Prognostic Value and Potential Regulatory Mechanism of Alternative Splicing in Geriatric Breast Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Process of Alternative Splicing Data Acquisition
2.2. A Preview of Survival-Related Alternative Splicing Events in Geriatric Breast Cancer
2.3. Prognostic Signatures for Alternative Splicing Events in Geriatric Breast Cancer
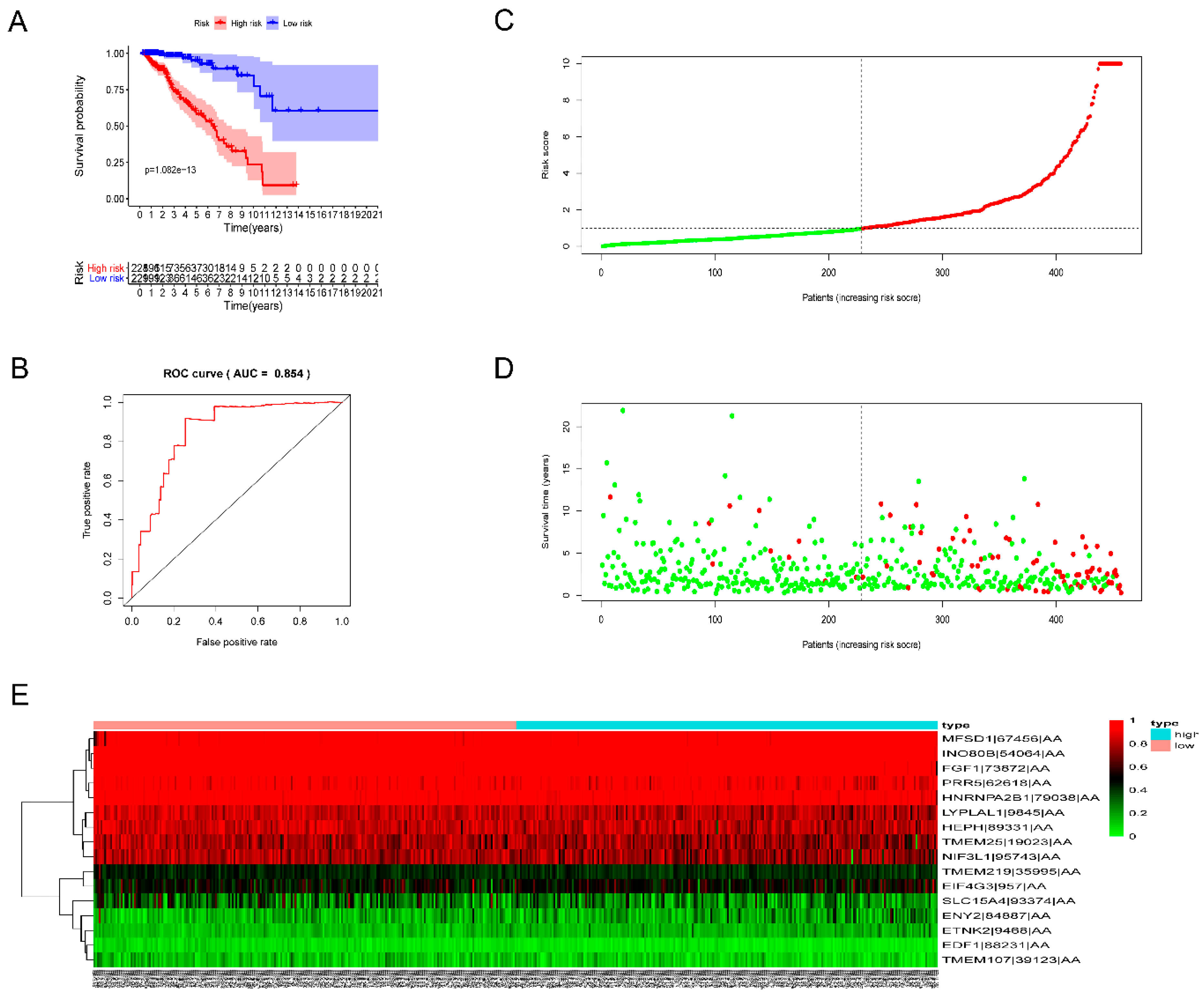
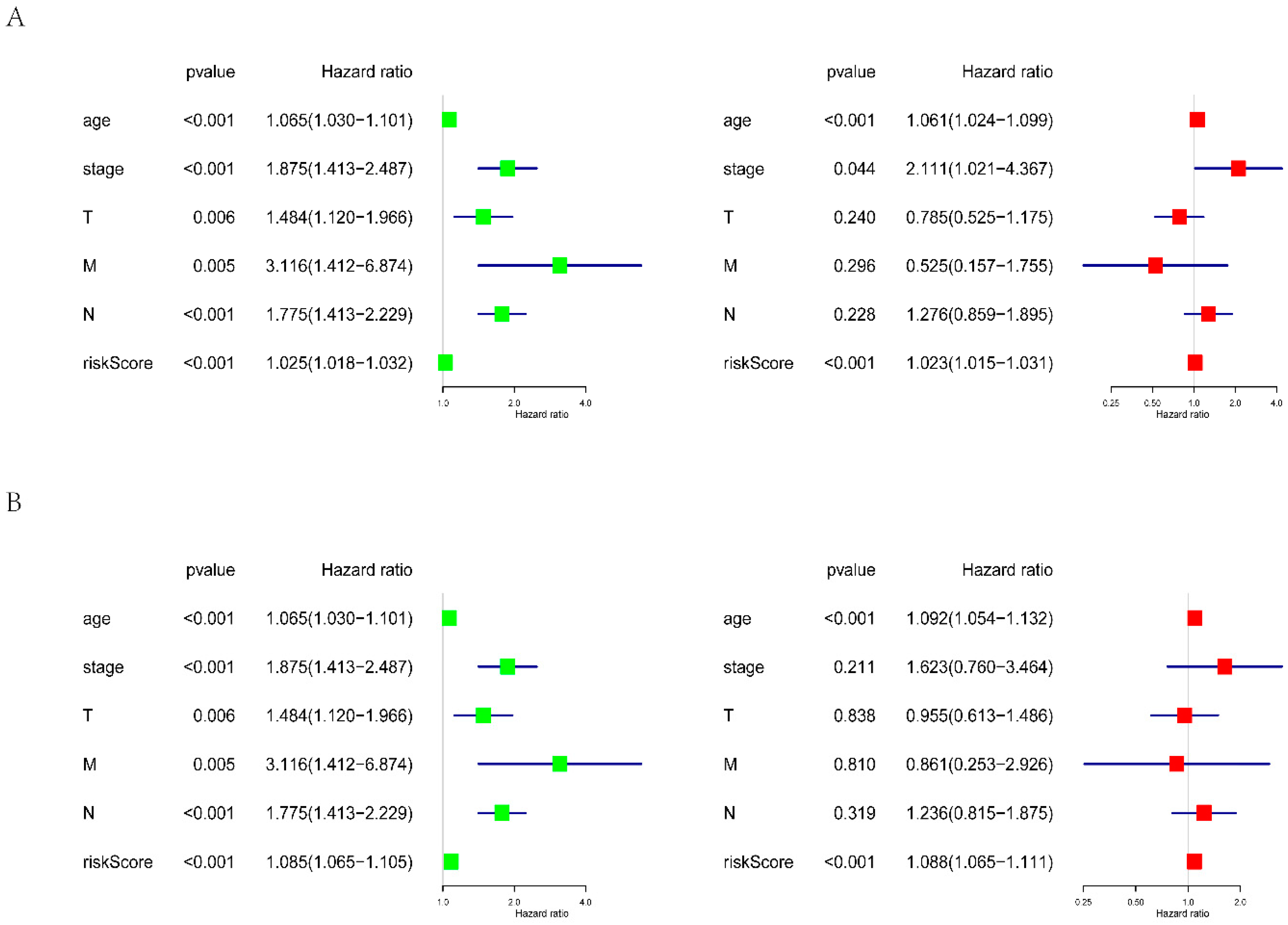
2.4. Evaluation of the Prognostic Value of the Risk Score
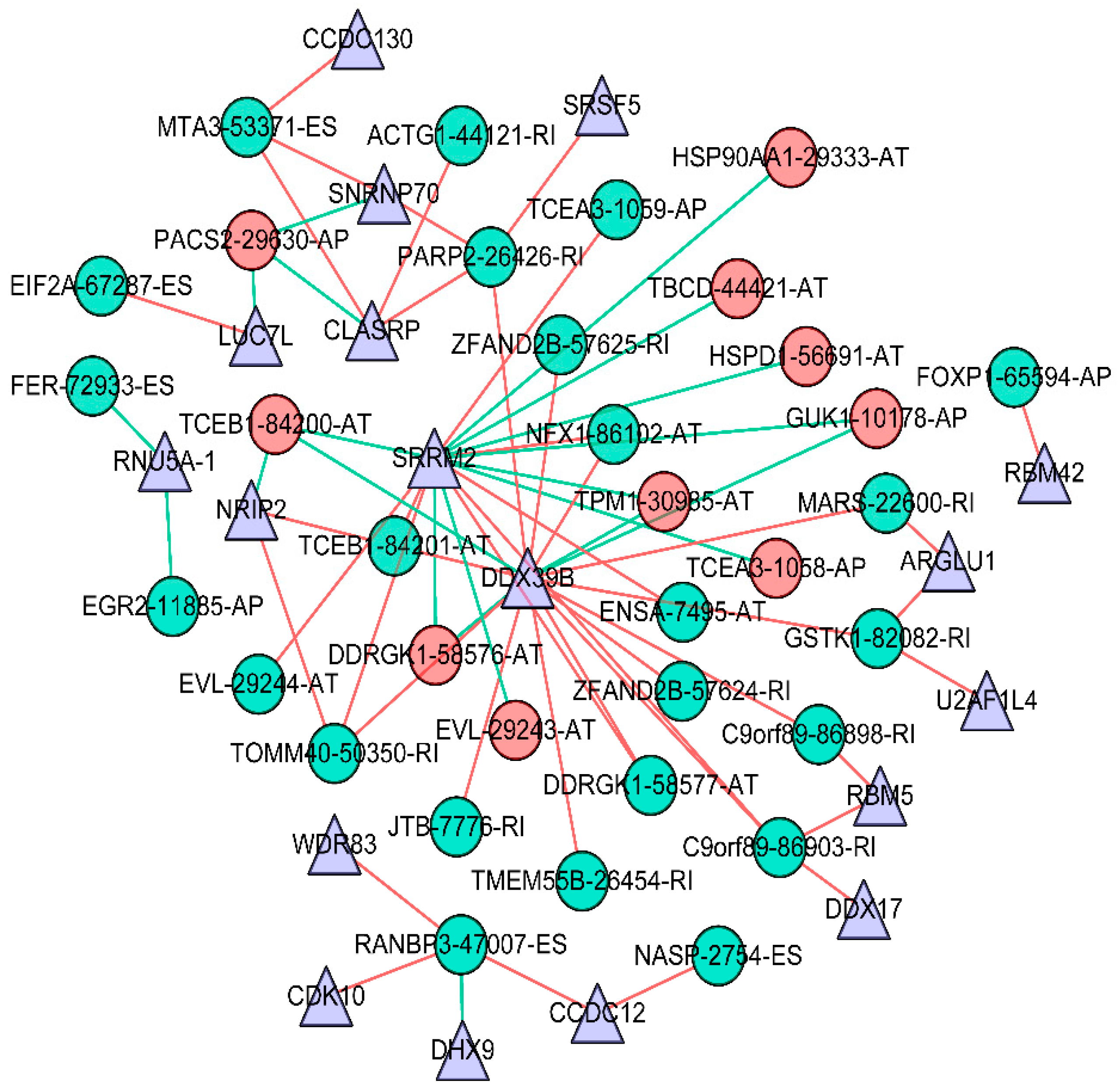
2.5. Building of the Potential Splicing Factor–Alternative Splicing Regulatory Network and Enrichment Analysis
3. Results
3.1. Information about Alternative Splicing Events
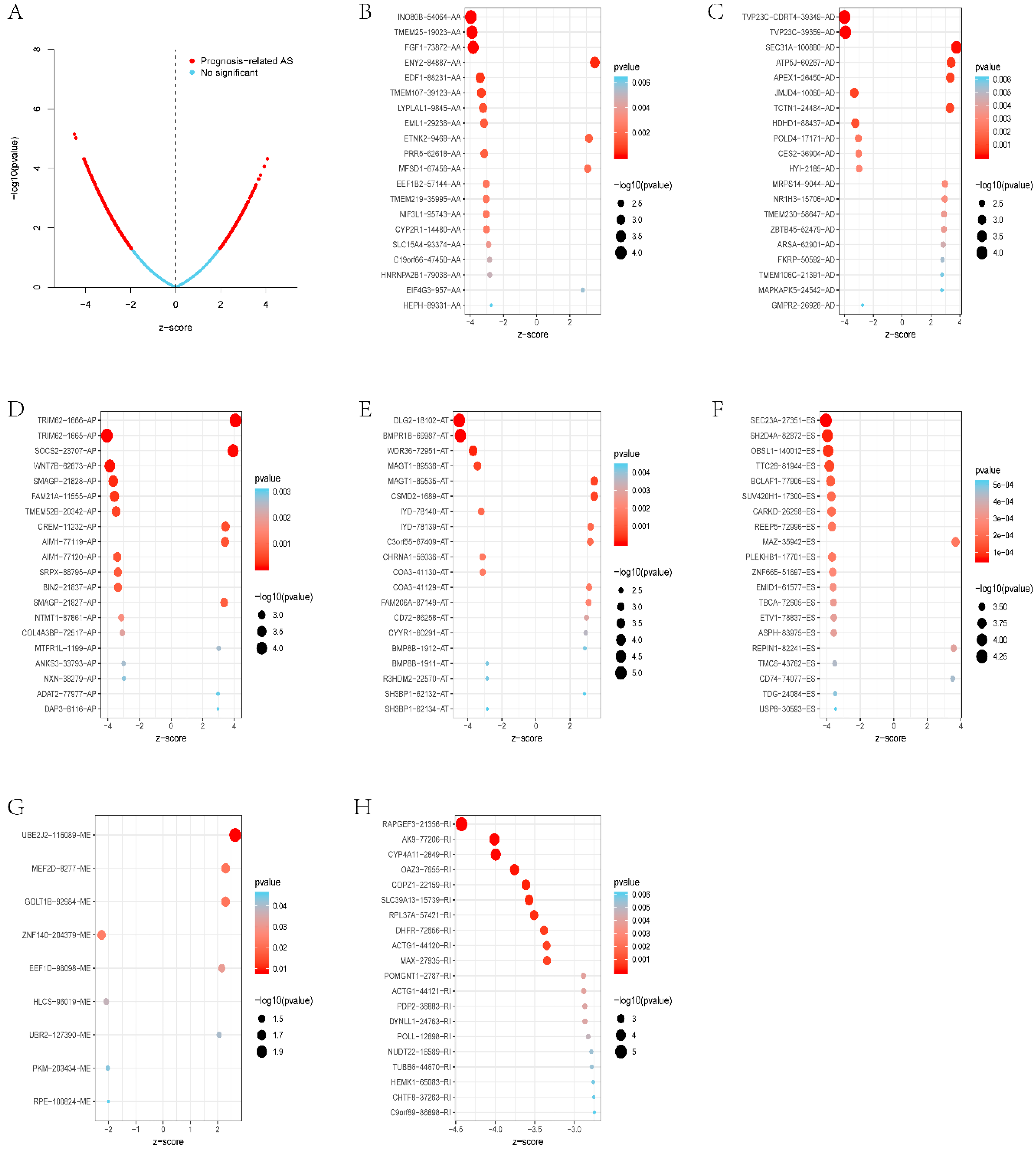
3.2. Survival-Related Alternative Splicing Events
3.3. Prognostic Signatures for Alternative Splicing Events in Breast Cancer
3.4. Survival-Associated Potential of the Splicing Factor–Alternative Splicing Regulatory Network and Enrichment Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Braithwaite, D.; Demb, J.; Henderson, L.M. Optimal breast cancer screening strategies for older women: Current perspectives. Clin. Interv. Aging 2016, 11, 111–125. [Google Scholar] [CrossRef] [PubMed]
- Gosain, R.; Pollock, Y.; Jain, D. Age-related Disparity: Breast Cancer in the Elderly. Curr. Oncol. Rep. 2016, 18, 69. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, C.E.; Lin, C.C.; Mariotto, A.B.; Siegel, R.L.; Stein, K.D.; Kramer, J.L.; Alteri, R.; Robbins, A.S.; Jemal, A. Cancer treatment and survivorship statistics, 2014. CA Cancer J. Clin. 2014, 64, 252–271. [Google Scholar] [CrossRef] [PubMed]
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef]
- de Kruijf, E.M.; Bastiaannet, E.; Ruberta, F.; de Craen, A.J.M.; Kuppen, P.J.K.; Smit, V.T.H.B.M.; van de Velde, C.J.H.; Liefers, G.J. Comparison of frequencies and prognostic effect of molecular subtypes between young and elderly breast cancer patients. Mol. Oncol. 2014, 8, 1014–1025. [Google Scholar] [CrossRef]
- Eppenberger-Castori, S.; Moore, D.H.; Thor, A.D.; Edgerton, S.M.; Kueng, W.; Eppenberger, U.; Benz, C.C. Age-associated biomarker profiles of human breast cancer. Int. J. Biochem. Cell Biol. 2002, 34, 1318–1330. [Google Scholar] [CrossRef]
