Ectopic Expression of Multiple Chrysanthemum (Chrysanthemum × morifolium) R2R3-MYB Transcription Factor Genes Regulates Anthocyanin Accumulation in Tobacco
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Isolation of TF Genes
2.3. Phylogenetic Clustering and Amino Acid Sequence Alignment
2.4. Expression Vector Construction and Tobacco Transformation
2.5. Anthocyanin Component Analysis
2.6. qRT-PCR Analysis
3. Results
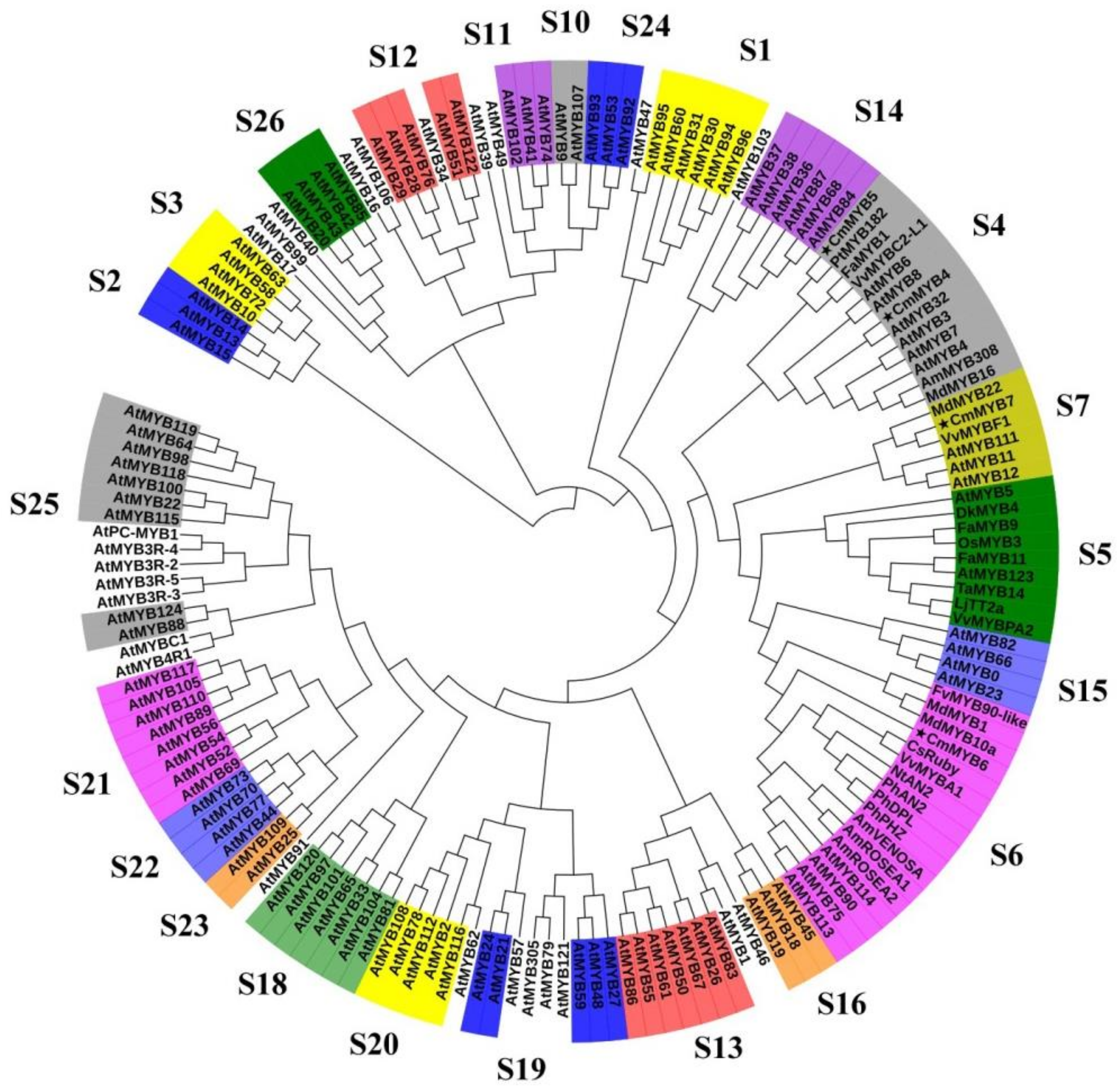
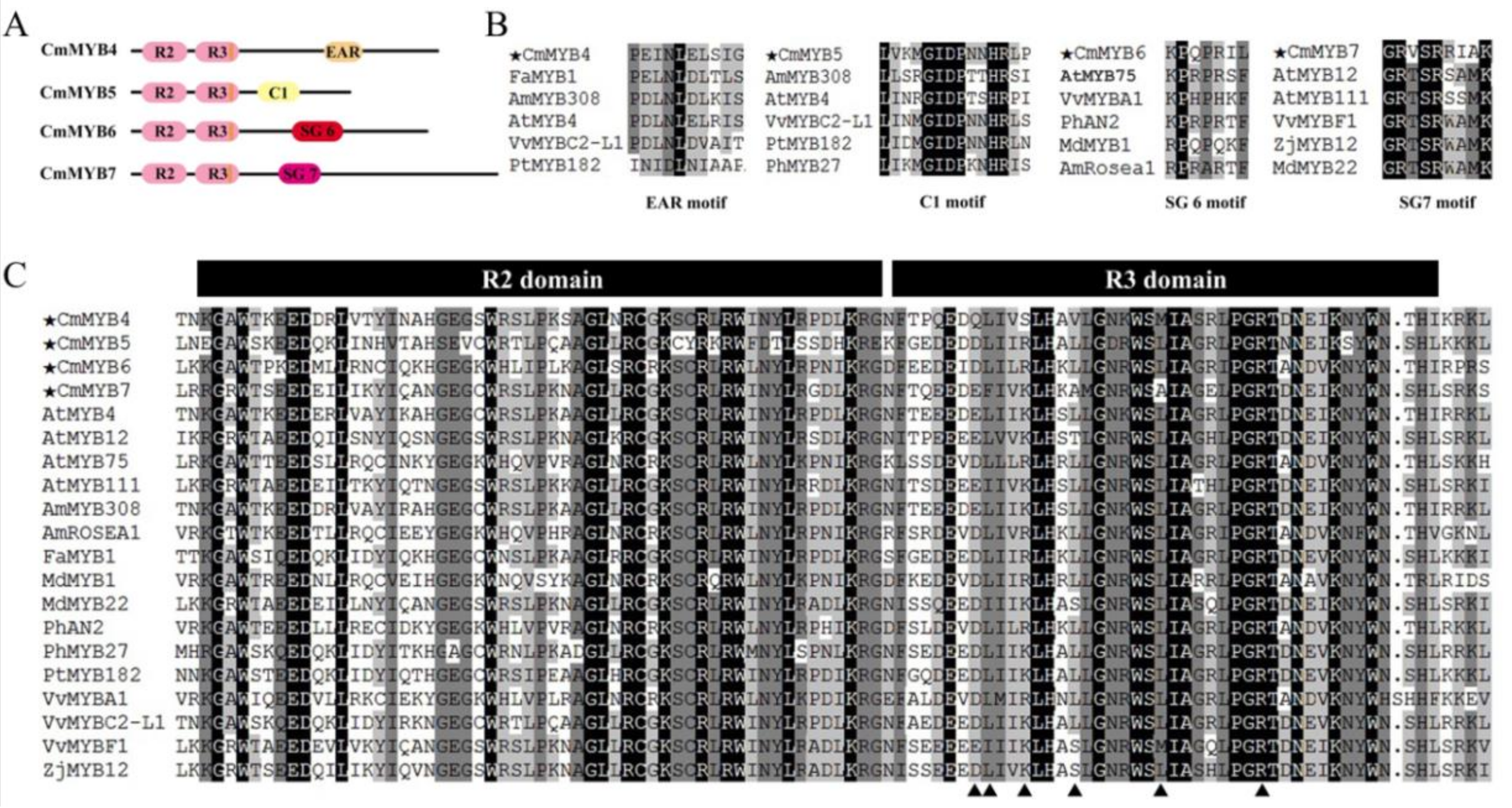
3.1. Phylogenetic and Sequence Alignment Analyses
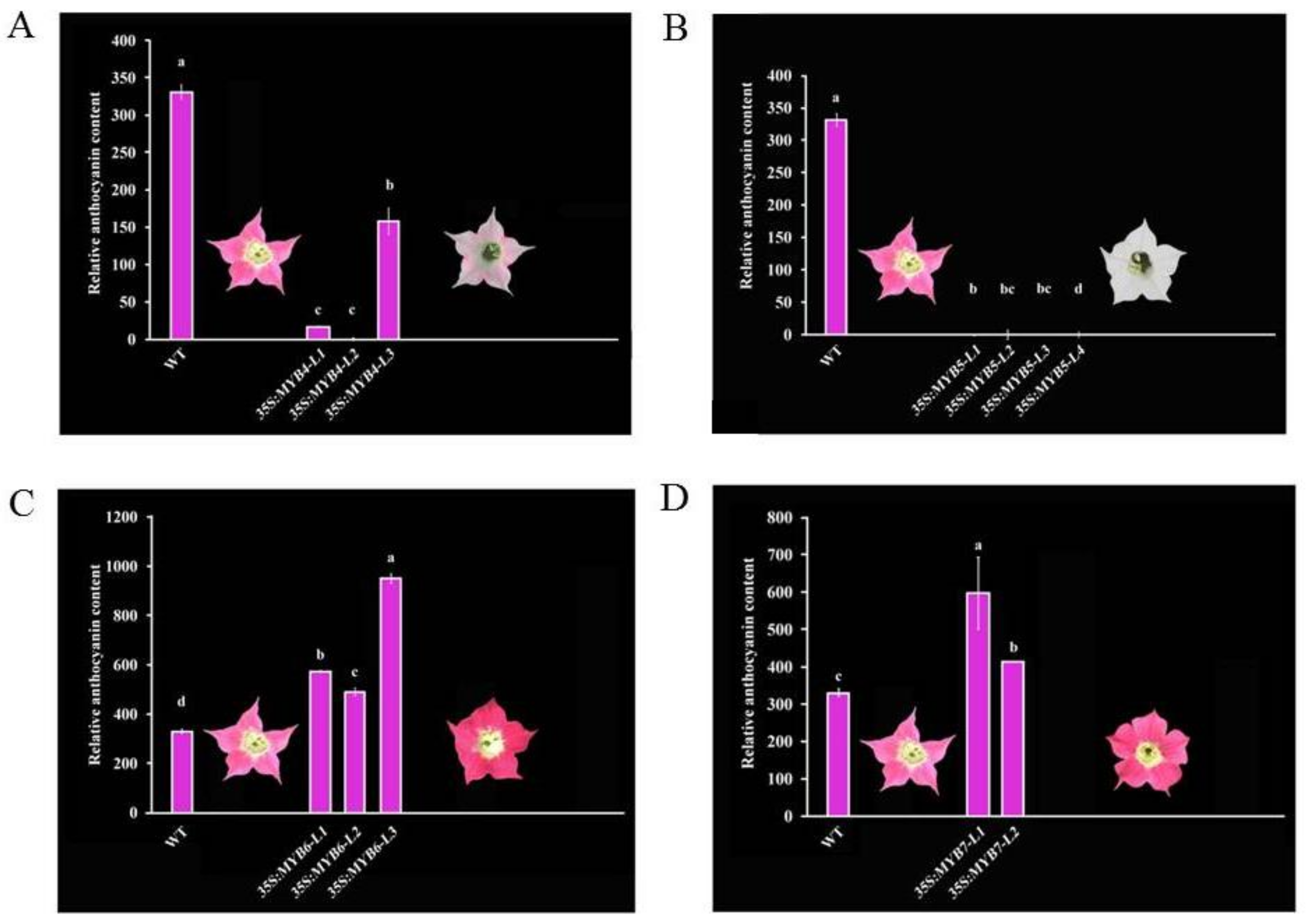
3.2. Pigmental and Phenotypic Analyses of Transgenic Tobacco Lines
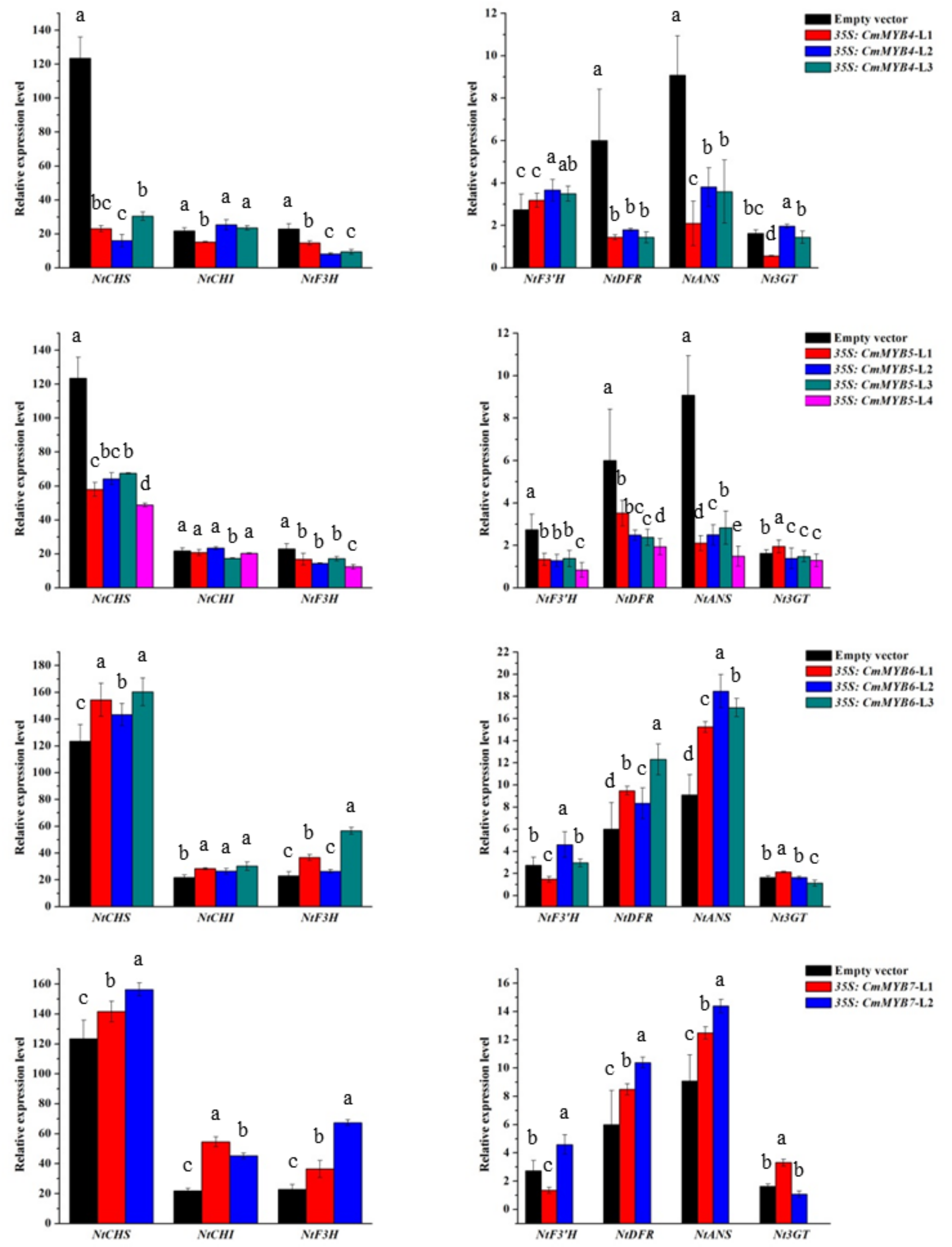
3.3. Expression of Anthocyanin Biosynthetic Genes in Transgenic Tobacco Lines
4. Discussion
4.1. All Four CmMYB TFs Are R2R3-MYBs with Diverse Functional Motifs in the C-Terminus
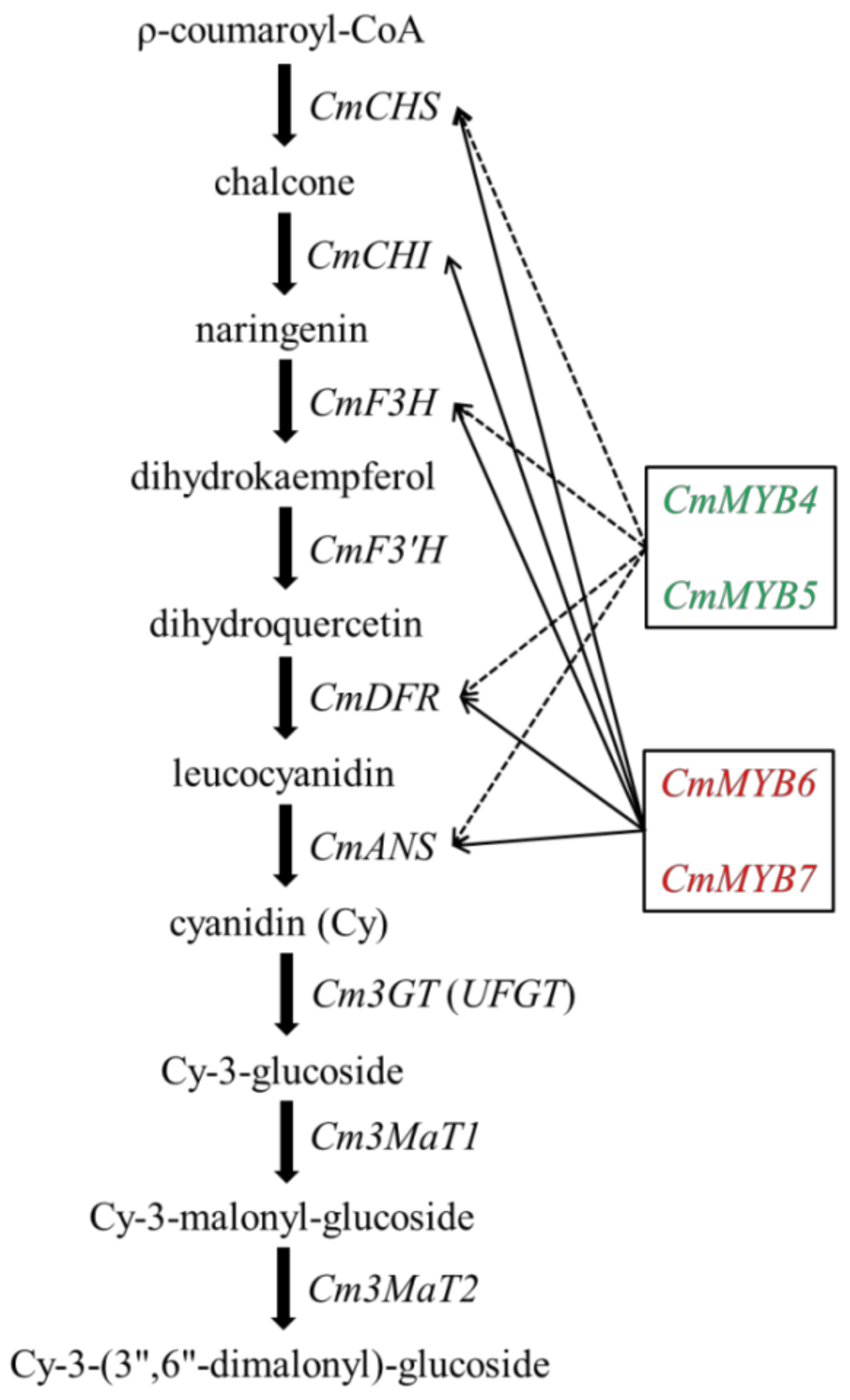
4.2. CmMYB4 and CmMYB5 Are Anthocyanin Repressors, while CmMYB6 and CmMYB7 Are Activators
4.3. The Combination of Protein–Protein Interactions among CmMYBs and bHLHs Needs to Be Further Studied
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Naing, A.H.; Park, K.I.; Ai, T.N.; Chung, M.Y.; Han, J.S.; Kang, Y.-W.; Lim, K.B.; Kim, C.K. Overexpression of snapdragon Delila (Del) gene in tobacco enhances anthocyanin accumulation and abiotic stress tolerance. BMC Plant Biol. 2017, 17, 65. [Google Scholar] [CrossRef] [PubMed]
- Holton, T.A.; Cornish, E.C. Genetics and Biochemistry of Anthocyanin Biosynthesis. Plant Cell 1995, 7, 1071–1083. [Google Scholar] [CrossRef] [PubMed]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Hichri, I.; Barrieu, F.; Bogs, J.; Kappel, C.; Delrot, S.; Lauvergeat, V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 2011, 62, 2465–2483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broun, P. Transcriptional control of flavonoid biosynthesis: A complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr. Opin. Plant Biol. 2005, 8, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Petroni, K.; Tonelli, C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011, 181, 219–229. [Google Scholar] [CrossRef]
