Effects of Fat and Fatty Acids on the Formation of Autolysosomes in the Livers from Yellow Catfish Pelteobagrus Fulvidraco
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification and Tissue Distribution of Gene Expression
2.2. Feed Preparation and Fish Culture
2.3. In Vitro Studies
2.3.1. Primary Hepatocytes Culture and Treatments
2.3.2. Cell Viability and Non-Esterified Fatty Acid (NEFA) Concentrations Detection
2.3.3. Flow Cytometry and the Laser Confocal Microscope for Detecting the Formation of Autolysosomes
2.3.4. Real-Time Quantitative Polymerase Chain Reaction (qPCR)
2.4. Statistical Analysis
3. Results
3.1. Molecular Characterization
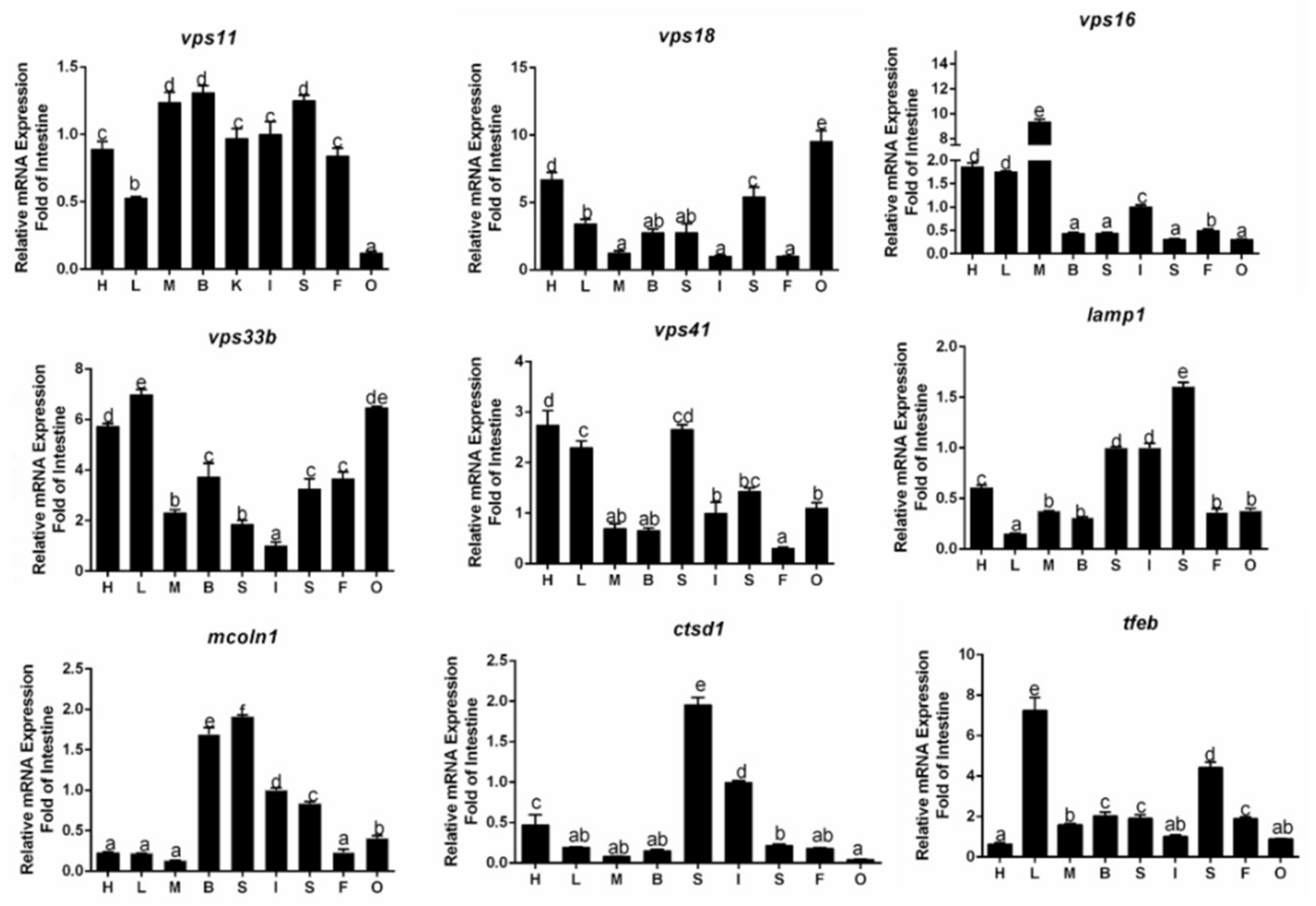
3.2. Tissue Distribution of Gene Expression
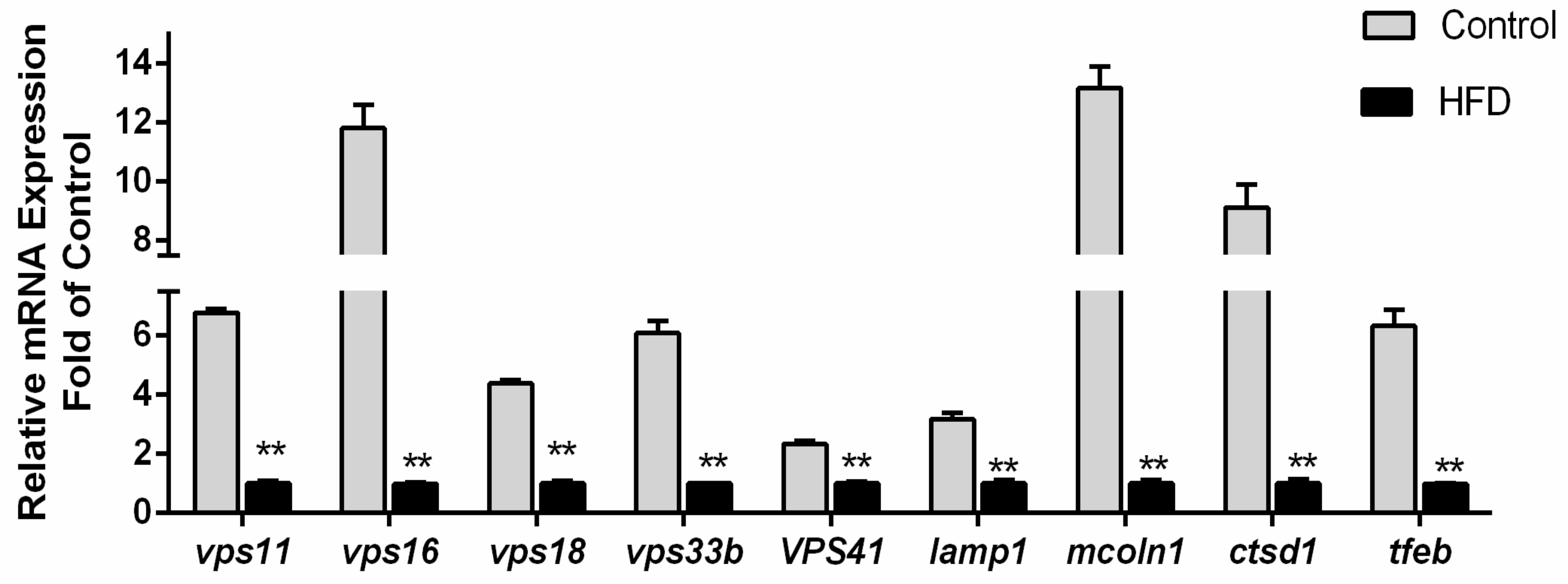
3.3. Transcriptional Responses of Nine Autolysosome-Related Genes by High-Fat Diet
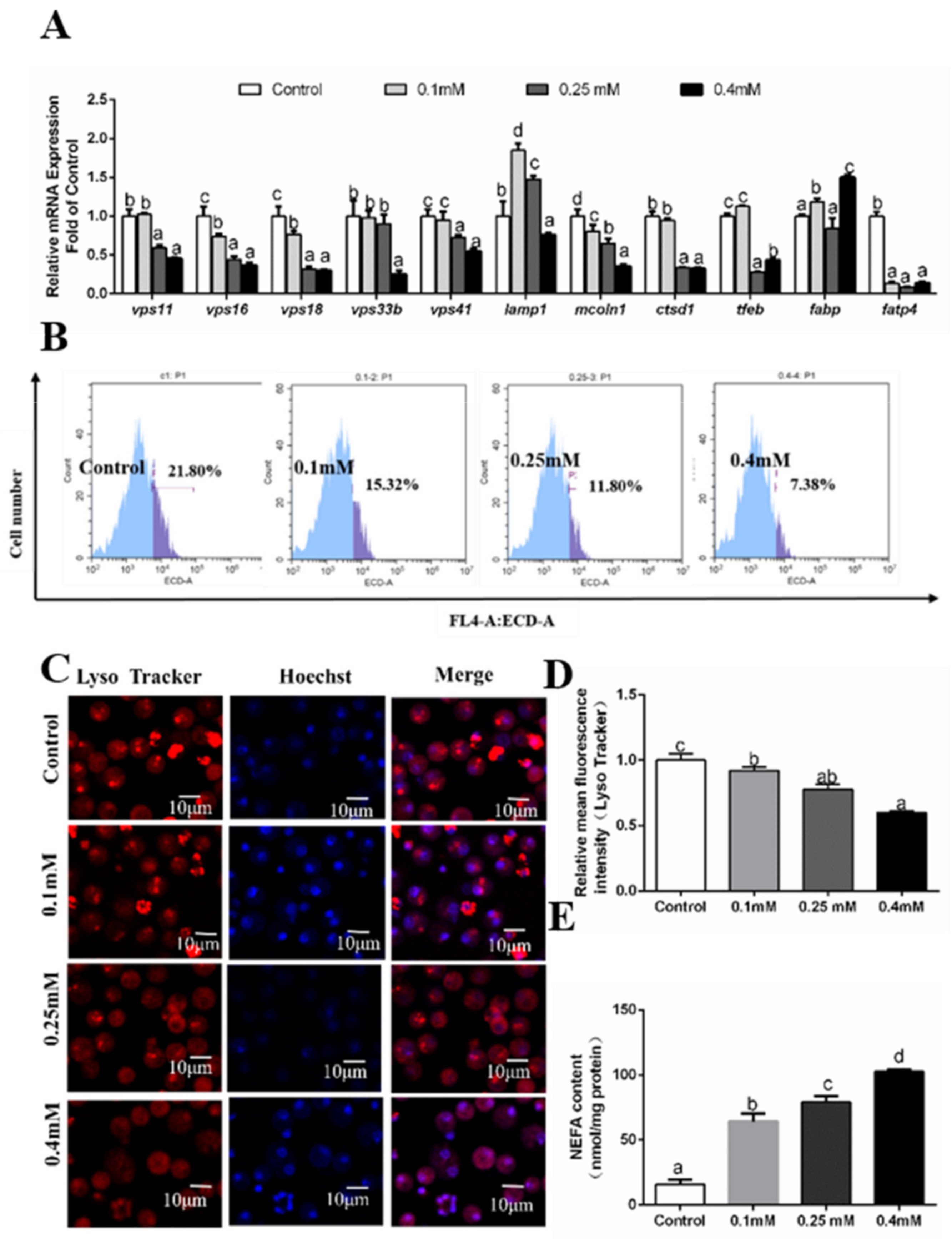
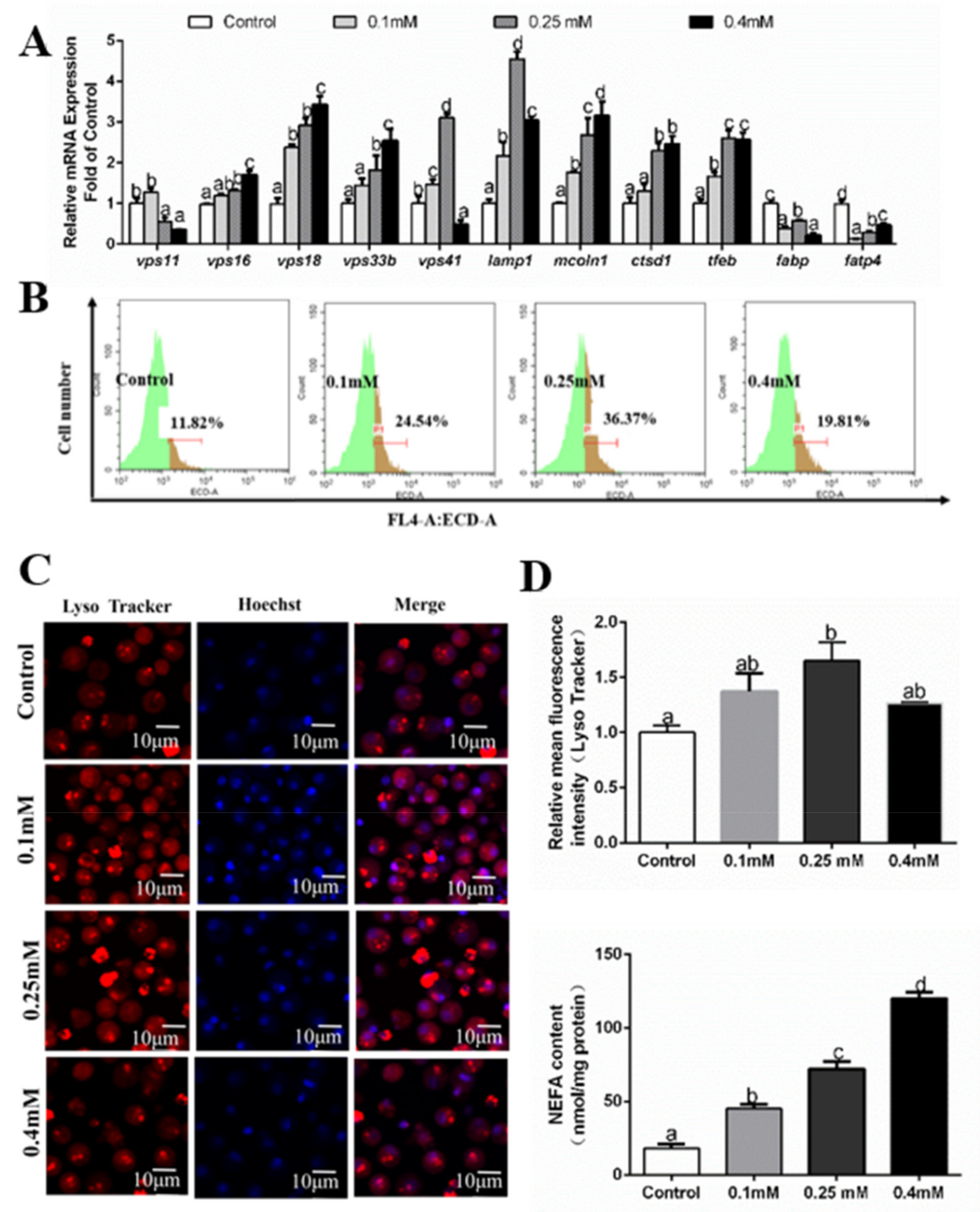
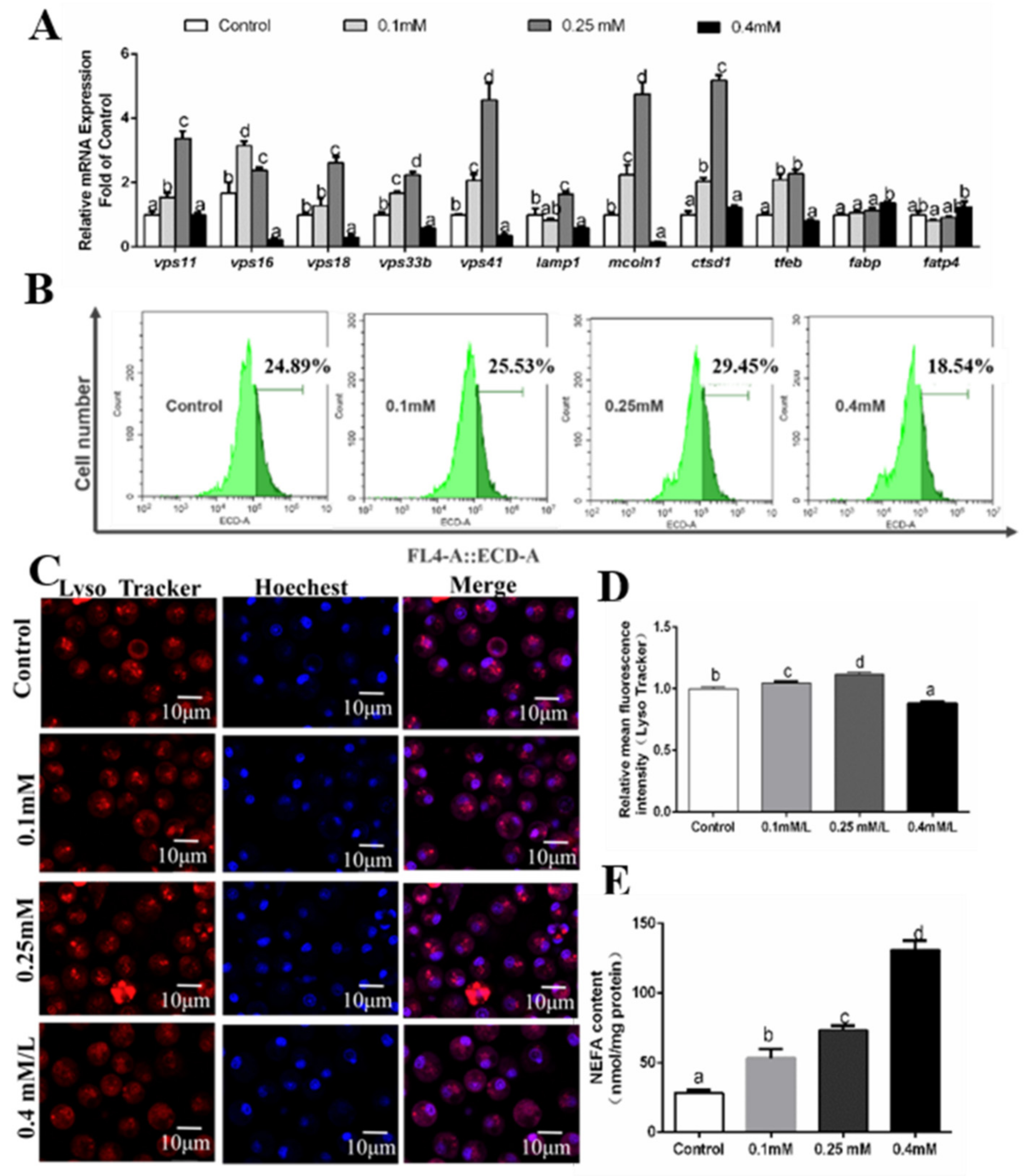
3.4. Effects of Palmitic Acid (PA) Incubations on the Formation of Autolysosomes
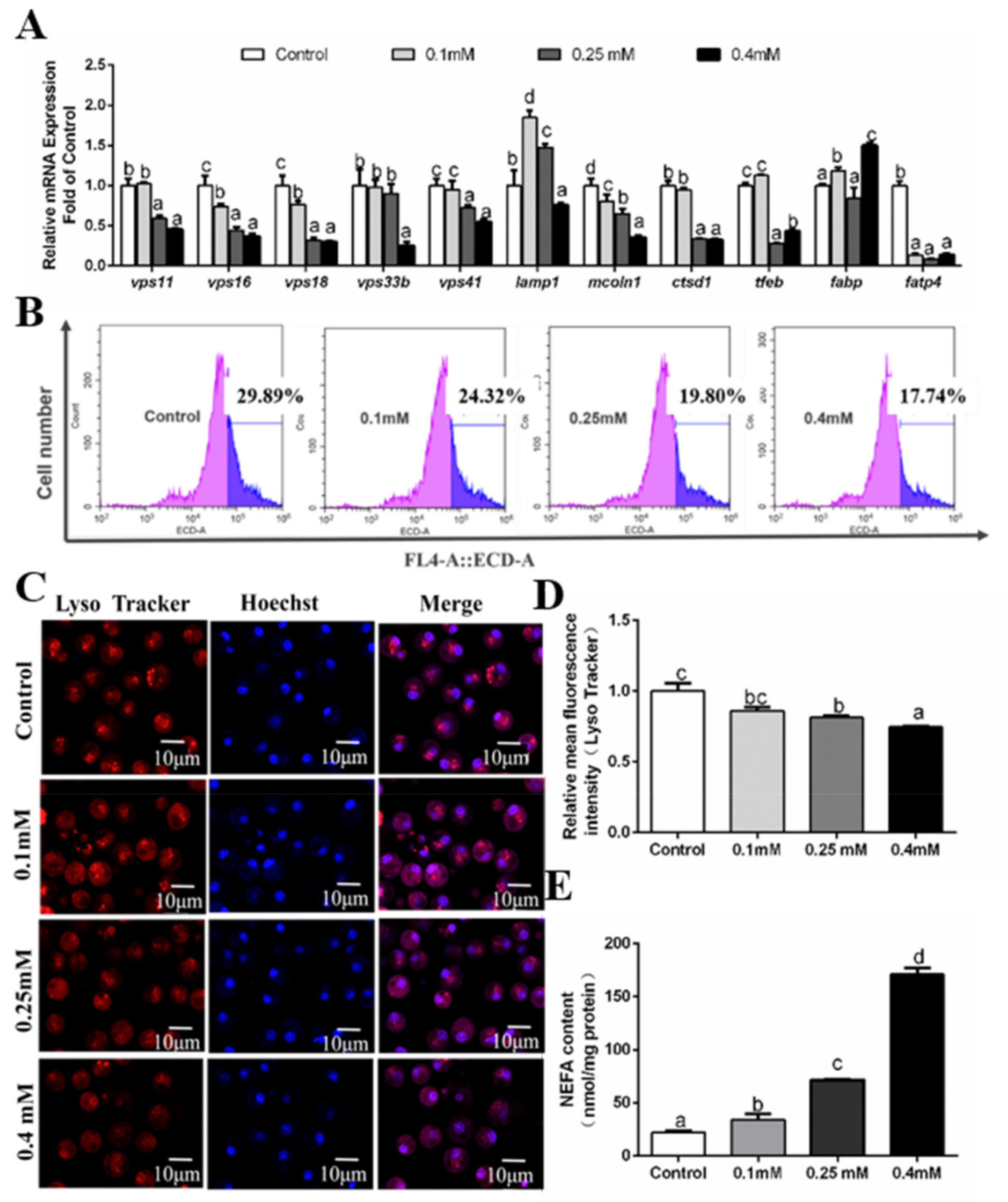
3.5. Effects of Oleic Acid (OA) Incubations on the Formation of Autolysosomes
4. Discussion
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Klionsky, D.J.; Emr, S.D. Autophagy as a regulated pathway of cellular degradation. Science 2000, 290, 1717–1721. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.Y.; Xu, X. Role of autophagy in metabolic syndrome-associated heart disease. Biochim. Biophys. Acta 2015, 1852, 225–231. [Google Scholar] [CrossRef]
- Kornfeld, S.; Mellman, I. The biogenesis of lysosomes. Annu. Rev. Cell. Dev. Biol. 2003, 5, 483–525. [Google Scholar] [CrossRef]
- Levine, B.; Klionsky, D.J. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004, 6, 463–477. [Google Scholar] [CrossRef]
- Nickerson, D.P.; Brett, C.L.; Merz, A.J. Vps-C complexes: Gatekeepers of endolysosomal traffic. Curr. Opin. Cell. Biol. 2009, 21, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.R. Transport vesicle docking: SNAREs and associates. Annu. Rev. Cell. Dev. Biol. 1996, 12, 441–461. [Google Scholar] [CrossRef]
- Jahn, R.; Sudhof, T.C. Membrane fusion and exocytosis. Annu. Rev. Biochem. 1999, 68, 863–911. [Google Scholar] [CrossRef]
- Seals, D.F.; Eitzen, G.; Margolis, N.; Wickner, W.T.; Price, A. A Ypt/Rab effector complex containing the Sec1 homolog VPS33p is required for homotypic vacuole fusion. Proc. Natl. Acad. Sci. USA 2000, 97, 9402–9407. [Google Scholar] [CrossRef]
- Sato, T.K.; Rehling, P.; Peterson, M.R.; Emr, S.D. Class C VPS protein complex regulates vacuolar SNARE pairing and is required for vesicle docking/fusion. Mol. Cell 2000, 6, 661–671. [Google Scholar] [CrossRef]
- Rieder, S.E.; Emr, S.D. A novel RING finger protein complex essential for a late step in protein transport to the yeast vacuole. Mol. Biol. Cell 1997, 8, 2307–2327. [Google Scholar] [CrossRef]
- Settembre, C. TFEB Links Autophagy to Lysosomal Biogenesis. Science 2011, 332, 1429–1433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liaudet-Coopman, E.; Beaujouin, M.; Derocq, D.; Garcia, M.; Glondu-Lassis, M.; Laurent-Matha, V. Newly discovered functions of a long-standing aspartic protease in cancer and apoptosis. Cancer. Lett. 2006, 237, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Wong, R.N.S. Evolution in the structure and function of aspartic proteases. J. Cell. Biochem. 