- Crivellari, D.; Aapro, M.; Leonard, R.; von Minckwitz, G.; Brain, E.; Goldhirsch, A.; Veronesi, A.; Muss, H. Breast cancer in the elderly. J. Clin. Oncol. 2007, 25, 1882–1890. [Google Scholar] [CrossRef]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.M.M.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef]
- Nassar, F.J.; Nasr, R.; Talhouk, R. MicroRNAs as biomarkers for early breast cancer diagnosis, prognosis and therapy prediction. Pharmacol. Ther. 2017, 172, 34–49. [Google Scholar] [CrossRef]
- Engelhardt, E.G.; Garvelink, M.M.; de Haes, J.H.C.J.M.; van der Hoeven, J.J.M.; Smets, E.M.A.; Pieterse, A.H.; Stiggelbout, A.M. Predicting and communicating the risk of recurrence and death in women with early-stage breast cancer: A systematic review of risk prediction models. J. Clin. Oncol. 2014, 32, 238–250. [Google Scholar] [CrossRef]
- Donegan, W.L. Tumor-related prognostic factors for breast cancer. CA Cancer J. Clin. 1997, 47, 28–51. [Google Scholar] [CrossRef] [PubMed]
- Bouchardy, C.; Rapiti, E.; Fioretta, G.R.; Laissue, P.; Neyroud-Caspar, I.; Schafer, P.; Kurtz, J.; Sappino, A.-P.; Vlastos, G. Undertreatment strongly decreases prognosis of breast cancer in elderly women. J. Clin. Oncol. 2003, 21, 3580–3587. [Google Scholar] [CrossRef] [PubMed]