- Albert, N.W.; Davies, K.M.; Lewis, D.H.; Zhang, H.; Montefiori, M.; Brendolise, C.; Boase, M.R.; Ngo, H.; Jameson, P.E.; Schwinn, K.E. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 2014, 26, 962–980. [Google Scholar] [CrossRef]
- Xie, Q.; Hu, Z.; Zhang, Y.; Tian, S.; Wang, Z.; Zhao, Z.; Yang, Y.; Chen, G. Accumulation and Molecular Regulation of Anthocyanin in Purple Tumorous Stem Mustard (Brassica juncea var. tumida Tsen et Lee). J. Agric. Food Chem. 2014, 62, 7813–7821. [Google Scholar] [CrossRef]
- Zhang, Q.; Hao, R.; Xu, Z.; Yang, W.; Wang, J.; Cheng, T.; Pan, H.; Zhang, Q. Isolation and functional characterization of a R2R3-MYB regulator of Prunus mume anthocyanin biosynthetic pathway. Plant Cell Tissue Organ Cult. 2017, 131, 417–429. [Google Scholar] [CrossRef]
- Schwinn, K.; Venail, J.; Shang, Y.; Mackay, S.; Alm, V.; Butelli, E.; Oyama, R.; Bailey, P.; Davies, K.; Martin, C. A Small Family of MYB-Regulatory Genes Controls Floral Pigmentation Intensity and Patterning in the Genus Antirrhinum. Plant Cell 2006, 18, 831–851. [Google Scholar] [CrossRef]
- Allan, A.C.; Hellens, R.P.; Laing, W.A. MYB transcription factors that colour our fruit. Trends Plant Sci. 2008, 13, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef] [PubMed]
- Davies, K.M.; Albert, N.W.; Schwinn, K.E. From landing lights to mimicry: The molecular regulation of flower colouration and mechanisms for pigmentation patterning. Funct. Plant Biol. 2012, 39, 619–638. [Google Scholar] [CrossRef]
- Zhang, Y.; Butelli, E.; Martin, C. Engineering anthocyanin biosynthesis in plants. Curr. Opin. Plant Biol. 2014, 19, 81–90. [Google Scholar] [CrossRef]
- Zhang, H.; Koes, R.; Shang, H.; Fu, Z.; Wang, L.; Dong, X.; Zhang, J.; Passeri, V.; Li, Y.; Jiang, H.; et al. Identification and functional analysis of three new anthocyanin R2R3-MYB genes in Petunia. Plant Direct 2019, 3, e00114. [Google Scholar] [CrossRef]
- Luo, X.; Zhao, H.; Yao, P.; Li, Q.; Huang, Y.; Li, C.; Chen, H.; Wu, Q. An R2R3-MYB transcription factor FtMYB15 involved in the synthesis of anthocyanin and Proanthocyanidins from Tartary buckwheat. J. Plant Growth Regul. 2018, 37, 76–84. [Google Scholar] [CrossRef]
- Feng, K.; Xu, Z.S.; Que, F.; Liu, J.X.; Wang, F.; Xiong, A.S. An R2R3-MYB transcription factor, OjMYB1, functions in anthocyanin biosynthesis in Oenanthe javanica. Planta 2018, 247, 301–315. [Google Scholar] [CrossRef]
- Tamagnone, L.; Mérida, Á.; Parr, A.; Mackay, S.; Culiáñez-Macià, F.A.; Roberts, K.; Martin, C. The AmMYB308 and AmMYB330 Transcription Factors from Antirrhinum Regulate Phenylpropanoid and Lignin Biosynthesis in Transgenic Tobacco. Plant Cell 1998, 10, 135–154. [Google Scholar] [CrossRef] [Green Version]
- Jin, H.; Cominelli, E.; Bailey, P.; Parr, A.; Mehrtens, F.; Jones, J.; Tonelli, C.; Weisshaar, B.; Martin, C. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 2000, 19, 6150–6161. [Google Scholar] [CrossRef] [Green Version]
- De Vos, C.H.R.; Wein, M.; Kroon, A.; O’Connell, A.P.; Aharoni, A.; Sun, Z.; Greco, R.; Mol, J.N.M. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 2001, 28, 319–332. [Google Scholar]
- Da Silva, J.A.T.; Shinoyama, H.; Aida, R.; Matsushita, Y.; Raj, S.K.; Chen, F. Chrysanthemum biotechnology: Quo vadis? Crit. Rev. Plant Sci. 2013, 32, 21–52. [Google Scholar] [CrossRef]
- Nakayama, M.; Koshioka, M.; Shibata, M.; Hiradate, S.; Sugie, H.; Yamaguchi, M.A. Identification of cyanidin 3-O-(3′′,6′′-O-dimalonyl-β-glucopyranoside) as a flower pigment of Chrysanthemum (Dendranthema grandiflorum). Biosci. Biotechnol. Biochem. 1997, 61, 1607–1608. [Google Scholar] [CrossRef]
- Sun, W.; Li, C.H.; Wang, L.S.; Dai, S.L. Analysis of anthocyanins and flavones in different-colored flowers of chrysanthemum. Chin. Bull. Bot. 2010, 45, 327–336. [Google Scholar]
- Shan, H.; Chen, S.; Jiang, J.; Chen, F.; Chen, Y.; Gu, C.; Li, P.; Song, A.; Zhu, X.; Gao, H. Heterologous expression of the chrysanthemum R2R3-MYB transcription factor CmMYB2 enhances drought and salinity tolerance, increases hypersensitivity to ABA and delays flowering in Arabidopsis thaliana. Mol. Biotechnol. 2012, 51, 160–173. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sheng, L.; Zhang, H.; Du, X.; An, C.; Xia, X.; Chen, F.; Jiang, J.; Chen, S. CmMYB19 Over-Expression Improves Aphid Tolerance in Chrysanthemum by Promoting Lignin Synthesis. Int. J. Mol. Sci. 2017, 18, 619. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Tang, X.; Huang, H.; Zhang, Y.; Dai, S. Transcriptomic analyses reveal species-specific light-induced anthocyanin biosynthesis in chrysanthemum. BMC Genom. 2015, 16, 202. [Google Scholar] [CrossRef]
- Sun, Y.-G.; Wang, B.; Jin, S.-H.; Qu, X.-X.; Li, Y.-J.; Hou, B.-K. Ectopic Expression of Arabidopsis Glycosyltransferase UGT85A5 Enhances Salt Stress Tolerance in Tobacco. PLoS ONE 2013, 8, e59924. [Google Scholar] [CrossRef]
- He, H.; Ke, H.; Keting, H.; Qiaoyan, X.; Silan, D. Flower colour modification of chrysanthemum by suppression of F3′H and overexpression of the exogenous Senecio cruentus F3′5′H gene. PLoS ONE 2013, 8, e74395. [Google Scholar] [CrossRef] [PubMed]
- Laitinen, R.A.E.; Ainasoja, M.; Broholm, S.K.; Teeri, T.H.; Elomaa, P. Identification of target genes for a MYB-type anthocyanin regulator in Gerbera hybrida. J. Exp. Bot. 2008, 59, 3691–3703. [Google Scholar] [CrossRef] [Green Version]
- Hong, Y.; Bai, X.; Sun, W.; Jia, F. The numerical classification of chrysanthemum flower color phenotype. Acta Horticult. Sin. 2012, 39, 1330–1340. [Google Scholar]
- Ramsay, N.A.; Glover, B.J. MYB–bHLH–WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005, 10, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Ohta, M.; Matsui, K.; Hiratsu, K.; Shinshi, H.; Ohme-Takagi, M. Repression Domains of Class II ERF Transcriptional Repressors Share an Essential Motif for Active Repression. Plant Cell 2001, 13, 1959–1968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paolocci, F.; Robbins, M.P.; Passeri, V.; Hauck, B.; Morris, P.; Rubini, A.; Arcioni, S.; Damiani, F. The strawberry transcription factor FaMYB1 inhibits the biosynthesis of proanthocyanidins in Lotus corniculatus leaves. J. Exp. Bot. 2010, 62, 1189–1200. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K. Negative regulation of defence and stress genes by EAR-motif-containing repressors. Trends Plant Sci. 2006, 11, 109–112. [Google Scholar] [CrossRef]
- Huang, Y.F.; Vialet, S.; Guiraud, J.L.; Torregrosa, L.; Bertrand, Y.; Cheynier, V.; This, P.; Terrier, N. A negative MYB regulator of proanthocyanidin accumulation, identified through expression quantitative locus mapping in the grape berry. New Phytol. 2014, 201, 795–809. [Google Scholar] [CrossRef] [PubMed]