1987, 33, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Eskelinen, E.L. Roles of LAMP-1 and LAMP-2 in lysosome biogenesis and autophagy. Mol. Aspects. Med. 2006, 27, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Medina, D.L.; Di Paola, S.; Peluso, I.; Armani, A.; De Stefani, D.; Venditti, R. Lysosomal calcium signaling regulates autophagy through calcineurin and TFEB. Nat. Cell. Biol. 2015, 17, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Sevrioukov, E.A.; He, J.P.; Moghrabi, N.; Sunio, A.; Kramer, H. A role for the deep orange and carnation eye color genes in lysosomal delivery in Drosophila. Mol. Cell 1999, 4, 479–486. [Google Scholar] [CrossRef]
- Maldonado, E.; Hernandez, F.; Lozano, C.; Castro, M.E.; Navarro, R.E. The zebrafish mutant vps18 as a model for vesicle-traffic related hypopigmentation diseases. Pigm. Cell. Melanoma 2010, 19, 315–326. [Google Scholar] [CrossRef]
- Thomas, J.L.; Vihtelic, T.S.; Dendekker, A.D.; Willer, G.; Luo, X.; Murphy, T.R.; Ronald, G.G.; David, R.H.; Thummel, R. The loss of vacuolar protein sorting 11 (vps11) causes retinal pathogenesis in a vertebrate model of syndromic albinism. Invest. Ophth. Vis. Sci. 2011, 52, 3119. [Google Scholar] [CrossRef]
- Lister, J.A.; Lane, B.M.; Nguyen, A.; Lunney, K. Embryonic expression of zebrafish MiT family genes tfe3b, tfeb, and tfec. Dev. Dyn. 2011, 240, 2529–2538. [Google Scholar] [CrossRef] [Green Version]
- Feng, T.; Zhang, H.; Liu, H.; Zhou, Z.C.; Niu, D.H.; Wong, L.L.; Kucuktas, H.; Liu, X.L.; Peatman, E.; Liu, Z.J. Molecular characterization and expression analysis of the channel catfish cathepsin D genes. Fish. Shellfish. Immun. 2011, 31, 164–169. [Google Scholar] [CrossRef]
- Jiang, S.T.; Wang, Y.T.; Chen, C.S. Purification and characterization of a proteinase identified as cathepsin D from tilapia muscle (Tilapia nilotica × Tilapia aurea). J. Agric. Food Chem. 1991, 39, 1597–1601. [Google Scholar] [CrossRef]