- Hodson, R. Precision medicine. Nature 2016, 537, S49. [Google Scholar] [CrossRef] [PubMed]
- Konig, I.R.; Fuchs, O.; Hansen, G.; von Mutius, E.; Kopp, M.V. What is precision medicine? Eur. Respir. J. 2017, 50, 1700391. [Google Scholar] [CrossRef] [PubMed]
- Le, K.-Q.; Prabhakar, B.S.; Hong, W.-J.; Li, L.-C. Alternative splicing as a biomarker and potential target for drug discovery. Acta Pharmacol. Sin. 2015, 36, 1212–1218. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Coulombe-Huntington, J.; Kang, S.; Sheynkman, G.M.; Hao, T.; Richardson, A.; Sun, S.; Yang, F.; Shen, Y.A.; Murray, R.R.; et al. Widespread Expansion of Protein Interaction Capabilities by Alternative Splicing. Cell 2016, 164, 805–817. [Google Scholar] [CrossRef] [PubMed]
- Kahles, A.; Lehmann, K.-V.; Toussaint, N.C.; Huser, M.; Stark, S.G.; Sachsenberg, T.; Stegle, O.; Kohlbacher, O.; Sander, C.; Network, C.G.A.R.; et al. Comprehensive Analysis of Alternative Splicing Across Tumors from 8705 Patients. Cancer Cell 2018, 34, 211–224. [Google Scholar] [CrossRef]
- Sebestyen, E.; Zawisza, M.; Eyras, E. Detection of recurrent alternative splicing switches in tumor samples reveals novel signatures of cancer. Nucleic Acids Res. 2015, 43, 1345–1356. [Google Scholar] [CrossRef]
