Characterization of GLOD4 in Leydig Cells of Tibetan Sheep during Different Stages of Maturity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Animals and Design
2.2. Total RNA Extraction and cDNA Synthesis
2.3. Cloning of Sheep GLOD4 Gene
2.4. Bioinformatics Analysis of the GLOD4 Gene
2.5. qRT-PCR
2.6. Western Blot
2.7. Immunohistochemistry and Immunofluorescence
2.8. Image Analysis and Data Statistics
3. Results
3.1. CDS Sequence Characterization of GLOD4
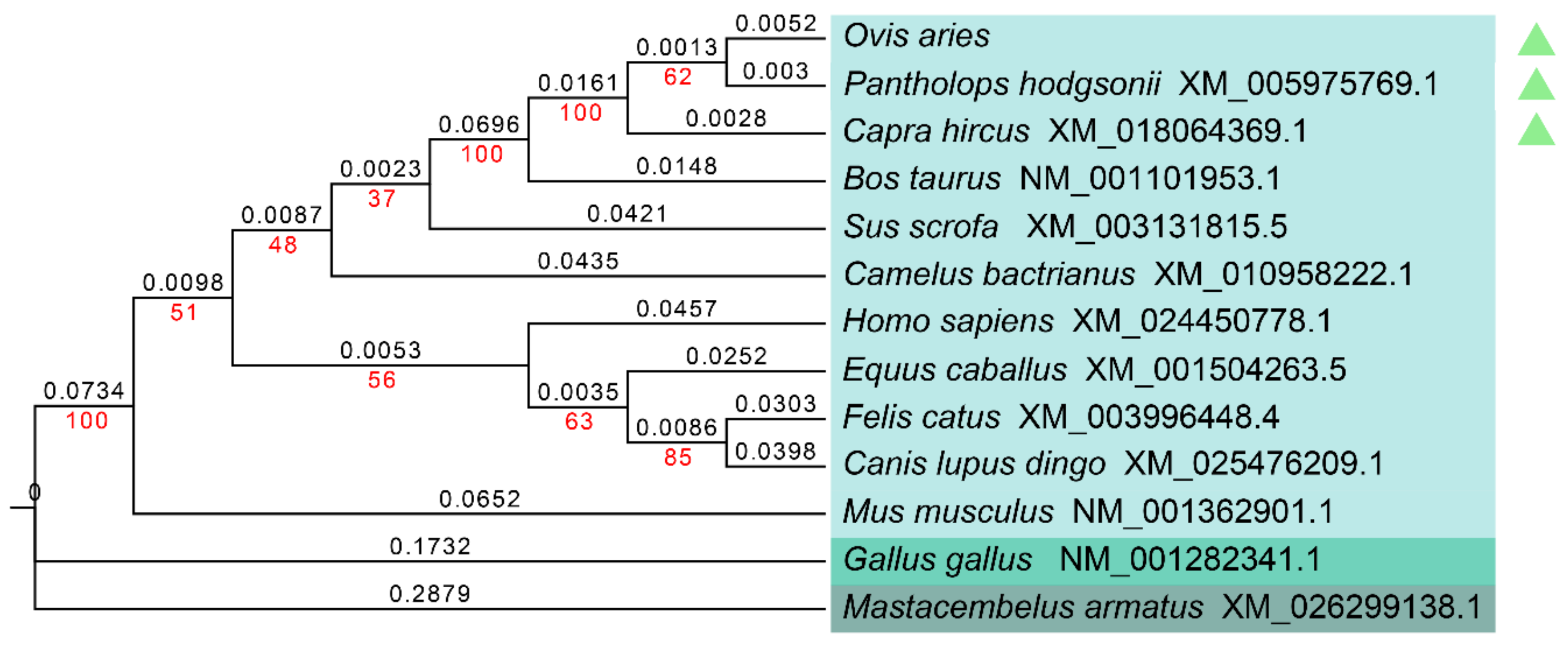
3.2. Homology Analysis and Evolutionary Relationships of the GLOD4 Gene among Different Species
3.3. Expression Patterns of the GLOD4 Transcript at Different Developmental Stages of Tibetan Sheep Testes
3.4. Localization of the GLOD4 Protein in Developmental Sheep Testes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Staub, C.; Johnson, L. Review: Spermatogenesis in the bull. Animal 2018, 12, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Chalmel, F.; Rolland, A.D. Linking transcriptomics and proteomics in spermatogenesis. Reproduction 2015, 150, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, Y.; Huang, L.; Yang, L.; Xing, X. SPAG4L/SPAG4Lβ interacts with Nesprin2 to participate in the meiosis of spermatogenesis. Acta Biochim. Biophys. Sin. 2019, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.X.; Wan, F.; Sun, F.Y.; Zhang, P.; Han, L.W.; Huang, Y.; Jiang, H.Q.; Zhao, X.T.; He, M.; Ye, Y.; et al. Cloning and characterization of a novel gene (C17orf25) from the deletion region on chromosome 17p13.3 in hepatocelular carcinoma. Cell Res. 2001, 11, 209–216. [Google Scholar] [CrossRef]
- Guo, J.Y.; Xu, J.; Mao, Q.; Fu, L.L.; Gu, J.R.; De, Z.J. The promoter analysis of the human C17orf25 gene, a novel chromosome 17p13.3 gene. Cell Res. 2002, 12, 339–352. [Google Scholar] [CrossRef]
- Yan, L.; Peng, M.H.; Chen, M.M.; Li, Y.P.; Zhang, Q.R.; Jiang, X.F.; Liu, Y.Q. Characterization of insect cytosolic juvenile hormone binding protein gene: Highly homology with vertebrate glyoxalase domain containing protein 4. Biochem. Syst. Ecol. 2015, 58, 227–234. [Google Scholar]
- Albee, A.J.; Kwan, A.L.; Lin, H.; Granas, D.; Stormo, G.D.; Dutcher, S.K. Identification of cilia genes that affect cell-cycle progression using whole-genome transcriptome analysis in Chlamydomonas reinhardtti. G3 Genes Genomes Genet. 2013, 3, 979–991. [Google Scholar]
- Zhang, H.T.; Yan, Z.Q.; Hu, X.B.; Yang, S.L.; Gong, Y. Interaction of C17orf25 with ADP-ribose pyrophosphatase NUDT9 detected via yeast two-hybrid method. Acta Biochim. Biophys. Sin. 2003, 35, 747–751. [Google Scholar]
- Dihazi, G.H.; Bibi, A.; Jahn, O.; Nolte, J.; Mueller, G.A.; Engel, W.; Dihazi, H. Impact of the antiproliferative agent ciclopirox olamine treatment on stem cells proteome. World J. Stem Cells 2013, 5, 9–25. [Google Scholar] [CrossRef]
- Suszyńska-Zajczyk, J.; Utyro, O.; Jakubowski, H. Methionine-induced hyperhomocysteinemia and bleomycin hydrolase deficiency alter the expression of mouse kidney proteins involved in renal disease. Mol. Genet. Metab. 2014, 112, 339–346. [Google Scholar] [CrossRef]
- Lu, Z.; Ma, Y.; Zhang, Q.; Zhao, X.; Zhang, Y.; Zhang, L. Proteomic analyses of ram (Ovis aries) testis during different developmental stages. Anim. Reprod. Sci. 2018, 189, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Li, Y. Rumen methanogen and protozoal communities of Tibetan sheep and Gansu Alpine Finewool sheep grazing on the Qinghai-Tibetan Plateau, China. BMC Microbiol. 2018, 18, 212. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ding, X.; Zeng, Y.; Yue, Y.; Guo, X.; Guo, T.; Chu, M.; Wang, F.; Han, J.; Feng, R.; et al. Genetic diversity and phylogenetic evolution of Tibetan sheep based on mtDNA D-loop sequences. PLoS ONE 2016, 11, e0159308. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.J.; Yang, J.; Xie, X.L.; Lv, F.H.; Cao, Y.H.; Li, W.R.; Liu, M.J.; Wang, Y.T.; Li, J.Q.; Liu, Y.G.; et al. The genome landscape of Tibetan sheep reveals adaptive introgression from argali and the history of early human settlements on the Qinghai-Tibetan Plateau. Mol. Biol. Evol. 2019, 36, 283–303. [Google Scholar] [CrossRef]
- Wang, G.; He, Y.; Luo, Y. Expression of OPA1 and Mic60 genes and their association with mitochondrial cristae morphology in Tibetan sheep. Cell Tissue Res. 2019, 376, 273–279. [Google Scholar] [CrossRef]
- Dong, S.K.; Long, R.J.; Kang, M.Y.; Pu, X.P.; Guo, Y.J. Effect of urea multinutritional molasses block supplementation on liveweight change of yak calves and productive and reproductive performances of yak cows. Can. J. Anim. Sci. 2003, 83, 141–145. [Google Scholar]
- Xin, G.S.; Long, R.J.; Guo, X.S.; Irvine, J.; Ding, L.M.; Ding, L.L.; Shang, Z.H. Blood mineral status of grazing Tibetan sheep in the Northeast of the Qinghai-Tibetan Plateau. Livest. Sci. 2011, 136, 102–107. [Google Scholar] [CrossRef]
- Xue, B.; Zhao, X.Q.; Zhang, Y.S. Seasonal changes in weight and body composition of yak grazing on alpine-meadow grassland in the Qinghai-Tibetan plateau of China. J. Anim. Sci. 2005, 83, 1908–1913. [Google Scholar] [CrossRef]
- Sun, Y.; Angerer, J.P.; Hou, F.J. Effects of grazing systems on herbage mass and liveweight gain of Tibetan sheep in Eastern Qinghai-Tibetan Plateau, China. Rangel. J. 2015, 37, 181–190. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Gao, P.; Hou, F.; Yan, T.; Chang, S.; Chen, X.; Wang, Z. Relationship between chemical composition of native forage and nutrient digestibility by Tibetan sheep on the Qinghai-Tibetan Plateau. J. Anim. Sci. 2018, 96, 1140–1149. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Deval, C.; Chaveroux, C.; Maurin, A.C.; Cherasse, Y.; Parry, L.; Carraro, V.; Milenkovic, D.; Ferrara, M.; Bruhat, A.; Jousse, C.; et al. Amino acid limitation regulates the expression of genes involved in several specific biological processes through GCN2-dependent and GCN2-independent pathways. FEBS J. 2009, 276, 707–718. [Google Scholar] [CrossRef]
- Jia, H.; Zhang, X.B.; Song, X.F. Correlation between half-life of human intracellular protein and its subcellular localization. Comput. Appl. Chem. 2011, 28, 411–414. [Google Scholar]
- Kumar, S.; Tsai, C.J.; Nussinov, R. Factors enhancing protein thermostability. Protein Eng. 2000, 13, 179–191. [Google Scholar] [CrossRef] [Green Version]
- Suszyńska-Zajczyk, J.; Wróblewski, J.; Utyro, O.; Luczak, M.; Marczak, L.; Jakubowski, H. Bleomycin hydrolase and hyperhomocysteinemia modulate the expression of mouse proteins involved in liver homeostasis. Amino Acids 2014, 46, 1471–1480. [Google Scholar] [CrossRef]
- Liu, S.; Liu, B.; He, S.; Zhao, Y.; Wang, Z. Cloning and characterization of zebra fish SPATA4 gene and analysis of its gonad specific expression. Biochem. Mosc. 2005, 70, 638–644. [Google Scholar] [CrossRef]
- Xie, M.C.; Ai, C.; Jin, X.M.; Liu, S.F.; Tao, S.X.; Li, Z.D.; Wang, Z. Cloning and characterization of chicken SPATA4 gene and analysis of its specific expression. Mol. Cell. Biochem. 2007, 306, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Li, L.; Xie, M.; Fuji, R.; Liu, S.; Yin, X.; Li, G.; Wang, Z. SPATA4 counteracts etoposide-induced apoptosis via modulating bcl-2 family proteins in HeLa cells. Biol. Pharm. Bull. 2015, 38, 1458–1463. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.M.; Murakami, N.; Kaseda, Y. Relationship between plasma testosterone concentrations and age, breeding season and harem size in Misaki feral horses. J. Vet. Med. Sci. 1998, 60, 643–645. [Google Scholar] [CrossRef] [PubMed]
- Zirkin, B.R.; Papadopoulos, V. Leydig cells: Formation, function, and regulation. Biol. Reprod. 2018, 99, 101–111. [Google Scholar] [CrossRef]
- Ho, H.J.; Shirakawa, H.; Giriwono, P.E.; Ito, A.; Komai, M. A novel function of geranylgeraniol in regulating testosterone production. Biosci. Biotechnol. Biochem. 2018, 82, 956–962. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, B.; Chakraborty, S.; Chakraborty, P.; Ghosh, D.; Jana, K. Protective effect of Resveratrol on Benzo(a)pyrene induced dysfunctions of steroidogenesis and steroidogenic acute regulatory gene cxpression in Leydig cells. Front. Endocrinol. 2019, 10, 272. [Google Scholar] [CrossRef]
- Miller, W.L.; Bose, H.S. Early steps in steroidogenesis: Intracellular cholesterol trafficking. J. Lipid Res. 2011, 52, 2111–2135. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, F.; Ye, L.; Zirkin, B.; Chen, H. Steroidogenesis in Leydig cells: Effects of aging and environmental factors. Reproduction 2017, 154, 111–122. [Google Scholar] [CrossRef]
- Stocco, D.M.; Clark, B.J. The role of the steroidogenic acute regulatory protein in steroidogenesis. Steroids 1997, 62, 29–36. [Google Scholar] [CrossRef]
- Mannervik, B. Molecular enzymology of the glyoxalase system. Drug Metabol. Drug Interact. 2008, 23, 13–28. [Google Scholar] [CrossRef]
- Duarte, A.; Poderoso, C.; Cooke, M.; Soria, G.; Cornejo Maciel, F.; Gottifredi, V.; Podestá, E.J. Mitochondrial fusion is essential for steroid biosynthesis. PLoS ONE 2012, 7, e45829. [Google Scholar] [CrossRef] [PubMed]
- Issop, L.; Fan, J.; Lee, S.; Rone, M.B.; Basu, K.; Mui, J.; Papadopoulos, V. Mitochondria-associated membrane formation in hormone-stimulated Leydig cell steroidogenesis: Role of ATAD3. Endocrinology 2015, 156, 334–345. [Google Scholar] [CrossRef] [PubMed]
- Hales, D.B.; Allen, J.A.; Shankara, T.; Janus, P.; Buck, S.; Diemer, T.; Hales, K.H. Mitochondrial function in Leydig cell steroidogenesis. Ann. N. Y. Acad. Sci. 2005, 1061, 120–134. [Google Scholar] [CrossRef] [PubMed]
- Midzak, A.S.; Chen, H.; Aon, M.A.; Papadopoulos, V.; Zirkin, B.R. ATP synthesis, mitochondrial function, and steroid biosynthesis in rodent primary and tumor Leydig cells. Biol. Reprod. 2011, 84, 976–985. [Google Scholar] [CrossRef]






| Gene | GenBank No. | Sequence(5′-3′) | Length/bp | Utilization |
|---|---|---|---|---|
| GLOD4 | XM_027975163.1 | F: ATGGTGGGGTATGGACCCG | 729 | Cloning |
| R: TTAACCTGTCGCCTTTGGTTTC | ||||
| GLOD4 | XM_027975163.1 | F: CTCCGTTGGTGAGTCTGG | 167 | qRT- PCR |
| R: ATCTCCTGCAATTGCGTCG | ||||
| β-actin | NM_001009784.1 | F: CTTCCAGCCTTCCTTCCTGG | 180 | qRT- PCR |
| R: GCCAGGGCAGTGATCTCTTT |
| Species | Genbank No. | Nucleotide Similarity % | Amino acid Similarity % |
|---|---|---|---|
| Pantholops hodgsonii (chiru) | XM_005975769.1 | 99.18 | 100 |
| Capra hircus (goat) | XM_018064369.1 | 99.18 | 100 |
| Bos Taurus (cattle) | NM_001101953.1 | 96.57 | 97.52 |
| Equus caballus (horse) | XM_001504263.5 | 87.79 | 85.95 |
| Sus scrofa (pig) | XM_003131815.5 | 87.52 | 85.54 |
| Camelus bactrianus (Bactrian camel) | XM_010958222.1 | 87.24 | 84.71 |
| Felis catus (domestic cat) | XM_003996448.4 | 86.15 | 85.95 |
| Canis lupus dingo (dingo) | XM_025476209.1 | 85.95 | 83.88 |
| Homo sapiens (human) | XM_024450778.1 | 85.23 | 84.3 |
| Mus musculus (house mouse) | NM_001362901.1 | 84.72 | 83.47 |
| Gallus gallus (chicken) | NM_001282341.1 | 74.54 | 76.76 |
| Mastacembelus armatus (zig-zag eel) | XM_026299138.1 | 69.74 | 68.18 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Li, T.; Liu, N.; Zhang, H.; Zhao, X.; Ma, Y. Characterization of GLOD4 in Leydig Cells of Tibetan Sheep during Different Stages of Maturity. Genes 2019, 10, 796. https://doi.org/10.3390/genes10100796
Wang X, Li T, Liu N, Zhang H, Zhao X, Ma Y. Characterization of GLOD4 in Leydig Cells of Tibetan Sheep during Different Stages of Maturity. Genes. 2019; 10(10):796. https://doi.org/10.3390/genes10100796
Chicago/Turabian StyleWang, Xia, Taotao Li, Ningbo Liu, Hongyu Zhang, Xingxu Zhao, and Youji Ma. 2019. "Characterization of GLOD4 in Leydig Cells of Tibetan Sheep during Different Stages of Maturity" Genes 10, no. 10: 796. https://doi.org/10.3390/genes10100796





