang, W.P.; Tan, T.; Li, Z.F.; OuYang, H.; Xu, X.; Zhou, B.; Feng, Y.L. Structural characterization and discrimination of Chimonanthus nitens Oliv. leaf from different geographical origins based on multiple chromatographic analysis combined with chemometric methods. J. Pharm. Biomed. Anal. 2018, 154, 236–244. [Google Scholar] [CrossRef]
- Tan, T.; Luo, Y.; Zhong, C.C.; Xu, X.; Feng, Y. Comprehensive profiling and characterization of coumarins from roots, stems, leaves, branches, and seeds of Chimonanthus nitens Oliv. using ultra-performance liquid chromatography/quadrupole-time-of-flight mass spectrometry combined with modified mass defect filter. J. Pharm. Biomed. Anal. 2017, 141, 140–148. [Google Scholar] [CrossRef]
- Yao, Y.; Huang, Z.X.; Shu, R.G.; Wan, Y.L.; Zhang, M. Studies on chemical component of leaves in Chimonanthus nitens (II). Zhong Yao Cai 2019, 42, 100–102. [Google Scholar] [CrossRef]
- Sun, Q.; Zhu, J.; Cao, F.; Chen, F. Anti-Inflammatory properties of extracts from Chimonanthus nitens Oliv. leaf. PLoS ONE 2017, 12, e0181094. [Google Scholar] [CrossRef]
- Chen, H.; Xiong, L.; Wang, N.; Liu, X.; Hu, W.; Yang, Z.; Jiang, Y.; Zheng, G.; Ouyang, K.; Wang, W. Chimonanthus nitens Oliv. leaf extract exerting anti-hyperglycemic activity by modulating GLUT4 and GLUT1 in the skeletal muscle of a diabetic mouse model. Food Funct. 2018, 9, 4959–4967. [Google Scholar] [CrossRef]
- Chen, H.; Jiang, Y.; Yang, Z.; Hu, W.; Xiong, L.; Wang, N.; Liu, X.; Zheng, G.; Ouyang, K.; Wang, W. Effects of Chimonanthus nitens Oliv. leaf extract on glycolipid metabolism and antioxidant capacity in diabetic model mice. Oxid. Med. Cell. Longev. 2017, 2017, 7648505. [Google Scholar] [CrossRef]
- Chen, H.; Ouyang, K.; Jiang, Y.; Yang, Z.; Hu, W.; Xiong, L.; Wang, N.; Liu, X.; Wang, W. Constituent analysis of the ethanol extracts of Chimonanthus nitens Oliv. leaves and their inhibitory effect on α-glucosidase activity. Int. J. Biol. Macromol. 2017, 98, 829–836. [Google Scholar] [CrossRef]
- Li, S.; Zou, Z. Toxicity of Chimonanthus nitens flower extracts to the golden apple snail, Pomacea canaliculata. Pestic. Biochem. Physiol. 2019, 160, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z. Primarly studies on phylogeography of Chimonanthus nitens Oliv. complex (Calycanthaceae). Nanchang Univ. 2014. [Google Scholar] [CrossRef]
- Liu, H.T.; Cao, M.P.; Zhang, X.F.; Si, J.P. HPLC fingerprints establishment of chemical constituents in genus Chimonanthus leaves. Zhongguo Zhong Yao Za Zhi 2013, 38, 1560–1563. [Google Scholar]
- Zhou, B.; Tan, M.; Lu, J.F.; Zhao, J.; Xie, A.F.; Li, S.P. Simultaneous determination of five active compounds in chimonanthus nitens by double-development HPTLC and scanning densitometry. Chem. Cent. J. 2012, 6, 46. [Google Scholar] [CrossRef]
- Qian, Z.; Zhang, S.J.; Jin, Y.Q.; Zhang, X.F. Transcriptome sequencing of leaves and flowers and screening and expression of differential genes in aroma synthesis in Chimonanthus salicifolius. J. Agric. Biotechnol. 2019, 5, 844–855. [Google Scholar] [CrossRef]
- Liang, X.; Zhao, C.; Su, W. A fast and reliable UPLC-PAD fingerprint analysis of Chimonanthus salicifolius combined with chemometrics methods. J. Chromatogr. Sci. 2016, 54, 1213–1219. [Google Scholar] [CrossRef][Green Version]
- Wu, Y.F.; Lv, G.Y.; Zhang, Z.R.; Yu, J.J.; Yan, M.Q.; Chen, S.H. The influence of storage time on content of volatile oil and cineole of Chimonanthus salicifolius. Zhongguo Zhong Yao Za Zhi 2013, 38, 2803–2806. [Google Scholar]
- Ling, Q. studies on chemical composition and bioactivity of leaf in Chimonanthus grammatus. Jiangxi Norm. Univ. 2014. [Google Scholar] [CrossRef]
- Liu, Y.; Yan, R.; Lu, S.; Zhang, Z.; Zou, Z.; Zhu, D. Chemical composition and antibacterial activity of the essential oil from leaves of Chimonanthus grammatus. Zhongguo Zhong Yao Za Zhi 2011, 36, 3149–3154. [Google Scholar]
- Ouyang, T.; Mai, X. Analysis of the chemical constituents of essential oil from Chimonanthus zhejiangensis by GC-MS. Zhong Yao Cai 2010, 33, 385–387. [Google Scholar]
- Luo, X.; Liu, J.; Zhao, A.; Chen, X.; Wan, W.; Chen, L. Karyotype analysis of Piptanthus concolor based on FISH with an oligonucleotide for rDNA 5S. Sci. Hortic. 2017, 226, 361–365. [Google Scholar] [CrossRef]