- Climente-Gonzalez, H.C.; Porta-Pardo, E.; Godzik, A.; Eyras, E. The Functional Impact of Alternative Splicing in Cancer. Cell Rep. 2017, 20, 2215–2226. [Google Scholar] [CrossRef]
- Mercatante, D.R.; Bortner, C.D.; Cidlowski, J.A.; Kole, R. Modification of alternative splicing of Bcl-x pre-mRNA in prostate and breast cancer cells analysis of apoptosis and cell death. J. Biol. Chem. 2001, 276, 16411–16417. [Google Scholar] [CrossRef]
- Basaran, G.A.; Twelves, C.; Dieras, V.R.; Cortes, J.; Awada, A. Ongoing unmet needs in treating estrogen receptor-positive/HER2-negative metastatic breast cancer. Cancer Treat. Rev. 2018, 63, 144–155. [Google Scholar] [CrossRef] [PubMed]
- Ponde, N.; Brandцёo, M.; El-Hachem, G.; Werbrouck, E.; Piccart, M. Treatment of advanced HER2-positive breast cancer: 2018 and beyond. Cancer Treat. Rev. 2018, 67, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Sveen, A.; Kilpinen, S.; Ruusulehto, A.; Lothe, R.A.; Skotheim, R.I. Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene 2016, 35, 2413–2427. [Google Scholar] [CrossRef] [PubMed]
