Aberrant Splicing Events Associated to CDH23 Noncanonical Splice Site Mutations in a Proband with Atypical Usher Syndrome 1
Abstract
:1. Introduction
2. Materials and Methods
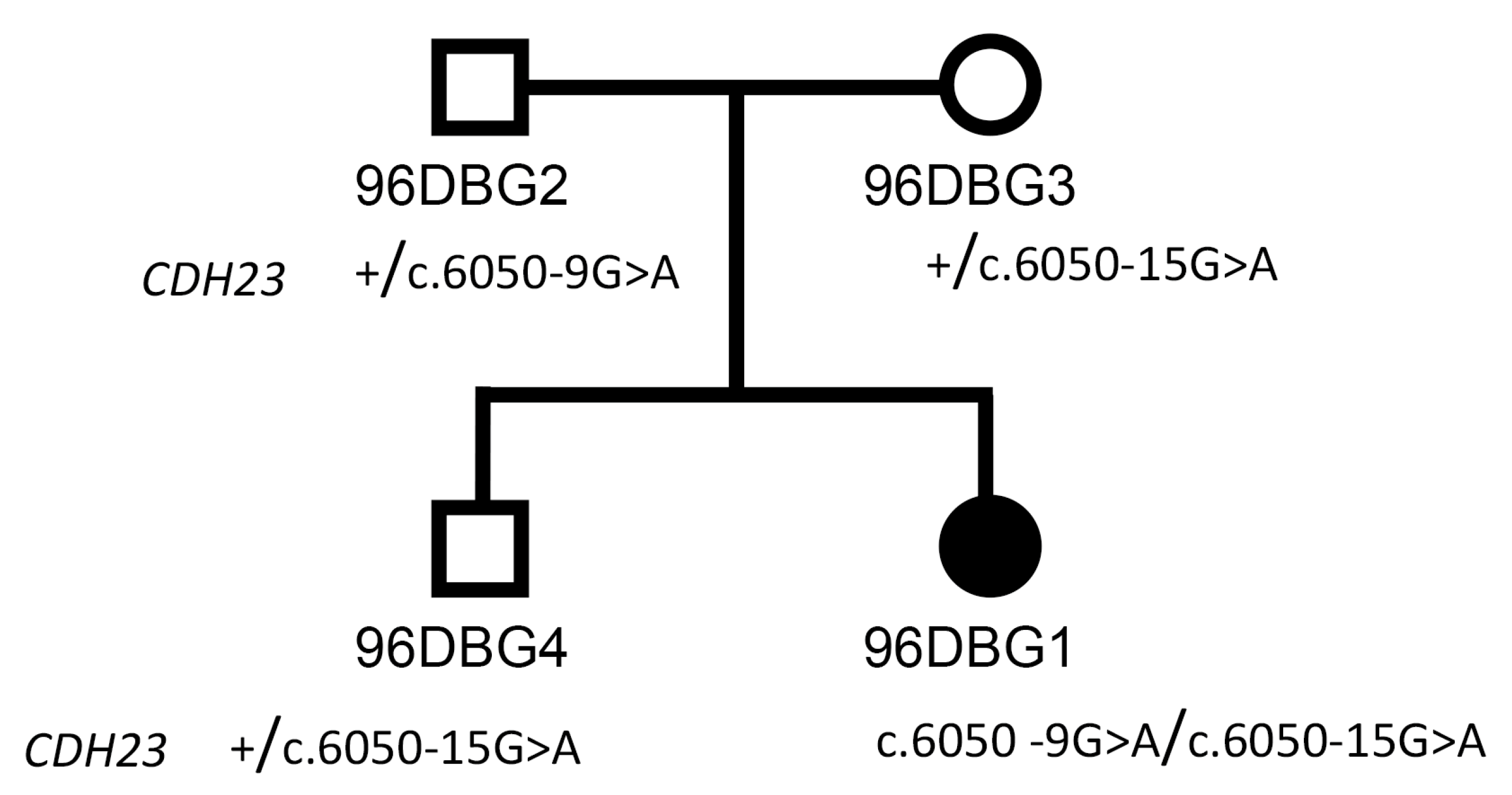
2.1. Clinical Diagnosis
2.2. Samples
2.3. In Silico Analysis of the Effect of Variants in Splicing
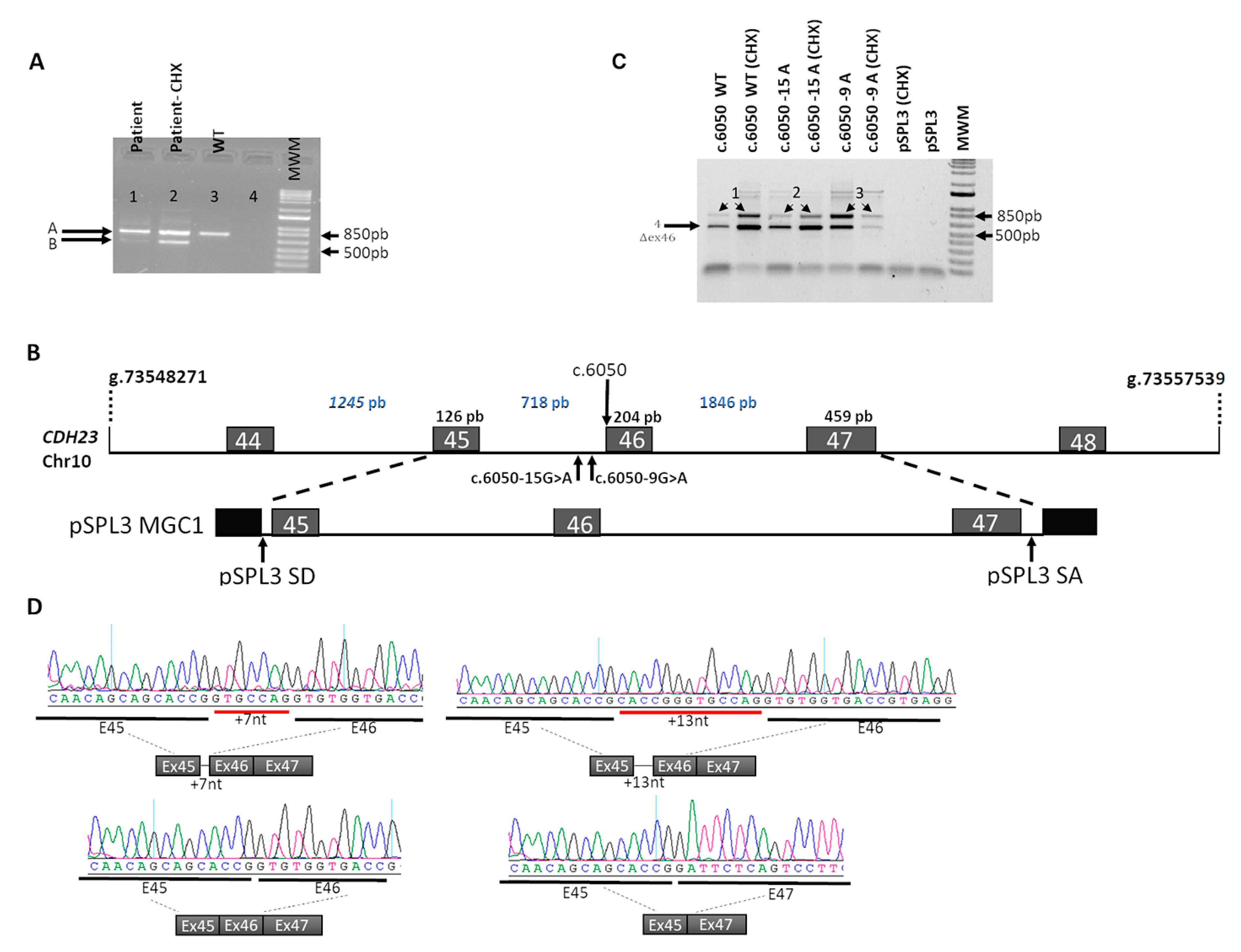
2.4. In Vivo Splicing Analysis of CDH23 Transcripts in the Patient
2.5. In Vitro Splicing Assays in HEK293T Cells
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mathur, P.D.; Yang, J. Usher syndrome and non-syndromic deafness: Functions of different whirlin isoforms in the cochlea, vestibular organs, and retina. Hear. Res. 2019, 375, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.W.; Johnson, R.D.; Langlo, C.S.; Cooper, R.F.; Razeen, M.M.; Russillo, M.C.; Dubra, A.; Connor, T.B.; Han, D.P.; Pennesi, M.E.; et al. Assessing Photoreceptor Structure in Retinitis Pigmentosa and Usher Syndrome. Investig. Ophthalmol. Vis. Sci. 2016, 57, 2428–2442. [Google Scholar] [Green Version]
- Yan, D.; Liu, X.Z. Genetics and pathological mechanisms of Usher syndrome. J. Hum. Genet. 2010, 55, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Astuto, L.M.; Bork, J.M.; Weston, M.D.; Askew, J.W.; Fields, R.R.; Orten, D.J.; Ohliger, S.J.; Riazuddin, S.; Morell, R.J.; Khan, S.; et al. CDH23 mutation and phenotype heterogeneity: A profile of 107 diverse families with Usher syndrome and nonsyndromic deafness. Am. J. Hum. Genet. 2002, 71, 262–275. [Google Scholar] [CrossRef] [PubMed]
- Bujakowska, K.M.; Consugar, M.; Place, E.; Harper, S.; Lena, J.; Taub, D.G.; White, J.; Navarro-Gomez, D.; Weigel DiFranco, C.; Farkas, M.H.; et al. Targeted Exon Sequencing in Usher Syndrome Type I. Invest. Ophthalmol. Vis. Sci. 2014, 55, 8488–8496. [Google Scholar] [Green Version]
