The Role of Insulation in Patterning Gene Expression
Abstract
1. Introduction
2. Discovery of Insulators and their Binding Proteins
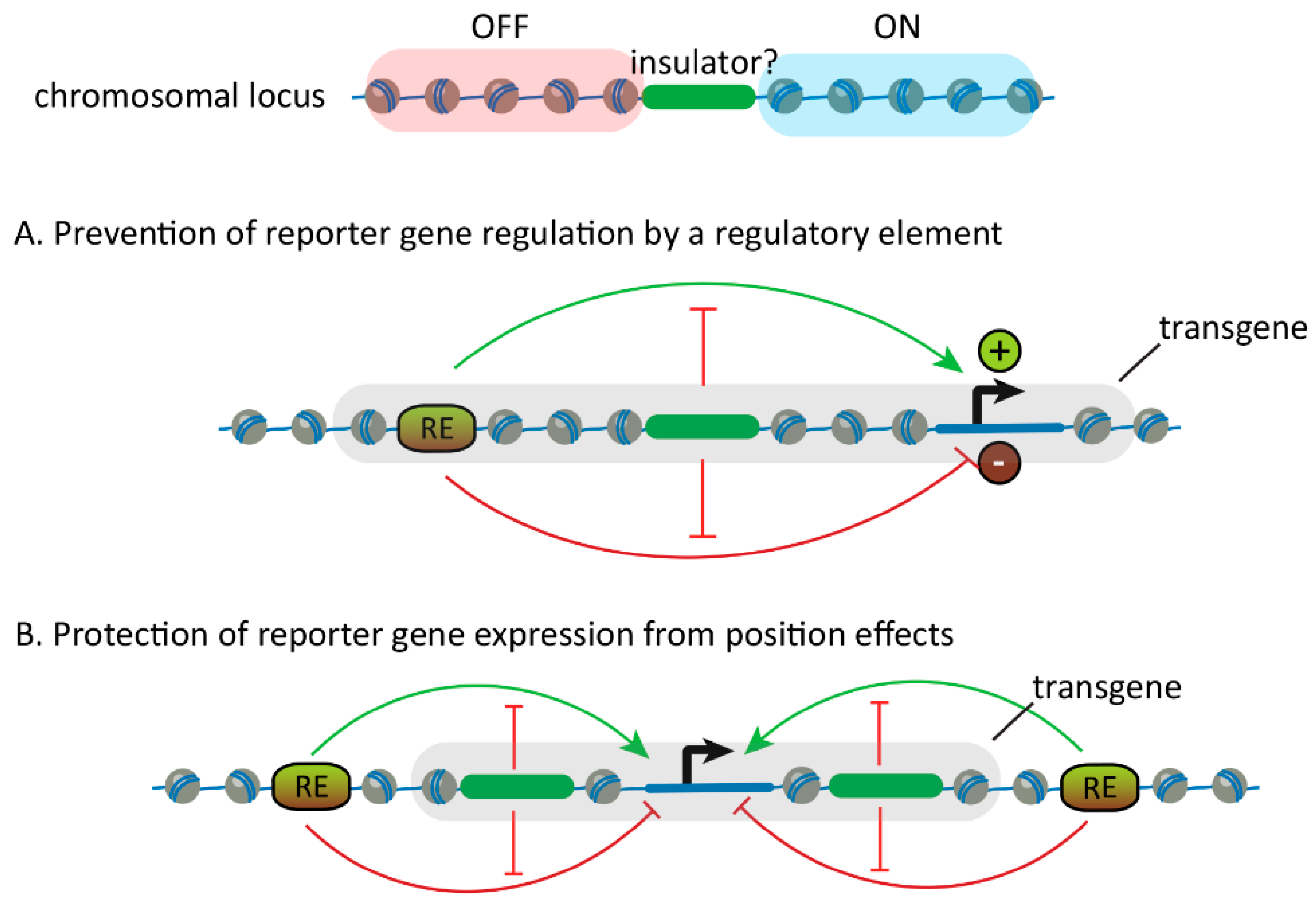
2.1. Discovery of Insulators
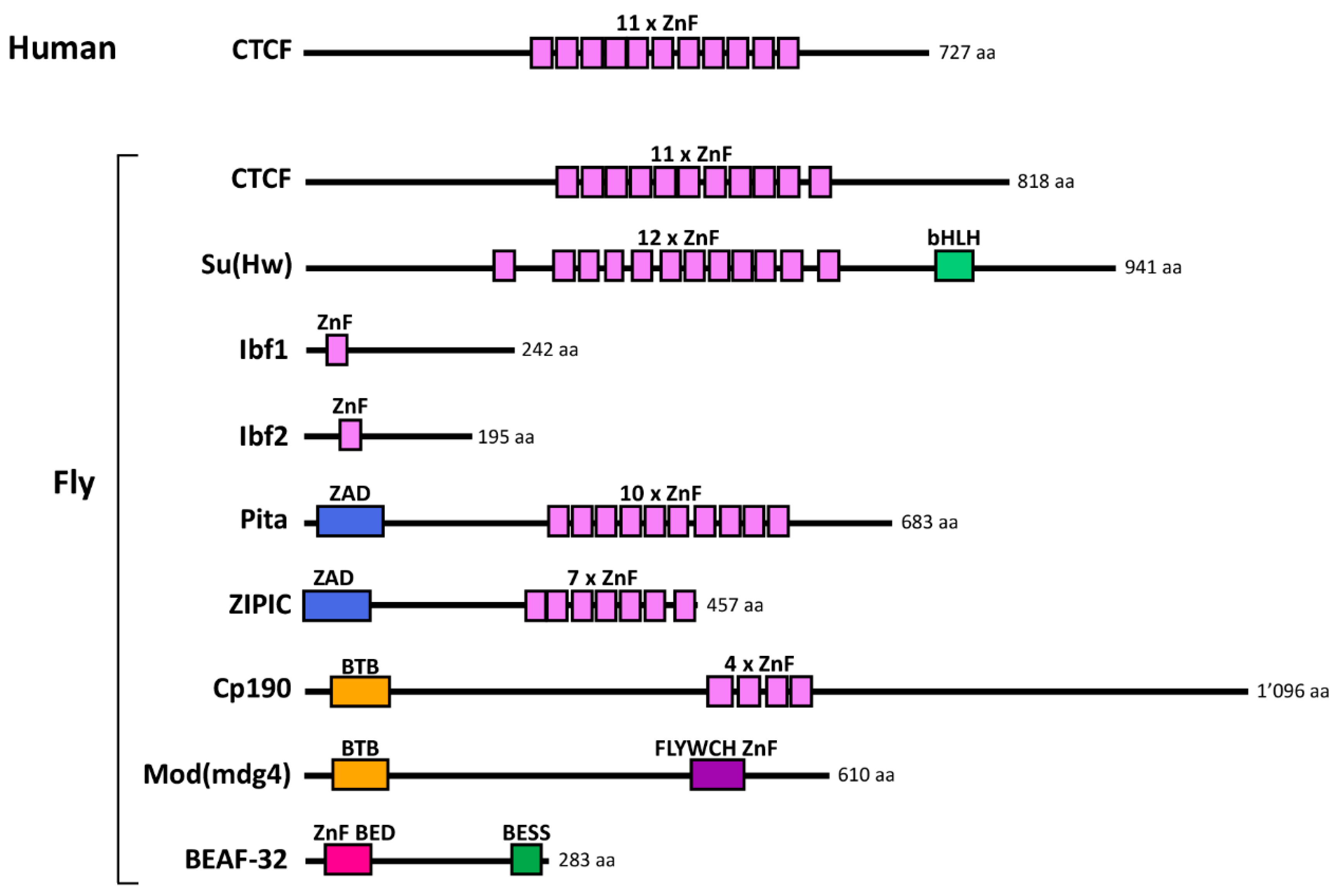
2.2. Discovery of Insulator-Binding Proteins (IBPs)
3. Properties of Insulators and their Binding Proteins
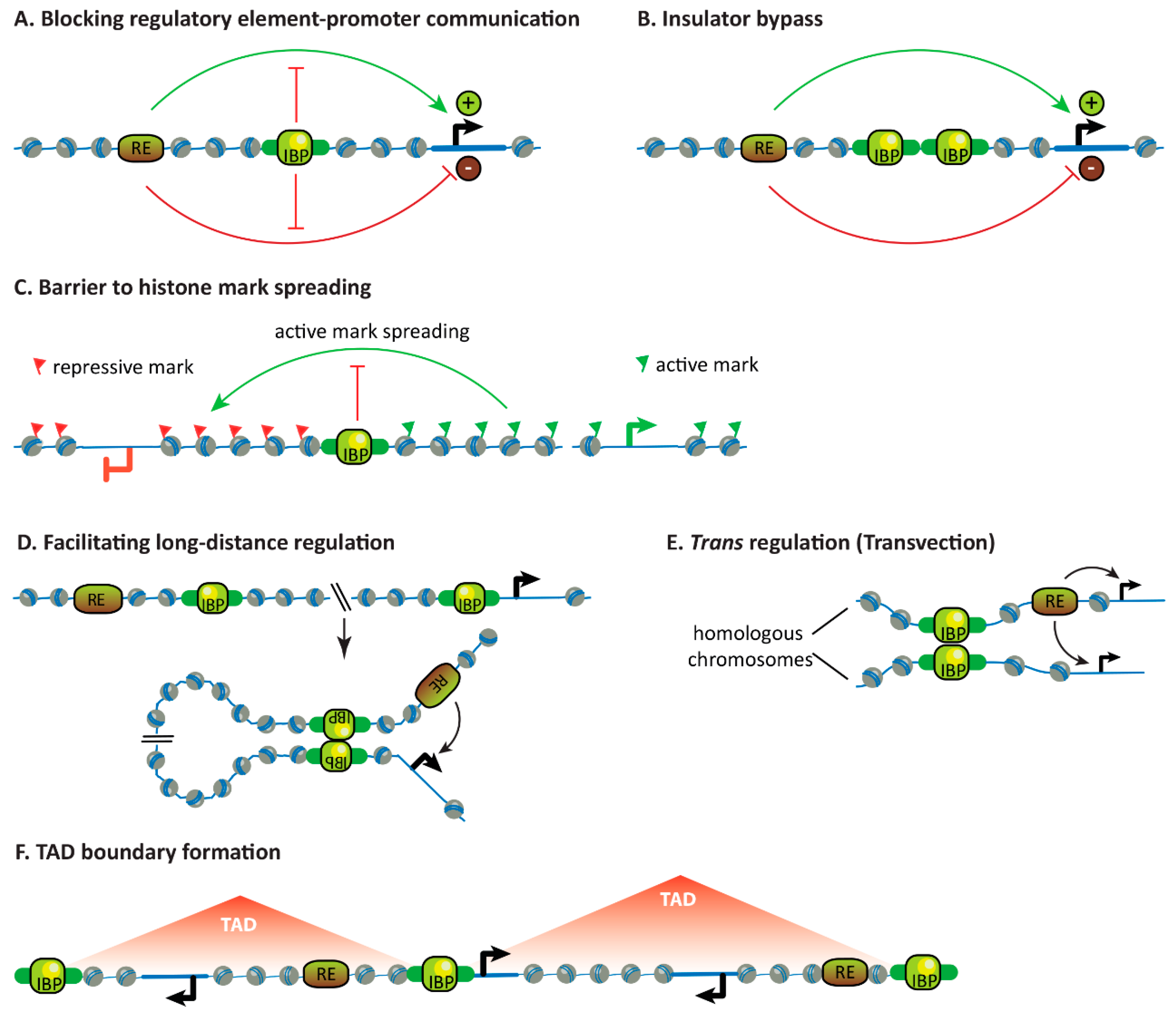
3.1. Blocking the Communication Between Regulatory Elements and Promoters
3.2. Insulator Bypass
3.3. Forming Barriers to Histone Mark Spreading
3.4. Facilitating Long-Distance Gene Regulation
3.5. Facilitating Trans-Regulation
3.6. Influencing Chromosome Topology
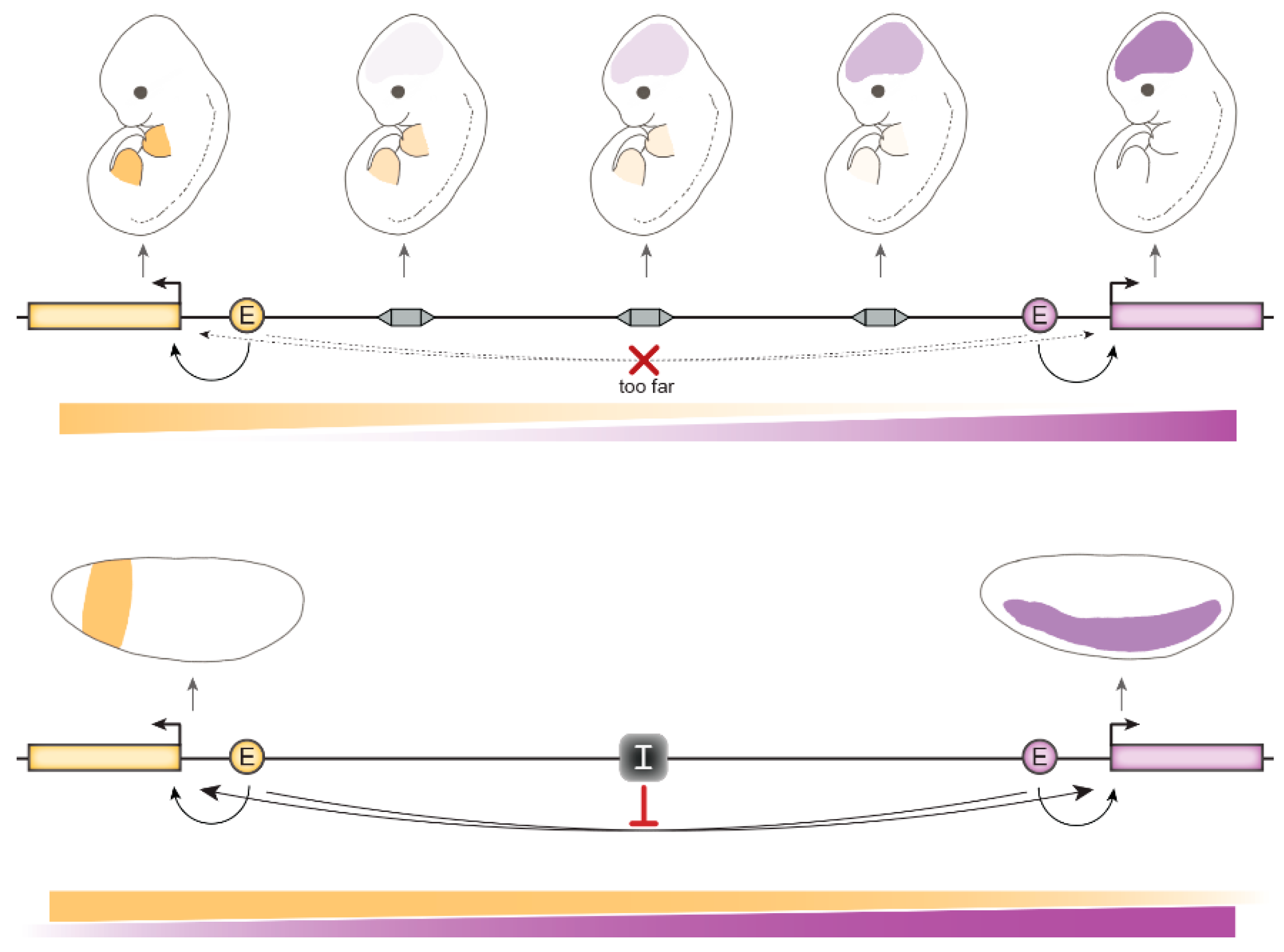
4. Relevance of gene insulation for animal development
4.1. In Mammals
4.1.1. CCCTC-Binding Factor (CTCF) is Essential in Mammalian Cells
4.1.2. CTCF-Mediated Chromosomal Loops Foster Long-Range Gene Regulation
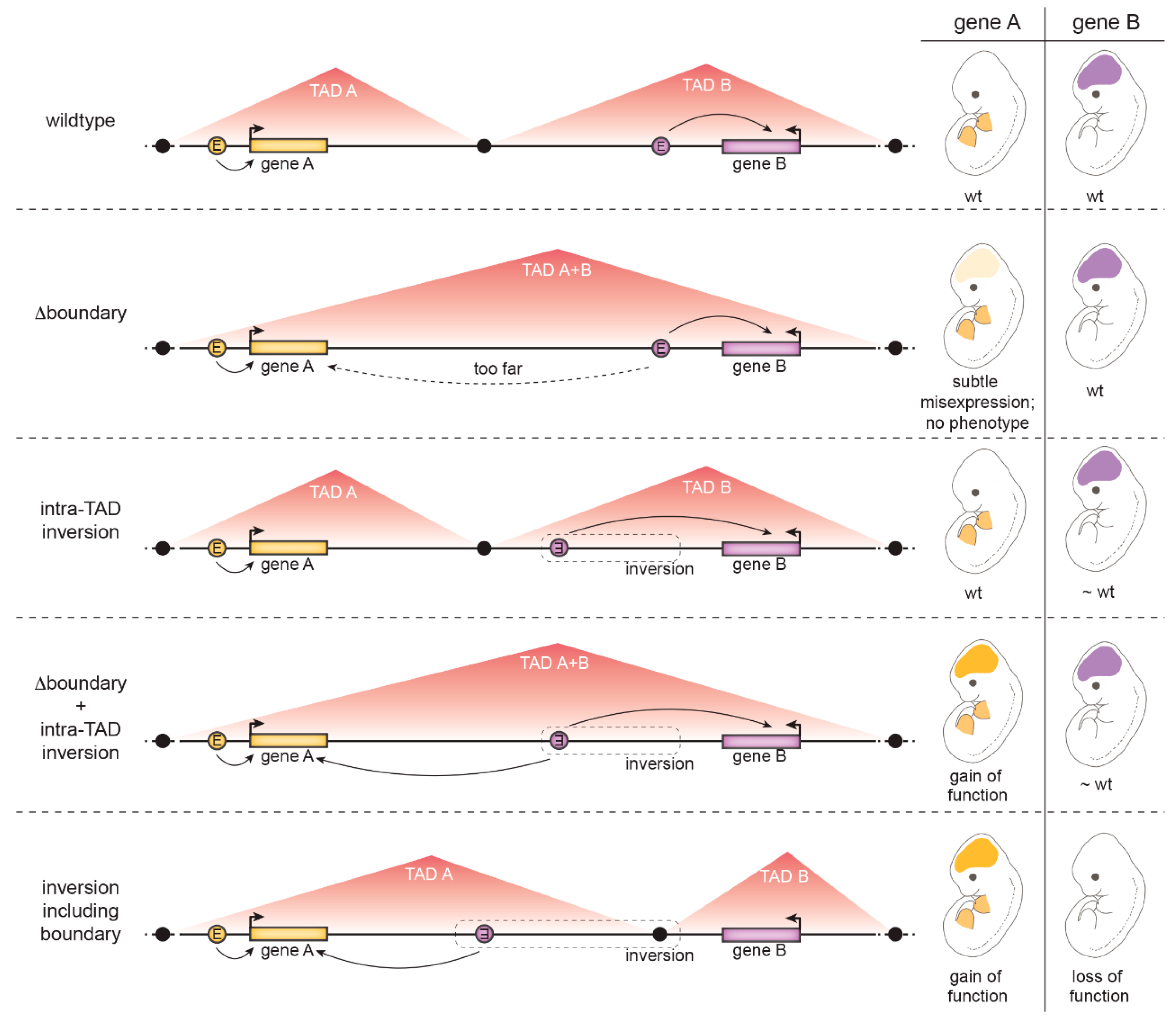
4.1.3. The Role of Insulation at Topologically Associated Domain (TAD) Boundaries
- TAD boundaries are in several cases composed of many clustered CTCF binding sites in convergent orientations facing opposite TAD borders. Several CTCF sites therefore have to be deleted to weaken the boundary. A clear example discussed above is the HoxD boundary in mice: only a ~400 kb deletion including the whole HoxD cluster resulted in TAD boundary loss and fusion of the flanking TADs [111].
- TADs are hierarchical structures composed of nested smaller CTCF-mediated loops [124]. Therefore, enhancers and promoters are not only connected by looping between TAD borders but additionally by intra-TAD loops. For example, at the Kcnj2/Sox9 locus, deletion of CTCF sites at the Kcnj2/Sox9 TAD boundary was not sufficient for the TADs to fuse – all major CTCF sites in the Sox9 TAD had to be additionally removed [118].
- Additional CTCF-independent forces drive genome compartmentalization, such as the segregation of transcriptionally active and silent chromatin or the yet not-well understood property of chromatin domains with specific histone marks to coalesce [69,81]. Thus, topology is not completely abrogated by CTCF manipulation [75].
- Activation of a gene by an ectopic enhancer may depend on its chromatin properties. For example, H3K27me3-decorated genes responded more strongly to an ectopic enhancer placed in proximity by a chromosomal inversion [119].
- The presence of a “decoy promoter” that competes for enhancer activity may mask ectopic activation of a gene [127].
- Even measurable changes in gene transcript levels are not always sufficient to cause an observable effect on gene function [117].
4.2. In Flies
4.2.1. Developmental Roles of Fly Insulators
4.2.2. Developmental Roles of Fly IBPs
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fukaya, T.; Lim, B.; Levine, M. Enhancer Control of Transcriptional Bursting. Cell 2016, 166, 358–368. [Google Scholar] [CrossRef]
- Bartman, C.R.; Hsu, S.C.; Hsiung, C.C.-S.; Raj, A.; Blobel, G.A. Enhancer Regulation of Transcriptional Bursting Parameters Revealed by Forced Chromatin Looping. Mol. Cell 2016, 62, 237–247. [Google Scholar] [CrossRef]
- Kvon, E.Z.; Kazmar, T.; Stampfel, G.; Yáñez-Cuna, J.O.; Pagani, M.; Schernhuber, K.; Dickson, B.J.; Stark, A. Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature 2014, 512, 91–95. [Google Scholar] [CrossRef]
- Cusanovich, D.A.; Hill, A.J.; Aghamirzaie, D.; Daza, R.M.; Pliner, H.A.; Berletch, J.B.; Filippova, G.N.; Huang, X.; Christiansen, L.; DeWitt, W.S.; et al. A Single-Cell Atlas of In Vivo Mammalian Chromatin Accessibility. Cell 2018, 174, 1309–1324.e18. [Google Scholar] [CrossRef] [PubMed]
- Gyurkovics, H.; Gausz, J.; Kummer, J.; Karch, F. A new homeotic mutation in the Drosophila bithorax complex removes a boundary separating two domains of regulation. EMBO J. 1990, 9, 2579–2585. [Google Scholar] [CrossRef] [PubMed]
- Peifer, M.; Bender, W. The anterobithorax and bithorax mutations of the bithorax complex. EMBO J. 1986, 5, 2293–2303. [Google Scholar] [CrossRef] [PubMed]
- Jack, J.; Dorsett, D.; Delotto, Y.; Liu, S. Expression of the cut locus in the Drosophila wing margin is required for cell type specification and is regulated by a distant enhancer. Development 1991, 113, 735–747. [Google Scholar] [PubMed]
- Geyer, P.K.; Corces, V.G. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 1992, 6, 1865–1873. [Google Scholar] [CrossRef]
- Kellum, R.; Schedl, P. A position-effect assay for boundaries of higher order chromosomal domains. Cell 1991, 64, 941–950. [Google Scholar] [CrossRef]
- Kellum, R.; Schedl, P. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol. Cell. Biol. 1992, 12, 2424–2431. [Google Scholar] [CrossRef]
- Chung, J.H.; Whiteley, M.; Felsenfeld, G. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell 1993, 74, 505–514. [Google Scholar] [CrossRef]
- Schwartz, Y.B.; Linder-Basso, D.; Kharchenko, P.V.; Tolstorukov, M.Y.; Kim, M.; Li, H.-B.; Gorchakov, A.A.; Minoda, A.; Shanower, G.; Alekseyenko, A.A.; et al. Nature and function of insulator protein binding sites in the Drosophila genome. Genome Res. 2012, 22, 2188–2198. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Maurano, M.T.; Wang, H.; Qi, H.; Song, C.-Z.; Navas, P.A.; Emery, D.W.; Stamatoyannopoulos, J.A.; Stamatoyannopoulos, G. Genomic discovery of potent chromatin insulators for human gene therapy. Nat. Biotechnol. 2015, 33, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Bell, A.C.; West, A.G.; Felsenfeld, G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 1999, 98, 387–396. [Google Scholar] [CrossRef]
- Raab, J.R.; Chiu, J.; Zhu, J.; Katzman, S.; Kurukuti, S.; Wade, P.A.; Haussler, D.; Kamakaka, R.T. Human tRNA genes function as chromatin insulators. EMBO J. 2012, 31, 330–350. [Google Scholar] [CrossRef] [PubMed]
- Moon, H.; Filippova, G.; Loukinov, D.; Pugacheva, E.; Chen, Q.; Smith, S.T.; Munhall, A.; Grewe, B.; Bartkuhn, M.; Arnold, R.; et al. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep. 2005, 6, 165–170. [Google Scholar] [CrossRef]
- Gómez-Marín, C.; Tena, J.J.; Acemel, R.D.; López-Mayorga, M.; Naranjo, S.; de la Calle-Mustienes, E.; Maeso, I.; Beccari, L.; Aneas, I.; Vielmas, E.; et al. Evolutionary comparison reveals that diverging CTCF sites are signatures of ancestral topological associating domains borders. Proc. Natl. Acad. Sci. USA 2015, 112, 7542–7547. [Google Scholar] [CrossRef]
- Heger, P.; Marin, B.; Bartkuhn, M.; Schierenberg, E.; Wiehe, T. The chromatin insulator CTCF and the emergence of metazoan diversity. Proc. Natl. Acad. Sci. USA 2012, 109, 17507–17512. [Google Scholar] [CrossRef]
- Zhao, K.; Hart, C.M.; Laemmli, U.K. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell 1995, 81, 879–889. [Google Scholar] [CrossRef]
- Sultana, H.; Verma, S.; Mishra, R.K. A BEAF dependent chromatin domain boundary separates myoglianin and eyeless genes of Drosophila melanogaster. Nucleic Acids Res. 2011, 39, 3543–3557. [Google Scholar] [CrossRef]
- Pai, C.-Y.; Lei, E.P.; Ghosh, D.; Corces, V.G. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol. Cell 2004, 16, 737–748. [Google Scholar] [CrossRef] [PubMed]
- Gerasimova, T.I.; Gdula, D.A.; Gerasimov, D.V.; Simonova, O.; Corces, V.G. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell 1995, 82, 587–597. [Google Scholar] [CrossRef]
- Cuartero, S.; Fresán, U.; Reina, O.; Planet, E.; Espinàs, M.L. Ibf1 and Ibf2 are novel CP190-interacting proteins required for insulator function. EMBO J. 2014, 33, 637–647. [Google Scholar] [CrossRef] [PubMed]
- Maksimenko, O.; Bartkuhn, M.; Stakhov, V.; Herold, M.; Zolotarev, N.; Jox, T.; Buxa, M.K.; Kirsch, R.; Bonchuk, A.; Fedotova, A.; et al. Two new insulator proteins, Pita and ZIPIC, target CP190 to chromatin. Genome Res. 2015, 25, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Zolotarev, N.; Maksimenko, O.; Kyrchanova, O.; Sokolinskaya, E.; Osadchiy, I.; Girardot, C.; Bonchuk, A.; Ciglar, L.; Furlong, E.E.M.; Georgiev, P. Opbp is a new architectural/insulator protein required for ribosomal gene expression. Nucleic Acids Res. 2017, 45, 12285–12300. [Google Scholar] [CrossRef]
- Schoborg, T.A.; Labrador, M. The phylogenetic distribution of non-CTCF insulator proteins is limited to insects and reveals that BEAF-32 is Drosophila lineage specific. J. Mol. Evol. 2010, 70, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Heger, P.; George, R.; Wiehe, T. Successive gain of insulator proteins in arthropod evolution. Evolution 2013, 67, 2945–2956. [Google Scholar] [CrossRef] [PubMed]
- Cusanovich, D.A.; Reddington, J.P.; Garfield, D.A.; Daza, R.M.; Aghamirzaie, D.; Marco-Ferreres, R.; Pliner, H.A.; Christiansen, L.; Qiu, X.; Steemers, F.J.; et al. The cis-regulatory dynamics of embryonic development at single-cell resolution. Nature 2018, 555, 538–542. [Google Scholar] [CrossRef]
- Cusanovich, D.A.; Reddington, J.P.; Garfield, D.A.; Daza, R.M.; Aghamirzaie, D.; Marco-Ferreres, R.; Pliner, H.A.; Christiansen, L.; Qiu, X.; Steemers, F.J.; et al. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 2012, 22, 1680–1688. [Google Scholar]
- Sigrist, C.J.; Pirrotta, V. Chromatin insulator elements block the silencing of a target gene by the Drosophila polycomb response element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics 1997, 147, 209–221. [Google Scholar]
- Mallin, D.R.; Myung, J.S.; Patton, J.S.; Geyer, P.K. Polycomb group repression is blocked by the Drosophila suppressor of Hairy-wing [su(Hw)] insulator. Genetics 1998, 148, 331–339. [Google Scholar] [PubMed]
- Cai, H.; Levine, M. Modulation of enhancer-promoter interactions by insulators in the Drosophila embryo. Nature 1995, 376, 533–536. [Google Scholar] [CrossRef] [PubMed]
- Spielmann, M.; Lupiáñez, D.G.; Mundlos, S. Structural variation in the 3D genome. Nat. Rev. Genet. 2018, 19, 453–467. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.N.; Shen, P. Effects of cis arrangement of chromatin insulators on enhancer-blocking activity. Science 2001, 291, 493–495. [Google Scholar] [CrossRef]
- Muravyova, E.; Golovnin, A.; Gracheva, E.; Parshikov, A.; Belenkaya, T.; Pirrotta, V.; Georgiev, P. Loss of insulator activity by paired Su(Hw) chromatin insulators. Science 2001, 291, 495–498. [Google Scholar] [CrossRef] [PubMed]
- Kyrchanova, O.; Toshchakov, S.; Podstreshnaya, Y.; Parshikov, A.; Georgiev, P. Functional interaction between the Fab-7 and Fab-8 boundaries and the upstream promoter region in the Drosophila Abd-B gene. Mol. Cell. Biol. 2008, 28, 4188–4195. [Google Scholar] [CrossRef] [PubMed]
- Narendra, V.; Rocha, P.P.; An, D.; Raviram, R.; Skok, J.A.; Mazzoni, E.O.; Reinberg, D. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science 2015, 347, 1017–1021. [Google Scholar] [CrossRef]
- Bowman, S.K.; Deaton, A.M.; Domingues, H.; Wang, P.I.; Sadreyev, R.I.; Kingston, R.E.; Bender, W. H3K27 modifications define segmental regulatory domains in the Drosophila bithorax complex. eLife 2014, 3, e02833. [Google Scholar] [CrossRef]
- Kahn, T.G.; Schwartz, Y.B.; Dellino, G.I.; Pirrotta, V. Polycomb complexes and the propagation of the methylation mark at the Drosophila Ubx gene. J. Biol. Chem. 2006, 281, 29064–29075. [Google Scholar] [CrossRef]
- Fujioka, M.; Sun, G.; Jaynes, J.B. The Drosophila eve insulator Homie promotes eve expression and protects the adjacent gene from repression by polycomb spreading. PLoS Genet. 2013, 9, e1003883. [Google Scholar] [CrossRef]
- Guelen, L.; Pagie, L.; Brasset, E.; Meuleman, W.; Faza, M.B.; Talhout, W.; Eussen, B.H.; de Klein, A.; Wessels, L.; de Laat, W.; et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 2008, 453, 948–951. [Google Scholar] [CrossRef] [PubMed]
- Cuddapah, S.; Jothi, R.; Schones, D.E.; Roh, T.-Y.; Cui, K.; Zhao, K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009, 19, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Van Bortle, K.; Ramos, E.; Takenaka, N.; Yang, J.; Wahi, J.E.; Corces, V.G. Drosophila CTCF tandemly aligns with other insulator proteins at the borders of H3K27me3 domains. Genome Res. 2012, 22, 2176–2187. [Google Scholar] [CrossRef] [PubMed]
- De Laat, W.; Duboule, D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature 2013, 502, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Furlong, E.E.M.; Levine, M. Developmental enhancers and chromosome topology. Science 2018, 361, 1341–1345. [Google Scholar] [CrossRef] [PubMed]
- Dekker, J.; Marti-Renom, M.A.; Mirny, L.A. Exploring the three-dimensional organization of genomes: Interpreting chromatin interaction data. Nat. Rev. Genet. 2013, 14, 390–403. [Google Scholar] [CrossRef] [PubMed]
- Benabdallah, N.S.; Williamson, I.; Illingworth, R.S.; Kane, L.; Boyle, S.; Sengupta, D.; Grimes, G.R.; Therizols, P.; Bickmore, W.A. Decreased Enhancer-Promoter Proximity Accompanying Enhancer Activation. Mol. Cell 2019. [Google Scholar] [CrossRef]
- Sanyal, A.; Lajoie, B.R.; Jain, G.; Dekker, J. The long-range interaction landscape of gene promoters. Nature 2012, 489, 109–113. [Google Scholar] [CrossRef]
- Ghavi-Helm, Y.; Klein, F.A.; Pakozdi, T.; Ciglar, L.; Noordermeer, D.; Huber, W.; Furlong, E.E.M. Enhancer loops appear stable during development and are associated with paused polymerase. Nature 2014, 512, 96–100. [Google Scholar] [CrossRef]
- Beagrie, R.A.; Scialdone, A.; Schueler, M.; Kraemer, D.C.A.; Chotalia, M.; Xie, S.Q.; Barbieri, M.; de Santiago, I.; Lavitas, L.-M.; Branco, M.R.; et al. Complex multi-enhancer contacts captured by genome architecture mapping. Nature 2017, 543, 519–524. [Google Scholar] [CrossRef]
- Quinodoz, S.A.; Ollikainen, N.; Tabak, B.; Palla, A.; Schmidt, J.M.; Detmar, E.; Lai, M.M.; Shishkin, A.A.; Bhat, P.; Takei, Y.; et al. Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus. Cell 2018, 174, 744–757.e24. [Google Scholar] [CrossRef]
- Fujioka, M.; Wu, X.; Jaynes, J.B. A chromatin insulator mediates transgene homing and very long-range enhancer-promoter communication. Development 2009, 136, 3077–3087. [Google Scholar] [CrossRef] [PubMed]
- Fujioka, M.; Mistry, H.; Schedl, P.; Jaynes, J.B. Determinants of Chromosome Architecture: Insulator Pairing in cis and in trans. PLoS Genet. 2016, 12, e1005889. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Levo, M.; Barinov, L.; Fujioka, M.; Jaynes, J.B.; Gregor, T. Dynamic interplay between enhancer-promoter topology and gene activity. Nat. Genet. 2018, 50, 1–1303. [Google Scholar] [CrossRef] [PubMed]
- Comet, I.; Savitskaya, E.; Schuettengruber, B.; Nègre, N.; Lavrov, S.; Parshikov, A.; Juge, F.; Gracheva, E.; Georgiev, P.; Cavalli, G. PRE-mediated bypass of two Su(Hw) insulators targets PcG proteins to a downstream promoter. Dev. Cell 2006, 11, 117–124. [Google Scholar] [CrossRef]
- Kyrchanova, O.; Chetverina, D.; Maksimenko, O.; Kullyev, A.; Georgiev, P. Orientation-dependent interaction between Drosophila insulators is a property of this class of regulatory elements. Nucleic Acids Res. 2008, 36, 7019–7028. [Google Scholar] [CrossRef]
- Comet, I.; Schuettengruber, B.; Sexton, T.; Cavalli, G. A chromatin insulator driving three-dimensional Polycomb response element (PRE) contacts and Polycomb association with the chromatin fiber. Proc. Natl. Acad. Sci. USA 2011, 108, 2294–2299. [Google Scholar] [CrossRef] [PubMed]
- Lewis, E.B. The theory and application of a new method of detecting chromosomal rearrangements in Drosophila melanogaster. Am. Soc. Nat. 1954, 88, 225–239. [Google Scholar] [CrossRef]
- Geyer, P.K.; Green, M.M.; Corces, V.G. Tissue-specific transcriptional enhancers may act in trans on the gene located in the homologous chromosome: The molecular basis of transvection in Drosophila. EMBO J. 1990, 9, 2247–2256. [Google Scholar] [CrossRef]
- Hiraoka, Y.; Dernburg, A.F.; Parmelee, S.J.; Rykowski, M.C.; Agard, D.A.; Sedat, J.W. The onset of homologous chromosome pairing during Drosophila melanogaster embryogenesis. J. Cell Biol. 1993, 120, 591–600. [Google Scholar] [CrossRef]
- Kravchenko, E.; Savitskaya, E.; Kravchuk, O.; Parshikov, A.; Georgiev, P.; Savitsky, M. Pairing between gypsy insulators facilitates the enhancer action in trans throughout the Drosophila genome. Mol. Cell. Biol. 2005, 25, 9283–9291. [Google Scholar] [CrossRef]
- Lim, B.; Heist, T.; Levine, M.; Fukaya, T. Visualization of Transvection in Living Drosophila Embryos. Mol. Cell 2018, 70, 287–296.e6. [Google Scholar] [CrossRef] [PubMed]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012, 485, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Nora, E.P.; Lajoie, B.R.; Schulz, E.G.; Giorgetti, L.; Okamoto, I.; Servant, N.; Piolot, T.; van Berkum, N.L.; Meisig, J.; Sedat, J.; et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 2012, 485, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Sexton, T.; Yaffe, E.; Kenigsberg, E.; Bantignies, F.; Leblanc, B.; Hoichman, M.; Parrinello, H.; Tanay, A.; Cavalli, G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 2012, 148, 458–472. [Google Scholar] [CrossRef]
- Sanborn, A.L.; Rao, S.S.P.; Huang, S.-C.; Durand, N.C.; Huntley, M.H.; Jewett, A.I.; Bochkov, I.D.; Chinnappan, D.; Cutkosky, A.; Li, J.; et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. USA 2015, 112, E6456–E6465. [Google Scholar] [CrossRef] [PubMed]
- Fudenberg, G.; Imakaev, M.; Lu, C.; Goloborodko, A.; Abdennur, N.; Mirny, L.A. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 2016, 15, 2038–2049. [Google Scholar] [CrossRef]
- Haarhuis, J.H.I.; van der Weide, R.H.; Blomen, V.A.; Yáñez-Cuna, J.O.; Amendola, M.; van Ruiten, M.S.; Krijger, P.H.L.; Teunissen, H.; Medema, R.H.; van Steensel, B.; et al. The Cohesin Release Factor WAPL Restricts Chromatin Loop Extension. Cell 2017, 169, 693–707.e14. [Google Scholar] [CrossRef]
- Rao, S.S.P.; Huntley, M.H.; Durand, N.C.; Stamenova, E.K.; Bochkov, I.D.; Robinson, J.T.; Sanborn, A.L.; Machol, I.; Omer, A.D.; Lander, E.S.; et al. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell 2014, 159, 1665–1680. [Google Scholar] [CrossRef]
- Tang, Z.; Luo, O.J.; Li, X.; Zheng, M.; Zhu, J.J.; Szalaj, P.; Trzaskoma, P.; Magalska, A.; Wlodarczyk, J.; Ruszczycki, B.; et al. CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell 2015, 163, 1611–1627. [Google Scholar] [CrossRef]
- Flavahan, W.A.; Drier, Y.; Liau, B.B.; Gillespie, S.M.; Venteicher, A.S.; Stemmer-Rachamimov, A.O.; Suvà, M.L.; Bernstein, B.E. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 2016, 529, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Hanssen, L.L.P.; Kassouf, M.T.; Oudelaar, A.M.; Biggs, D.; Preece, C.; Downes, D.J.; Gosden, M.; Sharpe, J.A.; Sloane-Stanley, J.A.; Hughes, J.R.; et al. Tissue-specific CTCF-cohesin-mediated chromatin architecture delimits enhancer interactions and function in vivo. Nat. Cell Biol. 2017, 13, 74. [Google Scholar] [CrossRef] [PubMed]
- De Wit, E.; Vos, E.S.M.; Holwerda, S.J.B.; Valdes-Quezada, C.; Verstegen, M.J.A.M.; Teunissen, H.; Splinter, E.; Wijchers, P.J.; Krijger, P.H.L.; de Laat, W. CTCF Binding Polarity Determines Chromatin Looping. Mol. Cell 2015, 60, 676–684. [Google Scholar] [CrossRef] [PubMed]
- Redolfi, J.; Zhan, Y.; Valdes-Quezada, C.; Kryzhanovska, M.; Guerreiro, I.; Iesmantavicius, V.; Pollex, T.; Grand, R.S.; Mulugeta, E.; Kind, J.; et al. DamC reveals principles of chromatin folding in vivo without crosslinking and ligation. Nat. Struct. Mol. Biol. 2019, 26, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Nora, E.P.; Goloborodko, A.; Valton, A.-L.; Gibcus, J.H.; Uebersohn, A.; Abdennur, N.; Dekker, J.; Mirny, L.A.; Bruneau, B.G. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell 2017, 169, 930–944.e22. [Google Scholar] [CrossRef] [PubMed]
- Schwarzer, W.; Abdennur, N.; Goloborodko, A.; Pekowska, A.; Fudenberg, G.; Loe-Mie, Y.; Fonseca, N.A.; Huber, W.; Haering, C.H.; Mirny, L.; et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature 2017, 551, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.S.P.; Huang, S.-C.; Hilaire, B.G.S.; Engreitz, J.M.; Perez, E.M.; Kieffer-Kwon, K.-R.; Sanborn, A.L.; Johnstone, S.E.; Bascom, G.D.; Bochkov, I.D.; et al. Cohesin Loss Eliminates All Loop Domains. Cell 2017, 171, 305–309.e24. [Google Scholar] [CrossRef]
- Wutz, G.; Várnai, C.; Nagasaka, K.; Cisneros, D.A.; Stocsits, R.R.; Tang, W.; Schoenfelder, S.; Jessberger, G.; Muhar, M.; Hossain, M.J.; et al. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 2017, 36, 3573–3599. [Google Scholar] [CrossRef]
- Phillips-Cremins, J.E.; Sauria, M.E.G.; Sanyal, A.; Gerasimova, T.I.; Lajoie, B.R.; Bell, J.S.K.; Ong, C.-T.; Hookway, T.A.; Guo, C.; Sun, Y.; et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 2013, 153, 1281–1295. [Google Scholar] [CrossRef]
- Shen, Y.; Yue, F.; McCleary, D.F.; Ye, Z.; Edsall, L.; Kuan, S.; Wagner, U.; Dixon, J.; Lee, L.; Lobanenkov, V.V.; et al. A map of the cis-regulatory sequences in the mouse genome. Nature 2012, 488, 116–120. [Google Scholar] [CrossRef]
- Rowley, M.J.; Nichols, M.H.; Lyu, X.; Ando-Kuri, M.; Rivera, I.S.M.; Hermetz, K.; Wang, P.; Ruan, Y.; Corces, V.G. Evolutionarily Conserved Principles Predict 3D Chromatin Organization. Mol. Cell 2017, 67, 837–852.e7. [Google Scholar] [CrossRef] [PubMed]
- Rowley, M.J.; Lyu, X.; Rana, V.; Ando-Kuri, M.; Karns, R.; Bosco, G.; Corces, V.G. Condensin II Counteracts Cohesin and RNA Polymerase II in the Establishment of 3D Chromatin Organization. Cell Rep. 2019, 26, 2890–2903.e3. [Google Scholar] [CrossRef] [PubMed]
- Blanton, J.; Gaszner, M.; Schedl, P. Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev. 2003, 17, 664–675. [Google Scholar] [CrossRef] [PubMed]
- Byrd, K.; Corces, V.G. Visualization of chromatin domains created by the gypsy insulator of Drosophila. J. Cell Biol. 2003, 162, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, F.; Bhardwaj, V.; Arrigoni, L.; Lam, K.C.; Grüning, B.A.; Villaveces, J.; Habermann, B.; Akhtar, A.; Manke, T. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat. Commun. 2018, 9, 189. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Sun, Q.; Czajkowsky, D.M.; Shao, Z. Sub-kb Hi-C in D. melanogaster reveals conserved characteristics of TADs between insect and mammalian cells. Nat. Commun. 2018, 9, 188. [Google Scholar] [CrossRef]
- Eagen, K.P.; Aiden, E.L.; Kornberg, R.D. Polycomb-mediated chromatin loops revealed by a subkilobase-resolution chromatin interaction map. Proc. Natl. Acad. Sci. USA 2017, 114, 8764–8769. [Google Scholar] [CrossRef]
- Stadler, M.R.; Haines, J.E.; Eisen, M.B. Convergence of topological domain boundaries, insulators, and polytene interbands revealed by high-resolution mapping of chromatin contacts in the early Drosophila melanogasterembryo. eLife 2017, 6, e29550. [Google Scholar] [CrossRef]
- Cubeñas-Potts, C.; Rowley, M.J.; Lyu, X.; Li, G.; Lei, E.P.; Corces, V.G. Different enhancer classes in Drosophila bind distinct architectural proteins and mediate unique chromatin interactions and 3D architecture. Nucleic Acids Res. 2017, 45, 1714–1730. [Google Scholar] [CrossRef]
- Ulianov, S.V.; Khrameeva, E.E.; Gavrilov, A.A.; Flyamer, I.M.; Kos, P.; Mikhaleva, E.A.; Penin, A.A.; Logacheva, M.D.; Imakaev, M.V.; Chertovich, A.; et al. Active chromatin and transcription play a key role in chromosome partitioning into topologically associating domains. Genome Res. 2016, 26, 70–84. [Google Scholar] [CrossRef]
- El-Sharnouby, S.; Fischer, B.; Magbanua, J.P.; Umans, B.; Flower, R.; Choo, S.W.; Russell, S.; White, R. Regions of very low H3K27me3 partition the Drosophila genome into topological domains. PLoS ONE 2017, 12, e0172725. [Google Scholar] [CrossRef] [PubMed]
- Chathoth, K.T.; Zabet, N.R. Chromatin architecture reorganisation during neuronal cell differentiation in Drosophila genome. Genome Res. 2019, 29, 613–625. [Google Scholar] [CrossRef] [PubMed]
- Hug, C.B.; Grimaldi, A.G.; Kruse, K.; Vaquerizas, J.M. Chromatin Architecture Emerges during Zygotic Genome Activation Independent of Transcription. Cell 2017, 169, 216–228.e19. [Google Scholar] [CrossRef] [PubMed]
- Ogiyama, Y.; Schuettengruber, B.; Papadopoulos, G.L.; Chang, J.-M.; Cavalli, G. Polycomb-Dependent Chromatin Looping Contributes to Gene Silencing during Drosophila Development. Mol. Cell 2018, 71, 73–88.e5. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Lyu, X.; Hou, C.; Takenaka, N.; Nguyen, H.Q.; Ong, C.-T.; Cubeñas-Potts, C.; Hu, M.; Lei, E.P.; Bosco, G.; et al. Widespread rearrangement of 3D chromatin organization underlies polycomb-mediated stress-induced silencing. Mol. Cell 2015, 58, 216–231. [Google Scholar] [CrossRef] [PubMed]
- Mateo, L.J.; Murphy, S.E.; Hafner, A.; Cinquini, I.S.; Walker, C.A.; Boettiger, A.N. Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature 2019, 167, 1–54. [Google Scholar] [CrossRef]
- Sleutels, F.; Soochit, W.; Bartkuhn, M.; Heath, H.; Dienstbach, S.; Bergmaier, P.; Franke, V.; Rosa-Garrido, M.; van de Nobelen, S.; Caesar, L.; et al. The male germ cell gene regulator CTCFL is functionally different from CTCF and binds CTCF-like consensus sites in a nucleosome composition-dependent manner. Epigenetics Chromatin 2012, 5, 8. [Google Scholar] [CrossRef]
- Soshnikova, N.; Montavon, T.; Leleu, M.; Galjart, N.; Duboule, D. Functional analysis of CTCF during mammalian limb development. Dev. Cell 2010, 19, 819–830. [Google Scholar] [CrossRef]
- Watson, L.A.; Wang, X.; Elbert, A.; Kernohan, K.D.; Galjart, N.; Bérubé, N.G. Dual effect of CTCF loss on neuroprogenitor differentiation and survival. J. Neurosci. 2014, 34, 2860–2870. [Google Scholar] [CrossRef]
- Moore, J.M.; Rabaia, N.A.; Smith, L.E.; Fagerlie, S.; Gurley, K.; Loukinov, D.; Disteche, C.M.; Collins, S.J.; Kemp, C.J.; Lobanenkov, V.V.; et al. Loss of maternal CTCF is associated with peri-implantation lethality of Ctcf null embryos. PLoS ONE 2012, 7, e34915. [Google Scholar] [CrossRef]
- Heath, H.; Ribeiro de Almeida, C.; Sleutels, F.; Dingjan, G.; van de Nobelen, S.; Jonkers, I.; Ling, K.-W.; Gribnau, J.; Renkawitz, R.; Grosveld, F.; et al. CTCF regulates cell cycle progression of alphabeta T cells in the thymus. EMBO J. 2008, 27, 2839–2850. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro de Almeida, C.; Stadhouders, R.; de Bruijn, M.J.W.; Bergen, I.M.; Thongjuea, S.; Lenhard, B.; van Ijcken, W.; Grosveld, F.; Galjart, N.; Soler, E.; et al. The DNA-binding protein CTCF limits proximal Vκ recombination and restricts κ enhancer interactions to the immunoglobulin κ light chain locus. Immunity 2011, 35, 501–513. [Google Scholar] [CrossRef] [PubMed]
- Wendt, K.S.; Yoshida, K.; Itoh, T.; Bando, M.; Koch, B.; Schirghuber, E.; Tsutsumi, S.; Nagae, G.; Ishihara, K.; Mishiro, T.; et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 2008, 451, 796–801. [Google Scholar] [CrossRef]
- Engel, N.; Thorvaldsen, J.L.; Bartolomei, M.S. CTCF binding sites promote transcription initiation and prevent DNA methylation on the maternal allele at the imprinted H19/Igf2 locus. Hum. Mol. Genet. 2006, 15, 2945–2954. [Google Scholar] [CrossRef] [PubMed]
- Narendra, V.; Bulajić, M.; Dekker, J.; Mazzoni, E.O.; Reinberg, D. CTCF-mediated topological boundaries during development foster appropriate gene regulation. Genes Dev. 2016, 30, 2657–2662. [Google Scholar] [CrossRef] [PubMed]
- Dowen, J.M.; Fan, Z.P.; Hnisz, D.; Ren, G.; Abraham, B.J.; Zhang, L.N.; Weintraub, A.S.; Schuijers, J.; Lee, T.I.; Zhao, K.; et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell 2014, 159, 374–387. [Google Scholar] [CrossRef]
- Ji, X.; Dadon, D.B.; Powell, B.E.; Fan, Z.P.; Borges-Rivera, D.; Shachar, S.; Weintraub, A.S.; Hnisz, D.; Pegoraro, G.; Lee, T.I.; et al. 3D Chromosome Regulatory Landscape of Human Pluripotent Cells. Cell Stem Cell 2016, 18, 262–275. [Google Scholar] [CrossRef]
- Symmons, O.; Uslu, V.V.; Tsujimura, T.; Ruf, S.; Nassari, S.; Schwarzer, W.; Ettwiller, L.; Spitz, F. Functional and topological characteristics of mammalian regulatory domains. Genome Res. 2014, 24, 390–400. [Google Scholar] [CrossRef]
- Montavon, T.; Soshnikova, N.; Mascrez, B.; Joye, E.; Thevenet, L.; Splinter, E.; de Laat, W.; Spitz, F.; Duboule, D. A regulatory archipelago controls Hox genes transcription in digits. Cell 2011, 147, 1132–1145. [Google Scholar] [CrossRef]
- Andrey, G.; Montavon, T.; Mascrez, B.; Gonzalez, F.; Noordermeer, D.; Leleu, M.; Trono, D.; Spitz, F.; Duboule, D. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science 2013, 340, 1234167. [Google Scholar] [CrossRef]
- Rodríguez-Carballo, E.; Lopez-Delisle, L.; Zhan, Y.; Fabre, P.J.; Beccari, L.; El-Idrissi, I.; Huynh, T.H.N.; Ozadam, H.; Dekker, J.; Duboule, D. The HoxD cluster is a dynamic and resilient TAD boundary controlling the segregation of antagonistic regulatory landscapes. Genes Dev. 2017, 31, 2264–2281. [Google Scholar] [CrossRef] [PubMed]
- Symmons, O.; Pan, L.; Remeseiro, S.; Aktas, T.; Klein, F.; Huber, W.; Spitz, F. The Shh Topological Domain Facilitates the Action of Remote Enhancers by Reducing the Effects of Genomic Distances. Dev. Cell 2016, 39, 529–543. [Google Scholar] [CrossRef] [PubMed]
- Lettice, L.A.; Heaney, S.J.H.; Purdie, L.A.; Li, L.; de Beer, P.; Oostra, B.A.; Goode, D.; Elgar, G.; Hill, R.E.; de Graaff, E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 2003, 12, 1725–1735. [Google Scholar] [CrossRef] [PubMed]
- Lonfat, N.; Montavon, T.; Darbellay, F.; Gitto, S.; Duboule, D. Convergent evolution of complex regulatory landscapes and pleiotropy at Hox loci. Science 2014, 346, 1004–1006. [Google Scholar] [CrossRef] [PubMed]
- Williamson, I.; Lettice, L.A.; Hill, R.E.; Bickmore, W.A. Shh and ZRS enhancer colocalisation is specific to the zone of polarising activity. Development 2016, 143, 2994–3001. [Google Scholar] [CrossRef] [PubMed]
- Williamson, I.; Kane, L.; Devenney, P.S.; Flyamer, I.M.; Anderson, E.; Kilanowski, F.; Hill, R.E.; Bickmore, W.A.; Lettice, L.A. Developmentally regulated Shh expression is robust to TAD perturbations. Development 2019. [Google Scholar] [CrossRef] [PubMed]
- Paliou, C.; Guckelberger, P.; Schöpflin, R.; Heinrich, V.; Esposito, A.; Chiariello, A.M.; Bianco, S.; Annunziatella, C.; Helmuth, J.; Haas, S.; et al. Preformed chromatin topology assists transcriptional robustness of Shh during limb development. Proc. Natl. Acad. Sci. USA 2019, 116, 12390–12399. [Google Scholar] [CrossRef]
- Despang, A.; Schöpflin, R.; Franke, M.; Ali, S.; Jerković, I.; Paliou, C.; Chan, W.-L.; Timmermann, B.; Wittler, L.; Vingron, M.; et al. Functional dissection of the Sox9-Kcnj2 locus identifies nonessential and instructive roles of TAD architecture. Nat. Genet. 2019, 51, 1263–1271. [Google Scholar] [CrossRef]
- Kraft, K.; Magg, A.; Heinrich, V.; Riemenschneider, C.; Schöpflin, R.; Markowski, J.; Ibrahim, D.M.; Acuna-Hidalgo, R.; Despang, A.; Andrey, G.; et al. Serial genomic inversions induce tissue-specific architectural stripes, gene misexpression and congenital malformations. Nat. Cell Biol. 2019, 21, 305–310. [Google Scholar] [CrossRef]
- Kokubu, C.; Horie, K.; Abe, K.; Ikeda, R.; Mizuno, S.; Uno, Y.; Ogiwara, S.; Ohtsuka, M.; Isotani, A.; Okabe, M.; et al. A transposon-based chromosomal engineering method to survey a large cis-regulatory landscape in mice. Nat. Genet. 2009, 41, 946–952. [Google Scholar] [CrossRef]
- Lupiáñez, D.G.; Kraft, K.; Heinrich, V.; Krawitz, P.; Brancati, F.; Klopocki, E.; Horn, D.; Kayserili, H.; Opitz, J.M.; Laxova, R.; et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 2015, 161, 1012–1025. [Google Scholar] [CrossRef] [PubMed]
- Hnisz, D.; Weintraub, A.S.; Day, D.S.; Valton, A.-L.; Bak, R.O.; Li, C.H.; Goldmann, J.; Lajoie, B.R.; Fan, Z.P.; Sigova, A.A.; et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 2016, 351, 1454–1458. [Google Scholar] [CrossRef]
- Franke, M.; Ibrahim, D.M.; Andrey, G.; Schwarzer, W.; Heinrich, V.; Schöpflin, R.; Kraft, K.; Kempfer, R.; Jerković, I.; Chan, W.-L.; et al. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature 2016, 538, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Mariani, L.; Barozzi, I.; Schulz, E.G.; Blüthgen, N.; Stadler, M.; Tiana, G.; Giorgetti, L. Reciprocal insulation analysis of Hi-C data shows that TADs represent a functionally but not structurally privileged scale in the hierarchical folding of chromosomes. Genome Res. 2017, 27, 479–490. [Google Scholar] [CrossRef]
- Ohtsuki, S.; Levine, M.; Cai, H.N. Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes Dev. 1998, 12, 547–556. [Google Scholar] [CrossRef] [PubMed]
- Haberle, V.; Arnold, C.D.; Pagani, M.; Rath, M.; Schernhuber, K.; Stark, A. Transcriptional cofactors display specificity for distinct types of core promoters. Nature 2019, 32, 801. [Google Scholar] [CrossRef]
- Kragesteen, B.K.; Spielmann, M.; Paliou, C.; Heinrich, V.; Schöpflin, R.; Esposito, A.; Annunziatella, C.; Bianco, S.; Chiariello, A.M.; Jerković, I.; et al. Dynamic 3D chromatin architecture contributes to enhancer specificity and limb morphogenesis. Nat. Genet. 2018, 50, 1463–1473. [Google Scholar] [CrossRef] [PubMed]
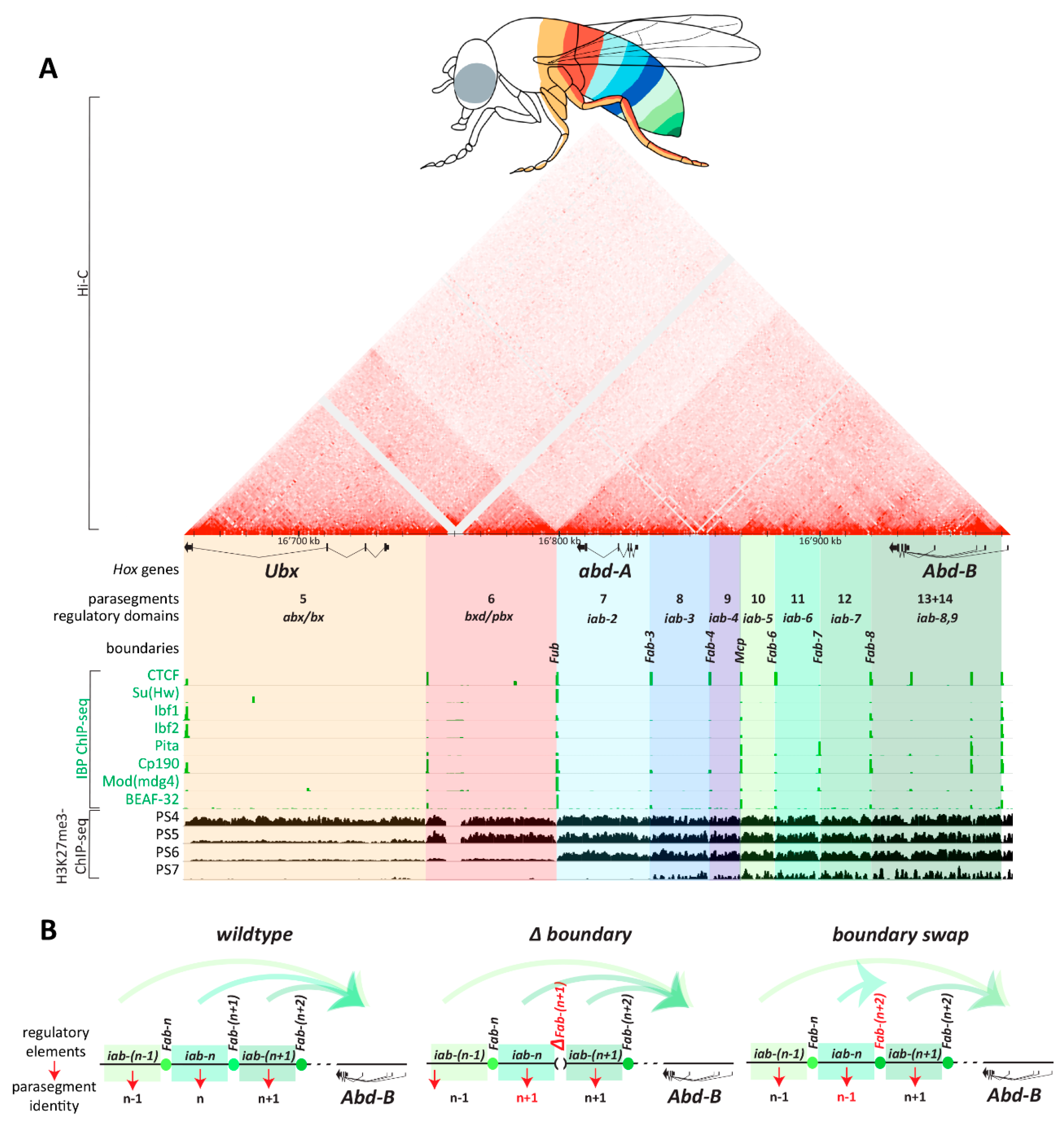
- Maeda, R.K.; Karch, F. The open for business model of the bithorax complex in Drosophila. Chromosoma 2015, 124, 293–307. [Google Scholar] [CrossRef]
- Mihaly, J.; Hogga, I.; Gausz, J.; Gyurkovics, H.; Karch, F. In situ dissection of the Fab-7 region of the bithorax complex into a chromatin domain boundary and a Polycomb-response element. Development 1997, 124, 1809–1820. [Google Scholar]
- Kyrchanova, O.; Mogila, V.; Wolle, D.; Deshpande, G.; Parshikov, A.; Cléard, F.; Karch, F.; Schedl, P.; Georgiev, P. Functional Dissection of the Blocking and Bypass Activities of the Fab-8 Boundary in the Drosophila Bithorax Complex. PLoS Genet. 2016, 12, e1006188. [Google Scholar] [CrossRef]
- Kyrchanova, O.; Zolotarev, N.; Mogila, V.; Maksimenko, O.; Schedl, P.; Georgiev, P. Architectural protein Pita cooperates with dCTCF in organization of functional boundaries in Bithorax complex. Development 2017, 144, 2663–2672. [Google Scholar] [CrossRef] [PubMed]
- Hogga, I.; Mihaly, J.; Barges, S.; Karch, F. Replacement of Fab-7 by the gypsy or scs insulator disrupts long-distance regulatory interactions in the Abd-B gene of the bithorax complex. Mol. Cell 2001, 8, 1145–1151. [Google Scholar] [CrossRef]
- Iampietro, C.; Cléard, F.; Gyurkovics, H.; Maeda, R.K.; Karch, F. Boundary swapping in the Drosophila Bithorax complex. Development 2008, 135, 3983–3987. [Google Scholar] [CrossRef] [PubMed]
- Postika, N.; Metzler, M.; Affolter, M.; Müller, M.; Schedl, P.; Georgiev, P.; Kyrchanova, O. Boundaries mediate long-distance interactions between enhancers and promoters in the Drosophila Bithorax complex. PLoS Genet. 2018, 14, e1007702. [Google Scholar] [CrossRef] [PubMed]
- Kyrchanova, O.; Sabirov, M.; Mogila, V.; Kurbidaeva, A.; Postika, N.; Maksimenko, O.; Schedl, P.; Georgiev, P. Complete reconstitution of bypass and blocking functions in a minimal artificial Fab-7 insulator from Drosophila bithorax complex. Proc. Natl. Acad. Sci. USA 2019, 9, 201907190. [Google Scholar] [CrossRef]
- Page, A.R.; Kovacs, A.; Deak, P.; Torok, T.; Kiss, I.; Dario, P.; Bastos, C.; Batista, P.; Gomes, R.; Ohkura, H.; et al. Spotted-dick, a zinc-finger protein of Drosophila required for expression of Orc4 and S phase. EMBO J. 2005, 24, 4304–4315. [Google Scholar] [CrossRef]
- Mohan, M.; Bartkuhn, M.; Herold, M.; Philippen, A.; Heinl, N.; Bardenhagen, I.; Leers, J.; White, R.A.H.; Renkawitz-Pohl, R.; Saumweber, H.; et al. The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. EMBO J. 2007, 26, 4203–4214. [Google Scholar] [CrossRef]
- Gerasimova, T.I.; Lei, E.P.; Bushey, A.M.; Corces, V.G. Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Mol. Cell 2007, 28, 761–772. [Google Scholar] [CrossRef]
- Bonchuk, A.; Maksimenko, O.; Kyrchanova, O.; Ivlieva, T.; Mogila, V.; Deshpande, G.; Wolle, D.; Schedl, P.; Georgiev, P. Functional role of dimerization and CP190 interacting domains of CTCF protein in Drosophila melanogaster. BMC Biol. 2015, 13, 63. [Google Scholar] [CrossRef]
- Hart, C.M. Do the BEAF insulator proteins regulate genes involved in cell polarity and neoplastic growth? Dev. Biol. 2014, 389, 121–123. [Google Scholar] [CrossRef][Green Version]
- Savitsky, M.; Kim, M.; Kravchuk, O.; Schwartz, Y.B. Distinct Roles of Chromatin Insulator Proteins in Control of the Drosophila Bithorax Complex. Genetics 2016, 202, 601–617. [Google Scholar] [CrossRef] [PubMed]
- Soshnev, A.A.; Baxley, R.M.; Manak, J.R.; Tan, K.; Geyer, P.K. The insulator protein Suppressor of Hairy-wing is an essential transcriptional repressor in the Drosophila ovary. Development 2013, 140, 3613–3623. [Google Scholar] [CrossRef] [PubMed]
- Bartkuhn, M.; Straub, T.; Herold, M.; Herrmann, M.; Rathke, C.; Saumweber, H.; Gilfillan, G.D.; Becker, P.B.; Renkawitz, R. Active promoters and insulators are marked by the centrosomal protein 190. EMBO J. 2009, 28, 877–888. [Google Scholar] [CrossRef] [PubMed]
- Ali, T.; Krüger, M.; Bhuju, S.; Jarek, M.; Bartkuhn, M.; Renkawitz, R. Chromatin binding of Gcn5 in Drosophila is largely mediated by CP190. Nucleic Acids Res. 2017, 45, 2384–2395. [Google Scholar] [CrossRef] [PubMed]
- Chodagam, S.; Royou, A.; Whitfield, W.; Karess, R.; Raff, J.W. The centrosomal protein CP190 regulates myosin function during early Drosophila development. Curr. Biol. 2005, 15, 1308–1313. [Google Scholar] [CrossRef] [PubMed]
- Gambetta, M.C.; Furlong, E.E.M. The Insulator Protein CTCF Is Required for Correct Hox Gene Expression, but Not for Embryonic Development in Drosophila. Genetics 2018, 210, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Bender, W.; Lucas, M. The border between the ultrabithorax and abdominal-A regulatory domains in the Drosophila bithorax complex. Genetics 2013, 193, 1135–1147. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Jiang, N.; Hart, C.M. Lack of the Drosophila BEAF insulator proteins alters regulation of genes in the Antennapedia complex. Mol. Genet. Genom. 2011, 285, 113–123. [Google Scholar] [CrossRef]
- Wood, A.M.; Van Bortle, K.; Ramos, E.; Takenaka, N.; Rohrbaugh, M.; Jones, B.C.; Jones, K.C.; Corces, V.G. Regulation of chromatin organization and inducible gene expression by a Drosophila insulator. Mol. Cell 2011, 44, 29–38. [Google Scholar] [CrossRef]
- Ong, C.-T.; Van Bortle, K.; Ramos, E.; Corces, V.G. Poly(ADP-ribosyl)ation regulates insulator function and intrachromosomal interactions in Drosophila. Cell 2013, 155, 148–159. [Google Scholar] [CrossRef]
- King, M.R.; Matzat, L.H.; Dale, R.K.; Lim, S.J.; Lei, E.P. The RNA-binding protein Rumpelstiltskin antagonizes gypsy chromatin insulator function in a tissue-specific manner. J. Cell. Sci. 2014, 127, 2956–2966. [Google Scholar] [CrossRef] [PubMed]
- Lewis, E.B. A gene complex controlling segmentation in Drosophila. Nature 1978, 276, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Bintu, B.; Mateo, L.J.; Su, J.-H.; Sinnott-Armstrong, N.A.; Parker, M.; Kinrot, S.; Yamaya, K.; Boettiger, A.N.; Zhuang, X. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science 2018, 362, eaau1783. [Google Scholar] [CrossRef] [PubMed]
- Sabari, B.R.; Dall’Agnese, A.; Boija, A.; Klein, I.A.; Coffey, E.L.; Shrinivas, K.; Abraham, B.J.; Hannett, N.M.; Zamudio, A.V.; Manteiga, J.C.; et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 2018, 361, eaar3958. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.M.; Guan, J.; Li, B.; Maliskova, L.; Song, M.; Shen, Y.; Huang, B.; Lomvardas, S.; Weiner, O.D. Live-cell imaging reveals enhancer-dependent Sox2 transcription in the absence of enhancer proximity. eLife 2019, 8, e41769. [Google Scholar] [CrossRef] [PubMed]
- Heist, T.; Fukaya, T.; Levine, M. Large distances separate coregulated genes in living Drosophila embryos. Proc. Natl. Acad. Sci. USA 2019, 116, 15062–15067. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Özdemir, I.; Gambetta, M.C. The Role of Insulation in Patterning Gene Expression. Genes 2019, 10, 767. https://doi.org/10.3390/genes10100767
Özdemir I, Gambetta MC. The Role of Insulation in Patterning Gene Expression. Genes. 2019; 10(10):767. https://doi.org/10.3390/genes10100767
Chicago/Turabian StyleÖzdemir, Isa, and Maria Cristina Gambetta. 2019. "The Role of Insulation in Patterning Gene Expression" Genes 10, no. 10: 767. https://doi.org/10.3390/genes10100767
APA StyleÖzdemir, I., & Gambetta, M. C. (2019). The Role of Insulation in Patterning Gene Expression. Genes, 10(10), 767. https://doi.org/10.3390/genes10100767