- Agafonov, D.E.; Deckert, J.; Wolf, E.; Odenwalder, P.; Bessonov, S.; Will, C.L.; Urlaub, H.; Luhrmann, R. Semiquantitative Proteomic Analysis of the Human Spliceosome via a Novel Two-Dimensional Gel Electrophoresis Method. Mol. Cell. Biol. 2011, 31, 2667–2682. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Ogawa, S. Splicing factor mutations and cancer. Wiley Interdiscip. Rev RNA 2014, 5, 445–459. [Google Scholar] [CrossRef]
- Koedoot, E.; Wolters, L.; Water, B.V.D.; Devedec, S.E.L. Splicing regulatory factors in breast cancer hallmarks and disease progression. Oncotarget 2019, 10, 6021. [Google Scholar]
- Tomczak, K.; Czerwinska, P.; Wiznerowicz, M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp. Oncol. 2015, 19, A68. [Google Scholar] [CrossRef]
- Li, Y.; Sun, N.; Lu, Z.; Sun, S.; Huang, J.; Chen, Z.; He, J. Prognostic alternative mRNA splicing signature in non-small cell lung cancer. Cancer Lett. 2017, 393, 40–51. [Google Scholar] [CrossRef]
- He, R.-Q.; Zhou, X.-G.; Yi, Q.-Y.; Deng, C.-W.; Gao, J.-M.; Chen, G.; Wang, Q.-Y. Prognostic Signature of Alternative Splicing Events in Bladder Urothelial Carcinoma Based on Spliceseq Data from 317 Cases. