1. Introduction
As the Earth’s climate continues to warm, the role that microbial ecosystems play in anthropogenic climate change is becoming increasingly apparent. As these microbial communities affect the climate, they, too, will be affected [
1] and may further amplify climate change [
2]. It is crucial that our understanding of this interaction improves to inform our collective knowledge of how climate is changing [
1]. Our understanding of these microbial communities naturally correlates well with the ease of access for sample collection [
3,
4]. In particular, polar environments suffer from logistically challenging sample collection and shipping. Hence, polar environments remain poorly understood at this microbial level despite the polar cryosphere representing approximately 14% of the Earth’s surface [
5]. Traditionally, samples must be collected on site using sterile equipment and transported back to a laboratory under appropriate conditions, such as under dry ice. Logistics are further hampered by clearing customs and other delays in transit. This often results in a significant length of time from sample collection to sample analysis. During this time, the composition of the microbial community can change or degrade.
One solution to these issues is to bring the laboratory closer to the sample collection site. This has been achieved by building remote institutions and research stations, for example networks of Arctic stations (
https://eu-interact.org/) and the many research stations on the Antarctic continent [
6]. Whilst this minimises the time and logistic requirements of sample analysis, this benefit is greatest for study sites in proximity to field stations [
7]. For example, a recent meta-analysis showed that 31% of Arctic ecology citations are derived from work within a 50 km radius of just two field stations [
7].
Conventional characterisation of microbial communities relies on DNA sequencing, since most microbes remain uncultured [
8]. Both high throughput metagenomic sequencing and amplicon sequencing are typically applied with the intention of revealing the proportions of microbial species present in a particular sample [
5,
9,
10]. Over the last decade, short read sequencing has been used predominantly. This requires a full genomics laboratory to house large, bulky equipment. In recent years, long read sequencing has seen increased use. Oxford Nanopore sequencing can produce significant amounts of read data (Mbp—Gbp) from a small device capable of being run directly from a laptop [
11]. Previous studies have capitalised on the small-form factor of this device to conduct sequencing efforts at the site of sample collection. For example, portable long-read sequencing was used to sequence plant virus genomes in real time to inform crop management in sub-Saharan Africa [
12]. It has also been proven to provide early detection and epidemiologic surveillance of Ebola and Zika virus’ during epidemic outbreaks [
13,
14]. The clear advantage of this technology is that the device can be run in low-resource environments. This is largely because sample preparation requires a minimal molecular biology set up and the sequencing run itself requires only a laptop, a Nanopore MinION device, and a Nanopore flowcell. This device has also been taken underground to demonstrate the ability to sequence at inhospitable sub-surface levels [
15]. It is a clear progression to then apply these minimal-infrastructure techniques to polar environments. Temperature regulation is a key challenge in these environments as nanopore flowcells are extremely sensitive to freezing. Teams have previously conducted DNA sequencing in polar environments using Oxford Nanopore devices. Johnson et al. conducted on-site calibrations in the Antarctic Dry Valleys and further sequencing from samples at a research station [
16]. Edwards et al. similarly performed sequencing at an Arctic research station following sample collection nearby [
10]. Edwards et al. further successfully achieved in-field sample-to-data collection at a field camp in Greenland (67° N) using a combination of a generator and battery power that was flown in [
10].
Thus far, the ability to sequence DNA off-grid has been limited by access to vehicles and generators. This study aimed to minimise the infrastructure required for off-grid DNA sequencing by utilising solar power alone for all DNA extraction and sequencing in a polar environment. Due to both long days in the summer and reflective ice and snow, solar energy is in no short supply in polar environments, and therefore provides the perfect compromise between portability and renewable power availability. This demonstrates that a sequencing effort could be successfully completed midway through an ongoing, unsupported expedition alongside the other physical and psychological challenges associated with survival in such a challenging environment. Data collection was conducted in a tent after the expedition team had conducted a ski traverse of Europe’s largest ice cap (100 km) over 11 days and used solar power alone. This study occurred at least 60 km from the nearest civilisation and at least 135 km from the nearest research facility. Crucially, everything required to conduct this metagenomic study was transported in a sledge along with all equipment required for long-term polar survival.