- Sonbol, F.-M.; Fornalè, S.; Capellades, M.; Encina, A.; Touriño, S.; Torres, J.-L.; Rovira, P.; Ruel, K.; Puigdomènech, P.; Rigau, J.; et al. The maize ZmMYB42 represses the phenylpropanoid pathway and affects the cell wall structure, composition and degradability in Arabidopsis thaliana. Plant Mol. Biol. 2009, 70, 283. [Google Scholar] [CrossRef]
- Kagale, S.; Rozwadowski, K. EAR motif-mediated transcriptional repression in plants: An underlying mechanism for epigenetic regulation of gene expression. Epigenetics 2011, 6, 141–146. [Google Scholar] [CrossRef]
- Kagale, S.; Links, M.G.; Rozwadowski, K. Genome-Wide Analysis of Ethylene-Responsive Element Binding Factor-Associated Amphiphilic Repression Motif-Containing Transcriptional Regulators in Arabidopsis. Plant Physiol. 2010, 152, 1109–1134. [Google Scholar] [CrossRef] [Green Version]
- Albert, N.W.; Lewis, D.H.; Zhang, H.; Schwinn, K.E.; Jameson, P.E.; Davies, K.M. Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 2011, 65, 771–784. [Google Scholar] [CrossRef]
- Yoshida, K.; Ma, D.; Constabel, C.P. The MYB182 protein down-regulates proanthocyanidin and anthocyanin biosynthesis in poplar by repressing both structural and regulatory flavonoid genes. Plant Physiol. 2015, 167, 693–710. [Google Scholar] [CrossRef] [PubMed]
- Matsui, K.; Umemura, Y.; Ohme-Takagi, M. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 2008, 55, 954–967. [Google Scholar] [CrossRef] [PubMed]
- Szemenyei, H.; Hannon, M.; Long, J.A. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 2008, 319, 1384–1386. [Google Scholar] [CrossRef]
- Pauwels, L.; Barbero, G.F.; Geerinck, J.; Tilleman, S.; Grunewald, W.; Pérez, A.C.; Chico, J.M.; Bossche, R.V.; Sewell, J.; Gil, E.; et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 2010, 464, 788. [Google Scholar] [CrossRef] [PubMed]
- Long, J.A.; Ohno, C.; Smith, Z.R.; Meyerowitz, E.M. TOPLESS regulates apical embryonic fate in Arabidopsis. Science 2006, 312, 1520–1523. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; He, X.; Poovaiah, C.R.; Wuddineh, W.A.; Ma, J.; Mann, D.G.; Wang, H.; Jackson, L.; Tang, Y.; Stewart, C.N., Jr.; et al. Functional characterization of the switchgrass (Panicum virgatum) R2R3-MYB transcription factor PvMYB4 for improvement of lignocellulosic feedstocks. New Phytol. 2012, 193, 121–136. [Google Scholar] [CrossRef]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Yamagishi, M.; Shimoyamada, Y.; Nakatsuka, T.; Masuda, K. Two R2R3-MYB Genes, Homologs of Petunia AN2, Regulate Anthocyanin Biosyntheses in Flower Tepals, Tepal Spots and Leaves of Asiatic Hybrid Lily. Plant Cell Physiol. 2010, 51, 463–474. [Google Scholar] [CrossRef] [Green Version]
- Schwinn, K.E.; Ngo, H.; Kenel, F.; Brummell, D.A.; Albert, N.W.; McCallum, J.A.; Pither-Joyce, M.; Crowhurst, R.N.; Eady, C.; Davies, K.M. The Onion (Allium cepa L.) R2R3-MYB Gene MYB1 Regulates Anthocyanin Biosynthesis. Front. Plant Sci. 2016, 7, 355. [Google Scholar] [CrossRef]
- Zou, K.; Wang, Y.; Zhao, M.; Zhao, L.; Xu, Z. Cloning and expression analysis of RrMYB113 gene related to anthocyanin biosynthesis in Rosa rugosa. Am. J. Plant Sci. 2018, 9, 701. [Google Scholar] [CrossRef]
- Plunkett, B.; Espley, R.V.; Dare, A.P.; Warren, B.A.W.; Grierson, E.; Cordiner, S.; Turner, J.L.; Allan, A.C.; Albert, N.W.; Davies, K.M.; et al. MYBA from blueberry (Vaccinium sect. Cyanococcus) is a subgroup 6 type R2R3MYB transcription factor that activates anthocyanin production. Front. Plant Sci. 2018, 9, 1300. [Google Scholar] [PubMed]
- Czemmel, S.; Stracke, R.; Weisshaar, B.; Cordon, N.; Harris, N.N.; Walker, A.R.; Robinson, S.P.; Bogs, J. The grapevine R2R3-MYB transcription factor VvMYBF1 regulates flavonol synthesis in developing grape berries. Plant Physiol. 2009, 151, 1513–1530. [Google Scholar] [CrossRef] [PubMed]
- Naing, A.H.; Kim, C.K. Roles of R2R3-MYB transcription factors in transcriptional regulation of anthocyanin biosynthesis in horticultural plants. Plant Mol. Biol. 2018, 98, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Sun, Z.; Wang, C.; Zhang, X.; Tang, Y.; Zhu, X.; Shao, J.; Wu, Y. Changing a conserved amino acid in R2R3-MYB transcription repressors results in their cytoplasmic accumulation and abolishes their repressive activity in Arabidopsis. Plant J. 2015, 84, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.-C.; Chen, Y.-Y.; Tsai, W.-C.; Chen, W.-H.; Chen, H.-H. Three R2R3-MYB transcription factors regulate distinct floral pigmentation patterning in Phalaenopsis spp. Plant Physiol. 2015, 168, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Peng, Q.; Zhao, J.; Owiti, A.; Ren, F.; Liao, L.; Wang, L.; Deng, X.; Jiang, Q.; Han, Y. Multiple R2R3-MYB Transcription Factors Involved in the Regulation of Anthocyanin Accumulation in Peach Flower. Front. Plant Sci. 2016, 7, 1557. [Google Scholar] [CrossRef]
- Xiang, L.; Liu, X.; Li, H.; Yin, X.; Grierson, D.; Li, F.; Chen, K. CmMYB#7, an R3 MYB transcription factor, acts as a negative regulator of anthocyanin biosynthesis in chrysanthemum. J. Exp. Bot. 2019, 70, 3111–3123. [Google Scholar] [PubMed]
- Spelt, C.; Quattrocchio, F.; Mol, J.N.M.; Koes, R. anthocyanin1 of Petunia Encodes a Basic Helix-Loop-Helix Protein That Directly Activates Transcription of Structural Anthocyanin Genes. Plant Cell 2000, 12, 1619–1631. [Google Scholar] [CrossRef]
- Elomaa, P.; Uimari, A.; Mehto, M.; Albert, V.A.; Laitinen, R.A.; Teeri, T.H. Activation of Anthocyanin Biosynthesis in Gerbera hybrida (Asteraceae) Suggests Conserved Protein-Protein and Protein-Promoter Interactions between the Anciently Diverged Monocots and Eudicots1. Plant Physiol. 2003, 133, 1831–1842. [Google Scholar] [CrossRef]
- Zimmermann, I.M.; Heim, M.A.; Weisshaar, B.; Uhrig, J.F. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like bHLH proteins. Plant J. 2004, 40, 22–34. [Google Scholar] [CrossRef]
- Ma, D.; Reichelt, M.; Yoshida, K.; Gershenzon, J.; Constabel, C.P. Two R2R3-MYB proteins are broad repressors of flavonoid and phenylpropanoid metabolism in poplar. Plant J. 2018, 96, 949–965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Primer Name | Primer Sequence (3′–5′) | Purpose |
|---|---|---|
| CmMYB4-F | ATGGTGAGATCACCTTGTTGTG | Gene isolation |
| CmMYB4-R | TCATCTCCATCTACTCGACAAG | Gene isolation |
| CmMYB5-F | ATGACAAAACCTTGTTGTGATT | Gene isolation |
| CmMYB5-R | TTAGGGCACCACTGGTCCACTC | Gene isolation |
| CmMYB6-F | ATGAGACCGAGTAGTAGTACAG | Gene isolation |
| CmMYB6-R | TCATAGTTGGTCCGAATTTAAA | Gene isolation |
| CmMYB7-F | ATGGGAAGAACACCGTGTTGTG | Gene isolation |
| CmMYB7-R | TTAATTCCATTGCCAAAGAAAC | Gene isolation |
| CmMYB4-F1 | CGGGATCCATGGTGAGATCACCTTGTTGTG | Vector construction |
| CmMYB4-R1 | CGGAATTCTCATCTCCATCTACTCGACAAG | Vector construction |
| CmMYB5-F1 | CGGGATCCATGACAAAACCTTGTTGTGATT | Vector construction |
| CmMYB5-R1 | CGGAATTCTTAGGGCACCACTGGTCCACTC | Vector construction |
| CmMYB6-F1 | CGGGATCCATGAGACCGAGTAGTAGTACAG | Vector construction |
| CmMYB6-R1 | CGGAATTCTCATAGTTGGTCCGAATTTAAA | Vector construction |
| CmMYB7-F1 | CGGGATCCATGGGAAGAACACCGTGTTGTG | Vector construction |
| CmMYB7-R1 | CGGAATTCTTAATTCCATTGCCAAAGAAAC | Vector construction |
| Ntα-Tub1-F | ATGAGAGAGTGCATATCGAT | qRT-PCR analysis |
| Ntα-Tub1-R | TTCACTGAAGAAGGTGTTGAA | qRT-PCR analysis |
| NtCHS-F | TTGTTCGAGCTTGTCTCTGC | qRT-PCR analysis |
| NtCHS-R | AGCCCAGGAACATCTTTGAG | qRT-PCR analysis |
| NtCHI-F | GTCAGGCCATTGAAAAGCTC | qRT-PCR analysis |
| NtCHI-R | CTAATCGTCAATGCCCCAAC | qRT-PCR analysis |
| NtF3H-F | CAAGGCATGTGTGGATATGG | qRT-PCR analysis |
| NtF3H-R | TGTGTCGTTTCAGTCCAAGG | qRT-PCR analysis |
| NtF3’H-F | AGGCTCAACACTTCTCGT | qRT-PCR analysis |
| NtF3’H-R | CATCAACTTTGGGCTTCT | qRT-PCR analysis |
| NtDFR-F | TCCCATCATGCGATCAT | qRT-PCR analysis |
| NtDFR-R | ATGGCTTCTTTGTCACGTC | qRT-PCR analysis |
| NtANS-F | TGGCGTTGAAGCTCATACTG | qRT-PCR analysis |
| NtANS-R | GGAATTAGGCACACACTTTGC | qRT-PCR analysis |
| Nt3GT-F | GAGTGCATTGGATGCCTTTT | qRT-PCR analysis |
| Nt3GT-R | CCAGCTCCATTAGGTCCTTG | qRT-PCR analysis |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hong, Y.; Li, M.; Dai, S. Ectopic Expression of Multiple Chrysanthemum (Chrysanthemum × morifolium) R2R3-MYB Transcription Factor Genes Regulates Anthocyanin Accumulation in Tobacco. Genes 2019, 10, 777. https://doi.org/10.3390/genes10100777
Hong Y, Li M, Dai S. Ectopic Expression of Multiple Chrysanthemum (Chrysanthemum × morifolium) R2R3-MYB Transcription Factor Genes Regulates Anthocyanin Accumulation in Tobacco. Genes. 2019; 10(10):777. https://doi.org/10.3390/genes10100777
Chicago/Turabian StyleHong, Yan, Mengling Li, and Silan Dai. 2019. "Ectopic Expression of Multiple Chrysanthemum (Chrysanthemum × morifolium) R2R3-MYB Transcription Factor Genes Regulates Anthocyanin Accumulation in Tobacco" Genes 10, no. 10: 777. https://doi.org/10.3390/genes10100777
APA StyleHong, Y., Li, M., & Dai, S. (2019). Ectopic Expression of Multiple Chrysanthemum (Chrysanthemum × morifolium) R2R3-MYB Transcription Factor Genes Regulates Anthocyanin Accumulation in Tobacco. Genes, 10(10), 777. https://doi.org/10.3390/genes10100777