- Carnevali, O. Molecular Cloning and Expression of Ovarian Cathepsin D in Seabream, Sparus aurata. Biol. Reprod. 1999, 61, 785–791. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, L.B.; Nielsen, H.H. Purification and characterization of cathepsin D from herring muscle (Clupea harengus). Comp. Biochem. Physiol. 2001, 128, 351–363. [Google Scholar] [CrossRef]
- Cho, J.H.; Park, I.Y.; Kim, H.S.; Lee, W.T.; Kim, M.S.; Kim, S.C. Cathepsin D produces antimicrobial peptide parasin I from histone H2A in the skin mucosa of fish. FASEB J. 2002, 16, 429–431. [Google Scholar] [CrossRef] [Green Version]
- Malhi, H.; Gores, G.J. Molecular mechanisms of lipotoxicity in nonalcoholic fatty liver disease. Semin. Liver. Dis. 2008, 28, 360–369. [Google Scholar] [CrossRef]
- Brookheart, R.T.; Michel, C.I.; Schaffer, J.E. As a matter of fat. Cell. Metab. 2009, 10, 9–12. [Google Scholar] [CrossRef]
- Wei, C.C.; Luo, Z.; Hogstrand, C.; Xu, Y.H.; Wu, L.X.; Chen, G.H.; Pan, Y.X.; Song, Y.F. Zinc reduces hepatic lipid deposition and activates lipophagy via Zn2+/MTF-1/PPARα and Ca2+/CaMKKß/AMPK pathways. FASEB J. 2018, 32, 6666–6680. [Google Scholar] [CrossRef]
- Chen, C.Y.; Liu, C.H.; Tsai, Y.C.; Hsu, H.C.; Chen, M.F. ER stress, but not autophagy, was activated for cadioprotection during short-term receiving the high/fat sucrose diet. Eur. Heart. J. 2013, 34, 693. [Google Scholar]
- Singh, R.; Kaushik, S.; Wang, Y.; Xiang, Y.; Novak, I.; Komatsu, M.; Tanaka, K.; Cuervo, A.M.; Czaja, M.J. Autophagy regulates lipid metabolism. Nature 2009, 458, 1131–1135. [Google Scholar] [CrossRef] [Green Version]
- Baylin, A.; Kabagambe, E.K.; Siles, X.; Campos, H. Adipose tissue biomarkers of fatty acid intake. Am. J. Clin. Nutr. 2002, 76, 750–757. [Google Scholar] [CrossRef] [Green Version]
- Listenberger, L.L.; Han, X.; Lewis, S.E.; Cases, S.; Farese, R.V., Jr.; Ory, D.S.; Schaffer, J.E. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc. Natl. Acad. Sci. USA 2003, 100, 3077–3082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mei, S.; Ni, H.M.; Manley, S.; Bockus, A.; Kassel, K.M.; Luyendyk, J.P.; Copple, B.L.; Ding, W.X. Differential roles of unsaturated and saturated fatty acids on autophagy and apoptosis in hepatocytes. J. Pharmacol. Exp. Ther. 2011, 339, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from proteins equences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [PubMed]
- Ling, S.C.; Wu, K.; Zhang, D.G.; Luo, Z. Endoplasmic reticulum stress mediated autophagy and apoptosis alleviate dietary fat-induced triglyceride accumulation in the intestinal and isolated intestinal epithelial cells of yellow catfish. J. Nutr. 2019. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Huang, C.; Shi, X.; Chen, F.; Xu, Y.H.; Pan, Y.X.; Luo, Z.; Liu, X. Role and mechanism of the AMPK pathway in waterborne Zn exposure influencing the hepatic energy metabolism of Synechogobius hasta. Sci. Rep. 2016, 6, 38716. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.C.; Luo, Z.; Song, Y.F.; Pan, Y.X.; Wu, K.; You, W.J. Identification of autophagy related genes LC3 and ATG4 from Pelteobagrus fulvidraco and their transcriptional responses to waterborne and dietborne zinc exposure. Chemosphere 2017, 175, 228–238. [Google Scholar] [CrossRef]
- Las, G.; Serada, S.B.; Wikstrom, J.D.; Twig, G.; Shirihai, O.S. Fatty acids suppress autophagic turnover in β-cells. J. Biol. Chem. 2011, 286, 42534–42544. [Google Scholar] [CrossRef]
- Yamamoto, T.; Takabatake, Y.; Takahashi, A.; Kimura, T.; Namba, T.; Matsuda, J. High-fat diet-induced lysosomal dysfunction and impaired autophagic flux contribute to lipotoxicity in the kidney. Jasn 2017, 28, 1534. [Google Scholar] [CrossRef]
- Radisky, D.C.; Snyder, W.B.; Emr, S.D.; Kaplan, J. Characterization of VPS41, a gene required for vacuolar trafficking and high-affinity iron transport in yeast. Proc. Natl. Acad. Sci. USA 1997, 94, 5662–5668. [Google Scholar] [CrossRef]
- Gerhardt, B.; Kordas, T.J.; Thompson, C.M.; Patel, P.; Vida, T. The vesicle transport protein vps33p is an atp-binding protein that localizes to the cytosol in an energy-dependent manner. J. Biol. Chem. 1998, 273, 15818–15829. [Google Scholar] [CrossRef]
- Huizing, M.; Didier, A.; Walenta, J.; Anikster, Y.; Gahl, W.A.; Krämer, H. Molecular cloning and characterization of human VPS18, VPS 11, VPS16, and VPS33. Gene 2001, 264, 241–247. [Google Scholar] [CrossRef]
- Borden, K.L.; Freemont, P.S. The RING finger domain: A recent example of a sequence-structure family. Current Opinion in Structural. Biology 1996, 6, 395–401. [Google Scholar]
- Lorick, K.L.; Jensen, J.P.; Fang, S.; Ong, A.M.; Hatakeyama, S.; Weissman, A.M. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA 1999, 96, 11364–11369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, B.Y.; Ueda, M.; Kominami, E.; Akagawa, K.; Kohsaka, S.; Akazawa, C. Identification of mouse vps16 and biochemical characterization of mammalian class c vps complex. Biochem. Biophys. Res. Commun. 2003, 311, 577–582. [Google Scholar] [CrossRef]
- Barriocanal, J.G.; Bonifacino, J.S.; Yuan, L.; Sandoval, I.V. Biosynthesis, glycosylation, movement through the Golgi system, and transport to lysosomes by an N-linked carbohydrate-independent mechanism of three lysosomal integral membrane proteins. J. Biol. Chem. 1986, 261, 16755–16763. [Google Scholar] [PubMed]
- Liu, X.; Shi, G.; Cui, D.; Wang, R.; Xu, T. Molecular cloning and comprehensive characterization of cathepsin D in the Miiuy croaker Miichthys miiuy. Fish. Shellfish. Immun. 2012, 32, 464–468. [Google Scholar] [CrossRef]
- Shi, G.P.; Munger, J.S.; Meara, J.P.; Rich, D.H.; Chapman, H.A. Molecular cloning and expression of human alveolar macrophage cathepsin S, an elastinolytic cysteine protease. J. Biol. Chem. 1992, 267, 7258e62. [Google Scholar]