- Gettelfinger, J.D.; Dahl, J.P. Syndromic Hearing Loss: A Brief Review of Common Presentations and Genetics. J. Pediatr. Genet. 2018, 7, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Feng, Y.; Liu, Y.; He, C.; Liu, J.; Chen, H.; Deng, Y.; Li, M.; Li, W.; Song, J.; et al. A novel ABHD12 nonsense variant in Usher syndrome type 3 family with genotype-phenotype spectrum review. Gene 2019, 704, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Licastro, D.; Mutarelli, M.; Peluso, I.; Neveling, K.; Wieskamp, N.; Rispoli, R.; Vozzi, D.; Athanasakis, E.; D’Eustacchio, A.; Pizzo, M.; et al. Molecular diagnosis of Usher syndrome: Application of two different next generation sequencing-based procedures. PLoS ONE 2012, 7, e43799. [Google Scholar] [CrossRef]
- Vastinsalo, H.; Isosomppi, J.; Aittakorpi, A.; Sankila, E.M. Two Finnish USH1B patients with three novel mutations in myosin VIIA. Mol. Vis. 2006, 12, 1093–1097. [Google Scholar]
- Bolz, H.J.; Roux, A.-F. Clinical utility gene card for: Usher syndrome. Eur. J. Hum. Genet. 2011, 19, 931. [Google Scholar] [CrossRef]
- Bork, J.M.; Peters, L.M.; Riazuddin, S.; Bernstein, S.L.; Ahmed, Z.M.; Ness, S.L.; Polomeno, R.; Ramesh, A.; Schloss, M.; Srisailpathy, C.R.; et al. Usher syndrome 1D and nonsyndromic autosomal recessive deafness DFNB12 are caused by allelic mutations of the novel cadherin-like gene CDH23. Am. J. Hum. Genet. 2001, 68, 26–37. [Google Scholar] [CrossRef]
- Mizutari, K.; Mutai, H.; Namba, K.; Miyanaga, Y.; Nakano, A.; Arimoto, Y.; Masuda, S.; Morimoto, N.; Sakamoto, H.; Kaga, K.; et al. High prevalence of CDH23 mutations in patients with congenital high-frequency sporadic or recessively inherited hearing loss. Orphanet J. Rare Dis. 2015, 10, 60. [Google Scholar] [CrossRef]
- Zhang, Q.; Peng, C.; Song, J.; Zhang, Y.; Chen, J.; Song, Z.; Shou, X.; Ma, Z.; Peng, H.; Jian, X.; et al. Germline Mutations in CDH23, Encoding Cadherin-Related 23, Are Associated with Both Familial and Sporadic Pituitary Adenomas. Am. J. Hum. Genet. 2017, 100, 817–823. [Google Scholar] [CrossRef]