2. Methods
2.1. Hardware Logistics
The entire laboratory setup was contained within two 9 L boxes in addition to a laptop (in waterproof/shockproof case) and 90 W solar panel (
Table 1). This was packed into the back of a 50 kg rated polar sledge (Aguille Alpine Trail Pulk, Cumbria, UK) and occupied ~30% of the sledge volume.
During the flight to Iceland, kit was stored in three different conditions: [cargo] Hold (ambient), Hold (polystyrene box with 4 °C ice packs), and hand-luggage (
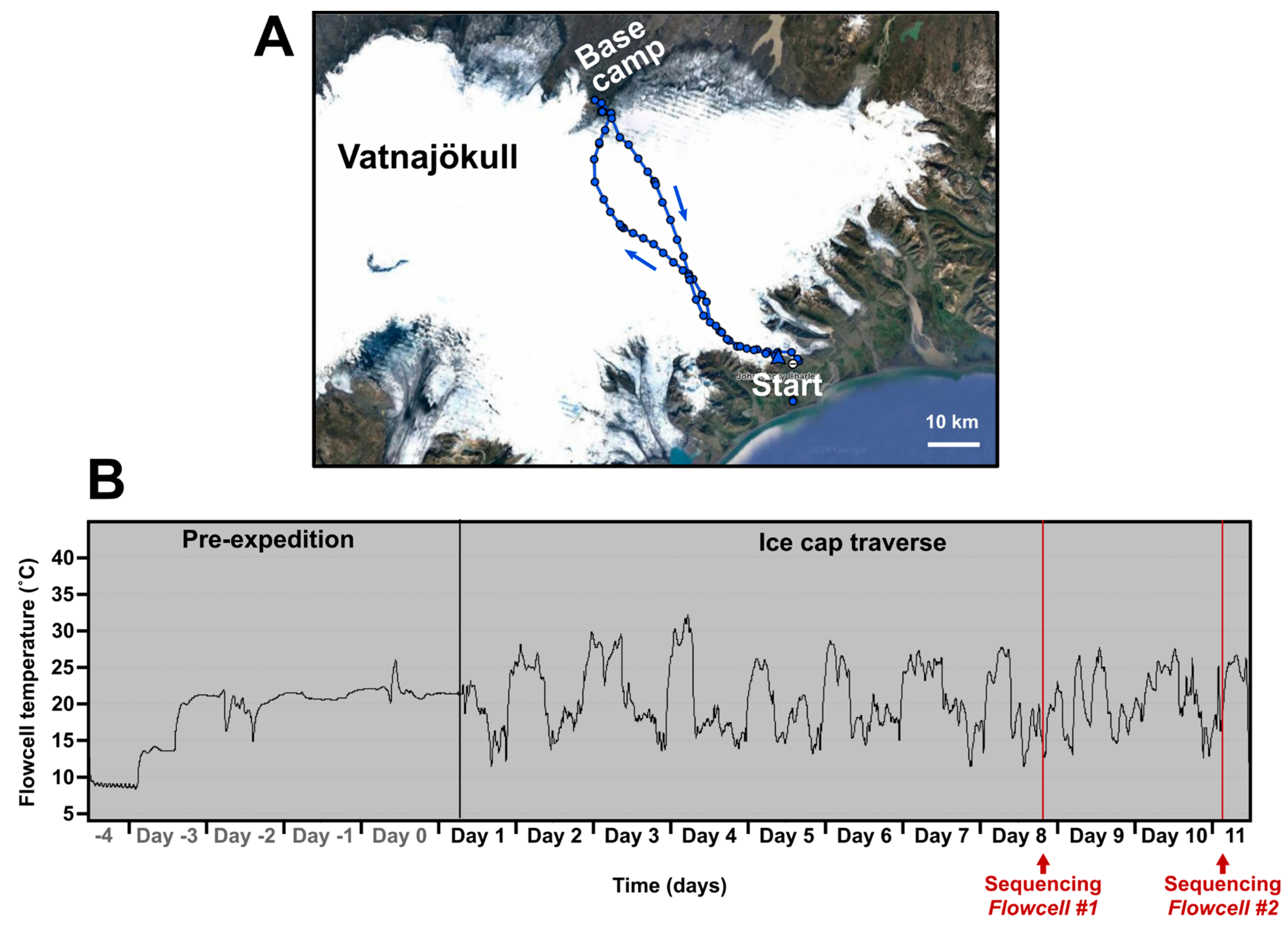
Table S1). During the ice cap traverse (route shown in
Figure 1A) three temperature zones were also established, kept on person (“Chest—warm”), taken into the tent each night (“Tent—box”), and left outside in the pulk (“Pulk—box”) (
Table S1). The temperature environment of “Chest—warm”, importantly, contained the nanopore flowcells that are destroyed if allowed to freeze. To control the temperature of the flowcells they were kept in the sleeping bag at night inside a Peli™ Case 1015 (Peli Products Ltd, Glossop, UK) and during the day items were kept inside a generic airtight plastic food container with foam padding for thermal insulation. This case was kept in a travel bag with a strap around the carrier’s neck. Temperature of the flowcells was constantly recorded (“Hold (polystyrene box)” during flight and “Chest-warm” during ski traverse) using a battery powered automatic temperature logger. This method maintained a temperature between ~+9 °C and ~+30 °C despite ambient outdoor temperatures reaching −17 °C (
Figure 1B). Overheating during exercise on warmer days became a potential issue. Temperature was continuously monitored using an aquarium thermometer taped to the outside of the plastic food container with the readout monitor clipped to a rucksack strap. Therefore, temperature control could be achieved by moving the case between clothing layers.
2.2. Sampling
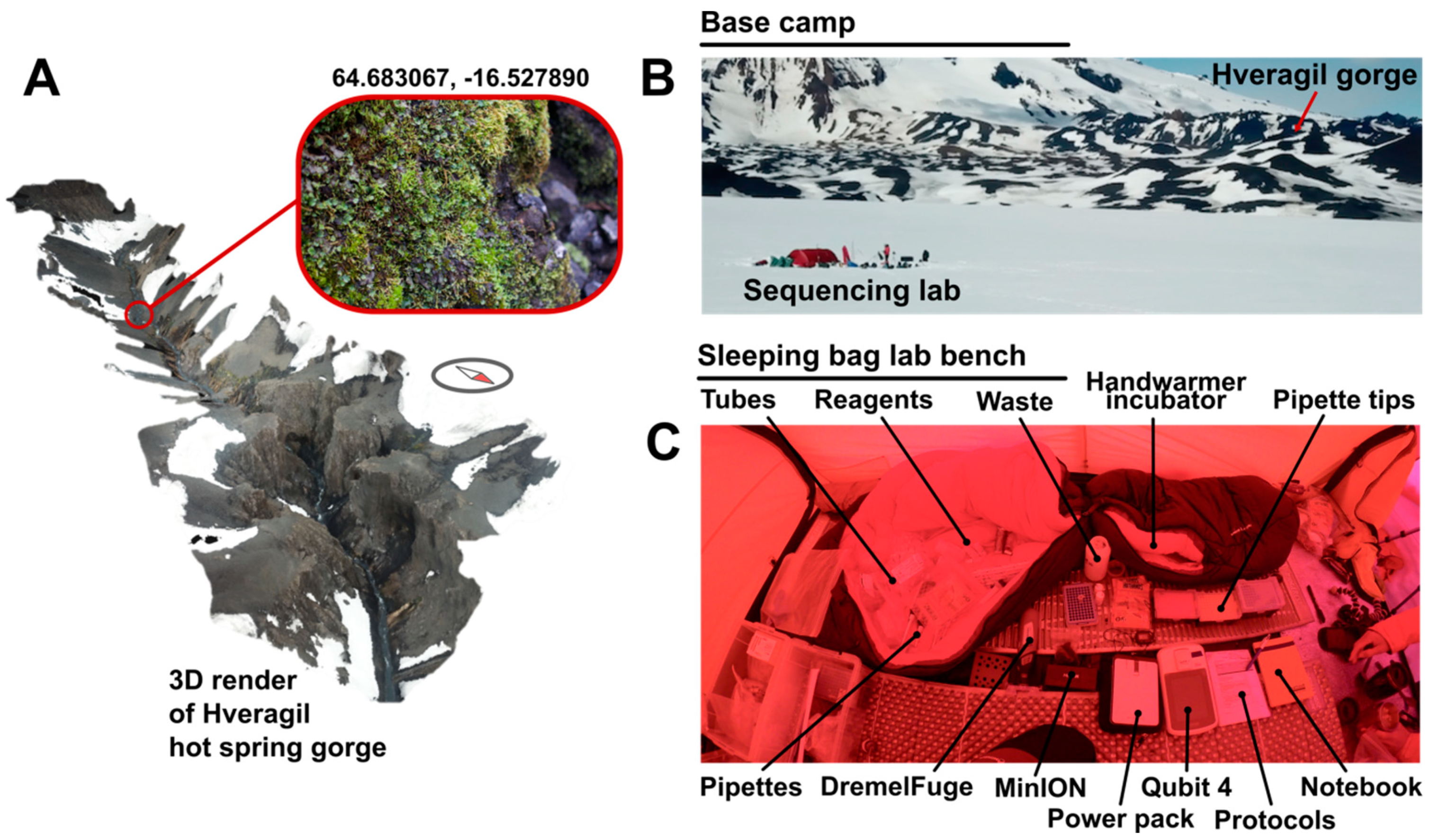
Samples were obtained from the Hveragil hot spring gorge, Iceland (decimal degree (DD) coordinates: 64.683067, −16.527890) following a ~60 km traverse of the Vatnajökull ice cap (starting decimal degree (DD) coordinates 64.255293, −15.863259) by ski and sledge (
Figure 2A). A metal spoon sterilised with ethanol was used to collect the sample into sterile sample bags. The sample consisted of soil approximately 3 cm under the surface of mosses and liverworts located on an overhanging rock 30 cm above the hot spring water surface (water at approximately +60 °C [
17]). Samples were then transported on foot and ski at ambient environmental temperature to the tent located 3 km from the sample collection site (
Figure 2B).
2.3. Battery-Powered DNA Extraction
DNA was extracted using the QIAGEN PowerSoil
® kit (Qiagen, Hilden, Germany) according to manufacturer’s instructions except for the following changes. Centrifugation steps were performed using a pre-charged Dremel™ 7750 battery-powered drill (Dremel, Prospect, IL, USA) with a 3D-printed tube holder (
https://www.thingiverse.com/thing:454962). Centrifugation steps were performed on “medium” speed (level 2) for bursts of 30 s, checking for the sufficient outcome each time to minimise battery usage. PowerBead tubes were vortexed for 30 s using a TerraLyzer™ (Zymo Research, Irvine, CA, USA) with a pre-charged 12 V battery. DNA was eluted using solution C6 (sterile elution buffer) pre-heated to body temperature by holding in a gloved hand. Extracted DNA was quantified using a Qubit™ 4 fluorometer (Thermo Fisher Scientific, Waltham, MA, USA) and dsDNA HS kit according to manufacturer’s instructions powered by a PowerAdd Pilot Pro2 20 Ah power pack. During sample preparation all solutions were kept around 20 °C using hand warmers inside a closed sleeping bag. DNA concentration was performed after extraction using 0.5× Ampure XP beads (Beckmann Coulter, Brea, CA, USA). Beads were pelleted using a strong magnet and washed with 80% ethanol. Elution was carried out with deionised water. The laboratory setup is shown in
Figure 2C.