- Qi, Z.X.; Zeng, H.; Li, X.L.; Chen, C.B.; Song, W.Q.; Chen, R.Y. The molecular characterization of maize B chromosome specific AFLPs. Cell Res. 2002, 12, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Matsui, Y.; Nakayama, Y.; Okamoto, M.; Fukumoto, Y.; Yamaguchi, N. Enrichment of cell populations in metaphase, anaphase, and telophase by synchronization using nocodazole and blebbistatin: A novel method suitable for examining dynamic changes in proteins during mitotic progression. Eur. J. Cell Biol. 2012, 91, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.M.; Liu, J.C. Fluorescence in situ hybridization (FISH) analysis of the locations of the oligonucleotides 5S rDNA, (AGGGTTT)3, and (TTG)6 in three genera of Oleaceae and their phylogenetic framework. Genes 2019, 10, 375. [Google Scholar] [CrossRef] [PubMed]
- Rho, R.I.; Hwang, Y.J.; Lee, H.I.; Lee, C.H.; Lim, K.B. Karyotype analysis using FISH (fluorescence in situ hybridization) in Fragaria. Sci. Hortic. 2012, 136, 95–100. [Google Scholar] [CrossRef]
- Luo, X.M.; Liu, J.C.; Wang, J.Y.; Gong, W.; Chen, L.; Wan, W.L. FISH analysis of Zanthoxylum armatum based on oligonucleotides for 5S rDNA and (GAA)6. Genome 2018, 61, 699–702. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, X.; Chen, Q.; Zhang, L.; Tang, H.; Luo, Y. Phylogenetic insight into subgenera Idaeobatus and Malachobatus (Rubus, Rosaceae) inferring from ISH analysis. Mol. Cytogenet. 2015, 8, 11–13. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Luo, X. First report of bicolour FISH of Berberis diaphana and B. soulieana reveals interspecific dierences and co-localization of (AGGGTTT)3 and rDNA 5S in B. diaphana. Hereditas 2019, 156, 13–21. [Google Scholar] [CrossRef]
- Luo, X.; Tinker, N.A.; Zhou, Y.; Wight, P.C.; Wan, W.; Chen, L.; Peng, Y. Genomic relationships among sixteen Avena taxa based on (ACT)6 trinucleotide repeat FISH. Genome 2018, 61, 63–70. [Google Scholar] [CrossRef]
- Cuadrado, A.; Cardoso, M.; Jouve, N. Increasing the physical markers of wheat chromosomes using SSRs as FISH probes. Genome 2008, 51, 809–815. [Google Scholar] [CrossRef]
- Nebel, B.R. Chromosome Structure. Bot. Rev. 1939, 5, 563–626. [Google Scholar] [CrossRef]
- Ris, H.; Kubai, D.F. Chromosome Structure. Annu. Rev. Genet. 1970, 4, 263–294. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.H.; Xiang, S.Q.; Guo, Q.G.; Jin, W.W.; Liang, G.L. Molecular cytogenetic analysis of genome-specific repetitive elements in Citrus clementina Hort. Ex Tan. and its taxonomic implications. BMC Plant Biol. 2019, 19, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Murray, B.G.; Friesen, N.; Heslop-Harrison, J.S. Molecular cytogenetic analysis of Podocarpus and comparison with other gymnosperm species. Ann. Bot. 2002, 89, 483–489. [Google Scholar] [CrossRef] [PubMed]
- Vasconcelos, E.V.; Vasconcelos, S.; Ribeiro, T.; Benko-Iseppon, A.M.; Brasileiro-Vidal, A.C. Karyotype heterogeneity in Philodendron s.l. (Araceae) revealed by chromosome mapping of rDNA loci. PLoS ONE 2018, 13, e0207318. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, X.; Chen, J. Physical Map of FISH 5S rDNA and (AG3T3)3 Signals Displays Chimonanthus campanulatus R.H. Chang & C.S. Ding Chromosomes, Reproduces its Metaphase Dynamics and Distinguishes Its Chromosomes. Genes 2019, 10, 904. https://doi.org/10.3390/genes10110904
Luo X, Chen J. Physical Map of FISH 5S rDNA and (AG3T3)3 Signals Displays Chimonanthus campanulatus R.H. Chang & C.S. Ding Chromosomes, Reproduces its Metaphase Dynamics and Distinguishes Its Chromosomes. Genes. 2019; 10(11):904. https://doi.org/10.3390/genes10110904
Chicago/Turabian StyleLuo, Xiaomei, and Jingyuan Chen. 2019. "Physical Map of FISH 5S rDNA and (AG3T3)3 Signals Displays Chimonanthus campanulatus R.H. Chang & C.S. Ding Chromosomes, Reproduces its Metaphase Dynamics and Distinguishes Its Chromosomes" Genes 10, no. 11: 904. https://doi.org/10.3390/genes10110904
APA StyleLuo, X., & Chen, J. (2019). Physical Map of FISH 5S rDNA and (AG3T3)3 Signals Displays Chimonanthus campanulatus R.H. Chang & C.S. Ding Chromosomes, Reproduces its Metaphase Dynamics and Distinguishes Its Chromosomes. Genes, 10(11), 904. https://doi.org/10.3390/genes10110904