- Aparisi, M.J.; García-García, G.; Aller, E.; Sequedo, M.D.; Martínez-Fernández de la Cámara, C.; Rodrigo, R.; Armengot, M.; Cortijo, J.; Milara, J.; Díaz-LLopis, M.; et al. Study of USH1 splicing variants through minigenes and transcript analysis from nasal epithelial cells. PLoS ONE 2013, 8, e57506. [Google Scholar] [CrossRef]
- Sangermano, R.; Khan, M.; Cornelis, S.S.; Richelle, V.; Albert, S.; Garanto, A.; Elmelik, D.; Qamar, R.; Lugtenberg, D.; van den Born, L.I.; et al. ABCA4 midigenes reveal the full splice spectrum of all reported noncanonical splice site variants in Stargardt disease. Genome Res. 2018, 28, 100–110. [Google Scholar] [CrossRef]
- Shaikh, S.S.; Nahorski, M.S.; Rai, H.; Woods, C.G. Before progressing from “exomes” to “genomes” don’t forget splicing variants. Eur. J. Hum. Genet. 2018, 26, 1559–1562. [Google Scholar] [CrossRef]
- Lim, K.H.; Ferraris, L.; Filloux, M.E.; Raphael, B.J.; Fairbrother, W.G. Using positional distribution to identify splicing elements and predict pre-mRNA processing defects in human genes. Proc. Natl. Acad. Sci. USA 2011, 108, 11093–11098. [Google Scholar] [CrossRef] [Green Version]
- Von Brederlow, B.; Bolz, H.; Janecke, A.; La O Cabrera, A.; Rudolph, G.; Lorenz, B.; Schwinger, E.; Gal, A. Identification and in vitro expression of novelCDH23 mutations of patients with Usher syndrome type 1D. Hum. Mutat. 2002, 19, 268–273. [Google Scholar] [CrossRef]
- Zernant, J.; Xie, Y.A.; Ayuso, C.; Riveiro-Alvarez, R.; Lopez-Martinez, M.-A.; Simonelli, F.; Testa, F.; Gorin, M.B.; Strom, S.P.; Bertelsen, M.; et al. Analysis of the ABCA4 genomic locus in Stargardt disease. Hum. Mol. Genet. 2014, 23, 6797–6806. [Google Scholar] [CrossRef]
- Zernant, J.; Lee, W.; Nagasaki, T.; Collison, F.T.; Fishman, G.A.; Bertelsen, M.; Rosenberg, T.; Gouras, P.; Tsang, S.H.; Allikmets, R. Extremely hypomorphic and severe deep intronic variants in the ABCA4 locus result in varying Stargardt disease phenotypes. Mol. Case Stud. 2018, 4, a002733. [Google Scholar] [CrossRef]
- Khan, M.; Cornelis, S.S.; Khan, M.I.; Elmelik, D.; Manders, E.; Bakker, S.; Derks, R.; Neveling, K.; van de Vorst, M.; Gilissen, C.; et al. Cost-effective molecular inversion probe-based ABCA4 sequencing reveals deep-intronic variants in Stargardt disease. Hum. Mutat. 2019. [Google Scholar] [CrossRef]
- Vaché, C.; Besnard, T.; Blanchet, C.; Baux, D.; Larrieu, L.; Faugère, V.; Mondain, M.; Hamel, C.; Malcolm, S.; Claustres, M.; et al. Nasal epithelial cells are a reliable source to study splicing variants in Usher syndrome. Hum. Mutat. 2010, 31, 734–741. [Google Scholar] [CrossRef] [Green Version]
- Schwander, M.; Lopes, V.; Sczaniecka, A.; Gibbs, D.; Lillo, C.; Delano, D.; Tarantino, L.M.; Wiltshire, T.; Williams, D.S.; Müller, U. A novel allele of myosin VIIa reveals a critical function for the C-terminal FERM domain for melanosome transport in retinal pigment epithelial cells. J. Neurosci. 2009, 29, 15810–15818. [Google Scholar] [CrossRef]

| Mutation | GRCh37/hg19 | Intron | Disease |
|---|---|---|---|
| c.145+6T>G | 73206158 | IVS-2 | Usher syndrome 1 |
| c.288+1G>C | 73269982 | IVS-3 | Usher syndrome 1 |
| c.336+1G>A | 73270759 | IVS-4 | Usher syndrome 1d |
| c.429+4G>A | 73270973 | IVS-5 | Non-syndromic autosomal recessive deafness |
| c.1134+1G>A | 73377151 | IVS-10 | Usher syndrome 1 |
| c.1135-1G>T | 73403617 | IVS-10 | Hearing loss |
| c.1987-2A>C | 73447402 | IVS-17 | Usher syndrome 1 |
| c.2176+1G>C | 73450342 | IVS-19 | Deafness, non-syndromic, autosomal recessive |
| c.2177-2A>G | 73453902 | IVS-19 | Usher syndrome 1 |
| c.2289+1G>A | 73454017 | IVS-20 | Usher syndrome 1d |
| c.2289+6T>G | 73454022 | IVS-20 | Hearing loss, non-syndromic |
| c.2398-1G>T | 73461778 | IVS-21 | Retinal disease |
| c.2587+1G>T | 73461969 | IVS-22 | Usher syndrome 1 |
| c.3580-1G>T | 73490225 | IVS-29 | Usher syndrome 1 |
| c.4104+4A>T | 73492136 | IVS-31 | Usher syndrome 1 |