2.4. Solar-Powered Nanopore Sequencing
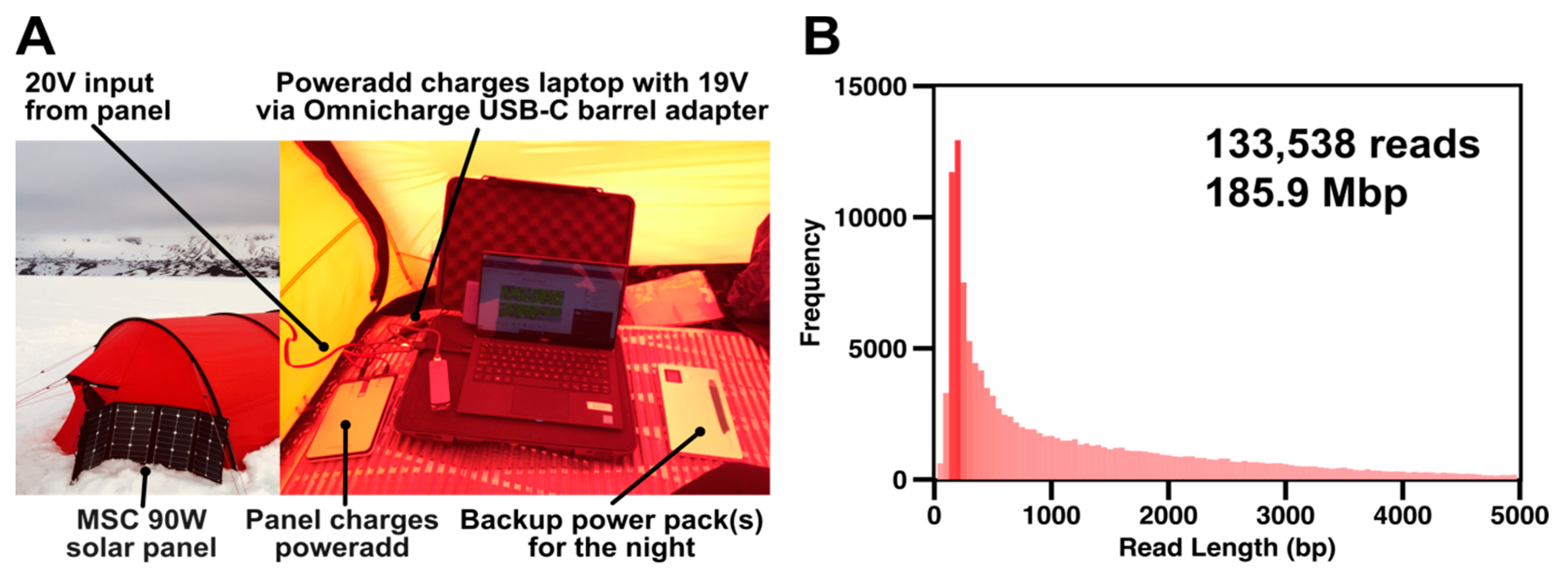
Extracted DNA was prepared using the field sequencing kit LRK001 (Oxford Nanopore Technologies) according to the manufacturer’s instructions. The 80 °C water bath was achieved by filling an insulated mug (Thermos L.L.C, Schaumburg, IL, USA) with boiling water and cooling it to 80 °C with ice as appropriate. Temperature was monitored using a standard meat thermometer. The prepared DNA library was loaded onto R9.4.1 flowcells. Sequencing was powered by one PowerAdd 20000 mAh battery and two Aceyoon 20000 mAh powerbanks, all fully charged using a 90 W Solar Panel (Mobile Solar Chargers, Somerset, UK), capable of generating approximately 20 V even in moderate cloud cover, prior to any sequencing run.
2.5. Data Analysis
Data from Flowcell #1 was locally basecalled by offline MinKNOW (v18.12.9) (Oxford Nanopore, Oxford, UK) during the run on a Dell XPS 13 i7 laptop with 16 GB RAM and 500 GB SSD. Data from Flowcell #2 was basecalled offline after the run using Guppy (v3.0.3) (Oxford Nanopore, Oxford, UK). FASTQ files from both sequencing runs were concatenated and uploaded to Kaiju (kaiju.binf.ku.dk) for taxonomic classification upon return to civilisation due to code errors running Kaiju offline in situ. Reads were compared to the “NCBI BLAST nr + euk” reference database using the “Greedy” setting, allowing mismatches with standard parameters (minimum match length 11, minimum match score 75, allowed mismatches 5).
4. Discussion
In this study the successful miniaturisation of a DNA sequencing laboratory such that can be hauled by human power within a larger self-supported expedition is demonstrated. The experimental outcomes prompt some observations that may aid future similar off-grid projects. First, the DremelFuge 3D printed attachment (FormLabs clear photopolymer resin) became brittle at lower temperatures and at one point cracked during centrifugation. This both posed a safety risk and nearly derailed the DNA extraction protocol. While a backup manual centrifuge was taken [
19] it too suffered from being brittle at lower temperatures. It is therefore recommended that future teams pick a more suitable resin for lower temperatures. Furthermore, there were issues keeping the Dremel drill charged using battery packs and, as such, the existing battery charge from before departure was relied upon. Since returning the Myspin 12 (Thermo Scientific, Waltham, MA, USA) has been used, which was successfully run from the PowerAdd power bank at 12 V. Due to its small footprint it is recommend future teams use this instead of the DremelFuge.
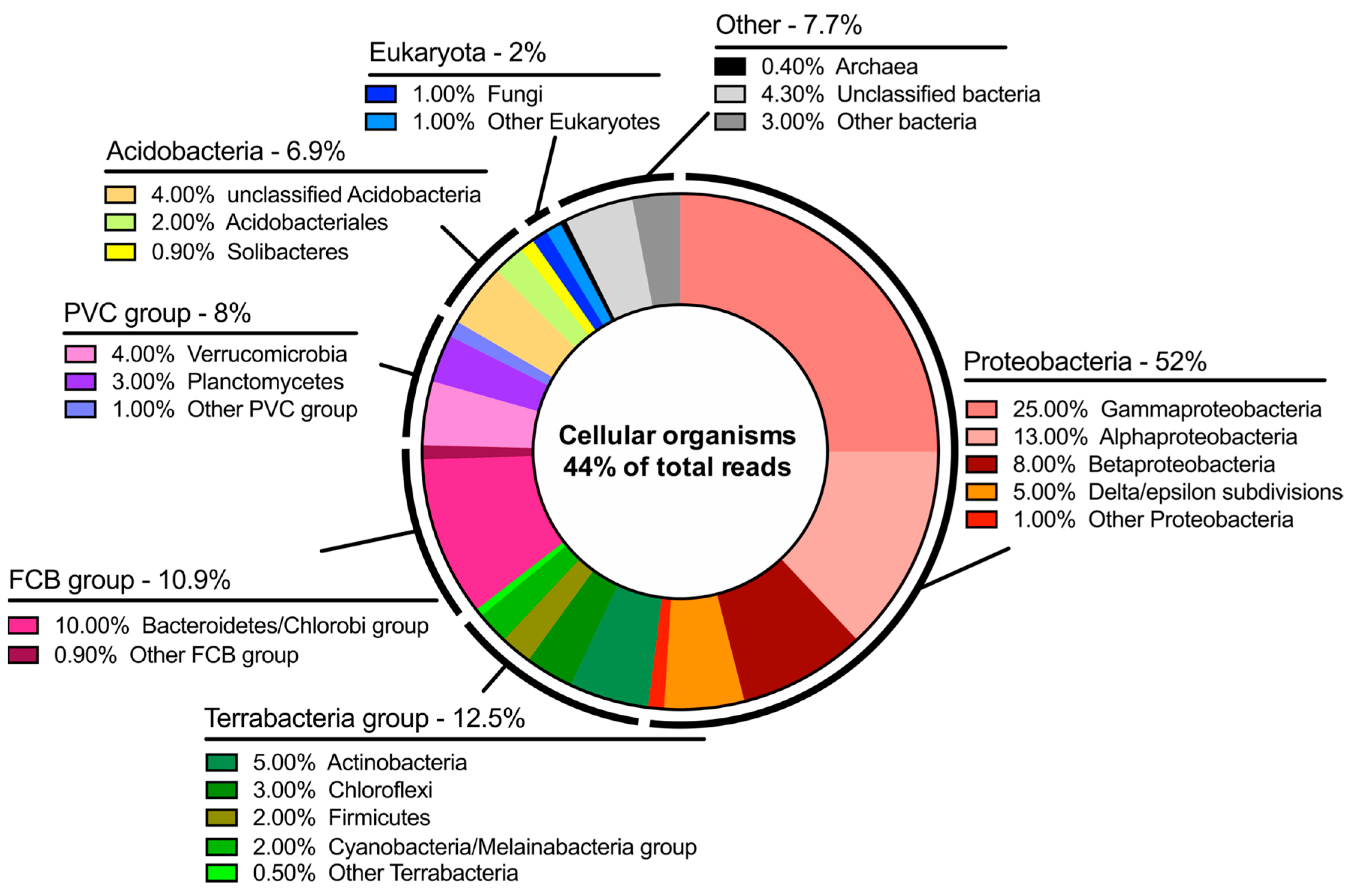
The dichotomy of microorganisms identified at Hveragil hot spring gorge was particularly intriguing. There was a sizable number of reads assigned to both cyanobacteria characteristic of the cold polar regions and thermophilic bacteria. It is believed that this indicates the interesting microenvironment present in hot spring streams fed by meltwater from the Vatnajökull ice cap [
17], resulting in a microbial population with presumably extremely diverse metabolisms. Naturally, this study demonstrates the methodology of entirely off-grid metagenome sequencing and future studies are needed to probe the microbial taxonomic distribution in this area further.
Additionally, it is worth noting that approximately 56% of the data generated could not be aligned to the “NCBI nr + euk” database. This number drops to 48% when a stringent quality threshold is applied to the reads. Therefore, it is expected most of this number represents organisms that have yet to be cultured and have their genomes characterized. While it may be expected that some of the data was from unknown organisms it is surprising that it was so high. Naturally, Hveragil gorge, on the northern edge of the ice cap, is among the most inaccessible places in Iceland. This, in combination with the interesting geochemistry of the area leads to the conclusion that 56% represents a pool of interesting organisms and metabolisms that could be later isolated and studied further.
This entire metagenomic study was conducted off-grid, on the ice, and all while over 135 km from the nearest research facility as the crow flies. Off-grid sequencing reduced the time from sample-to-data to under 5 h compared to >days/weeks for sample collection and transport back to a research institution. In this way potential bias that may be introduced due to lengthy transport times often under suboptimal storage conditions is avoided [
10]. This off-grid DNA sequencing method requires minimal infrastructure, and therefore lends itself to being integrated into a scientific program of the many expeditions that set out to the most remote corners of the polar world. This work represents a blueprint for a minimal and sustainable remote laboratory that, permitting technological advancements, could be adapted to perform other types of in situ sample extractions and downstream transcriptomics, metabolomics, or proteomics studies. By aiding the wider distribution of DNA sequencing capabilities, we hope to improve our collective understanding of the microbial communities that exist in some of the planet’s most remote environments and the role that they play in our changing climate.