- Fisher, D.E.; Carr, C.S.; Parent, L.A.; Sharp, P.A. TFEB has DNA-binding and oligomerization properties of a unique helix-loop-helix/leucine-zipper family. Genes. Dev. 1991, 5, 2342–2352. [Google Scholar] [CrossRef]
- O’Shea, E.K.; Rutkowski, R.; Kim, P.S. Evidence that the leucine zipper is a coiled coil. Science 1989, 243, 538–542. [Google Scholar] [CrossRef] [PubMed]
- Baricos, W.H.; Zhou, Y.W.; Fuerst, R.S.; Barrett, A.J.; Shah, S.V. The role of aspartic and cysteine proteinases in albumin degradation by rat kidney cortical lysosomes. Arch. Biochem. Biophys. 1987, 256, 687–691. [Google Scholar] [CrossRef]
- Ebato, C.; Uchida, T.; Arakawa, M.; Komatsu, M.; Ueno, T.; Komiya, K.; Azuma, K.; Hirose, T.; Tanaka, K.; Kominami, E.; et al. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell. Metab. 2008, 8, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Koga, H.; Kaushik, S.; Cuervo, A.M. Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 2010, 24, 3052–3065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, S.H.; Shui, G.; Zhou, J.; Li, J.J.E.; Bay, B.H.; Wenk, M.R.; Shen, H.M. Induction of autophagy by palmitic acid via protein kinase C-mediated signaling pathway independent of mTOR (mammalian target of rapamycin). J. Biol. Chem. 2012, 287, 14364–14376. [Google Scholar] [CrossRef] [PubMed]


| Gene | Accession No. | 5′ Untranslated Regions (UTR)(bp) | ORF (bp) | 3′ UTR (bp) | Full Length (bp) | Protein (aa) |
|---|---|---|---|---|---|---|
| vps11 | MH301091 | 22 | 2784 | 274 | 3080 | 928 |
| vps16 | MH301092 | 48 | 2508 | 589 | 3144 | 835 |
| vps18 | MH301093 | 305 | 2979 | 98 | 3379 | 992 |
| vps33b | MH301094 | 69 | 1853 | 230 | 2230 | 617 |
| vps41 | MH301095 | 67 | 2564 | 427 | 2991 | 854 |
| lamp1 | MH301096 | 84 | 1245 | 167 | 1496 | 415 |
| mcoln1 | MH301097 | 217 | 1745 | 196 | 2158 | 581 |
| ctsd1 | MH301098 | 86 | 1190 | 569 | 1845 | 396 |
| tfeb | MH459004 | 181 | 1475 | 367 | 2023 | 491 |
| Genes | Ictalurus punctatus | Danio rerio | Xenopus tropicalis | Rattus norvegicus | Homo sapiens |
|---|---|---|---|---|---|
| vps11 | 95.1 | 89.3 | 84.1 | 84.6 | 84.4 |
| vps16 | 94.73 | 85.87 | 62.80 | 65.67 | 67.71 |
| vps18 | 92.04 | 85.99 | 70.67 | 63.61 | 64.61 |
| vps33b | 93.19 | 88.01 | 75.12 | 74.64 | 63.65 |
| vps41 | 96.02 | 91.80 | 77.52 | 80.09 | 80.56 |
| lamp1 | 76.68 | 86.8 | 27.61 | 42.61 | 42.24 |
| mcoln1 | 87.44 | 79.9 | 65.82 | 60.91 | 60.90 |
| ctsd1 | 92.93 | 85.93 | 63.26 | 63.83 | 67.08 |
| tfeb | 86.60 | 69.82 | 44.20 | 48.65 | 46.55 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, L.-X.; Wei, C.-C.; Yang, S.-B.; Zhao, T.; Luo, Z. Effects of Fat and Fatty Acids on the Formation of Autolysosomes in the Livers from Yellow Catfish Pelteobagrus Fulvidraco. Genes 2019, 10, 751. https://doi.org/10.3390/genes10100751
Wu L-X, Wei C-C, Yang S-B, Zhao T, Luo Z. Effects of Fat and Fatty Acids on the Formation of Autolysosomes in the Livers from Yellow Catfish Pelteobagrus Fulvidraco. Genes. 2019; 10(10):751. https://doi.org/10.3390/genes10100751
Chicago/Turabian StyleWu, Li-Xiang, Chuan-Chuan Wei, Shui-Bo Yang, Tao Zhao, and Zhi Luo. 2019. "Effects of Fat and Fatty Acids on the Formation of Autolysosomes in the Livers from Yellow Catfish Pelteobagrus Fulvidraco" Genes 10, no. 10: 751. https://doi.org/10.3390/genes10100751
APA StyleWu, L.-X., Wei, C.-C., Yang, S.-B., Zhao, T., & Luo, Z. (2019). Effects of Fat and Fatty Acids on the Formation of Autolysosomes in the Livers from Yellow Catfish Pelteobagrus Fulvidraco. Genes, 10(10), 751. https://doi.org/10.3390/genes10100751