| c.4105-4_4105-2delGCAinsTCT | 73493993 | IVS-31 | Usher syndrome |
| c.4489-2A>C | 73500577 | IVS-35 | Usher syndrome |
| c.4846-3C>G | 73537434 | IVS-37 | Hearing loss, autosomal recessive |
| c.5068-2A>T | 73537944 | IVS-38 | Usher syndrome 1 |
| c.5187+2T>C | 73538067 | IVS-39 | Usher syndrome 1 |
| c.5368+1G>A | 73539205 | IVS-40 | Usher syndrome 1 |
| c.5820+5G>A | 73545500 | IVS-43 | Sector retinitis pigmentosa and hearing loss |
| c.5821-2A>G | 73548695 | IVS-43 | Usher syndrome 1 |
| c.5923+1G>A | 73548800 | IVS-44 | Usher syndrome |
| c.5924-2A>C | 73550043 | IVS-44 | Hearing loss |
| c.6049G>A | 73550170 | E-44 NCSS IVS-45 | Usher syndrome 1 |
| c.6049+1G>A | 73550171 | IVS-45 | Usher syndrome 1 |
| c.6050-1G>C | 73550888 | IVS-45 | Usher syndrome 1 |
| c.6050-9G>A | 73550880 | IVS-45 | Usher syndrome 1 |
| c.6050-15>A | 73550874 | IVS-45 | Usher syndrome 1 |
| c.6712+1G>A | 73553398 | IVS-47 | Usher syndrome 1 |
| c.6829+1G>A | 73556978 | IVS-48 | Usher syndrome 1 |
| c.6829+2T>C | 73556979 | IVS-48 | Usher syndrome 1 |
| c.6830-2_6830delAGC | 73558109 | IVS-48 | Usher syndrome 1d |
| c.7225-2A>G | 73559247 | IVS-50 | Usher syndrome 1 |
| c.7362+5G>A | 73559391 | IVS-51 | Usher syndrome 1 |
| c.7482+1G>A | 73560513 | IVS-52 | Usher syndrome |
| c.7660+1G>T | 73562833 | IVS-53 | Usher syndrome 1 |
| c.7660+5G>A | 73562837 | IVS-53 | Hearing loss |
| c.8064+2T>C | 73565756 | IVS-55 | Usher syndrome 1 |
| c.8722+1delG | 73567765 | IVS-59 | Usher syndrome 1 |
| c.9199-4G>A | 73571264 | IVS-62 | Usher syndrome 1 |
| c.9278+5G>C | 73571352 | IVS-63 | Usher syndrome 1 |
| c.9510+19_9510+25delGGCATCA | 73572385 | IVS-66 | Usher syndrome 1 |
| c.9510+1G>A | 73572367 | IVS-66 | Usher syndrome 1 |
| Nucleotide Sequence | Splice Site | Splicing Predictor | Observed In Vivo Splicing Events in Control and Patient (Double Heterozygote) | Observed In Vitro Splicing Events | |||
|---|---|---|---|---|---|---|---|
| SSF (0–100) | Max Ent (0–16) | NNSPLICE (0–1) | Genesplicer (0–15) | ||||
| WT | WT acceptor | NR | 2.45 | NR | NR | WT transcript Skipping of exon 46 | WT transcript Skipping of exon 46 |
| c.6050-9 G>A | New acceptor | NR | 3.54 | NR | 1.96 | Extension of exon 46 due to the upstream addition of 7nt Skipping of exon 46. WT transcript | New acceptor site: + 7nt intronic included in extended exon 46 Skipping of exon 46 |
| c.6050-15G>A | New acceptor | 75.34 | 6.23 | NR | 7.84 | Extension of exon 46 due to the upstream addition of 13 nt Skipping of exon 46.WT transcript | New acceptor site: + 13nt intronic included in extended exon 46 Skipping of exon 46 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valero, R.; de Castro-Miró, M.; Jiménez-Ochoa, S.; Rodríguez-Ezcurra, J.J.; Marfany, G.; Gonzàlez-Duarte, R. Aberrant Splicing Events Associated to CDH23 Noncanonical Splice Site Mutations in a Proband with Atypical Usher Syndrome 1. Genes 2019, 10, 732. https://doi.org/10.3390/genes10100732
Valero R, de Castro-Miró M, Jiménez-Ochoa S, Rodríguez-Ezcurra JJ, Marfany G, Gonzàlez-Duarte R. Aberrant Splicing Events Associated to CDH23 Noncanonical Splice Site Mutations in a Proband with Atypical Usher Syndrome 1. Genes. 2019; 10(10):732. https://doi.org/10.3390/genes10100732
Chicago/Turabian StyleValero, Rebeca, Marta de Castro-Miró, Sofía Jiménez-Ochoa, Juan José Rodríguez-Ezcurra, Gemma Marfany, and Roser Gonzàlez-Duarte. 2019. "Aberrant Splicing Events Associated to CDH23 Noncanonical Splice Site Mutations in a Proband with Atypical Usher Syndrome 1" Genes 10, no. 10: 732. https://doi.org/10.3390/genes10100732
APA StyleValero, R., de Castro-Miró, M., Jiménez-Ochoa, S., Rodríguez-Ezcurra, J. J., Marfany, G., & Gonzàlez-Duarte, R. (2019). Aberrant Splicing Events Associated to CDH23 Noncanonical Splice Site Mutations in a Proband with Atypical Usher Syndrome 1. Genes, 10(10), 732. https://doi.org/10.3390/genes10100732





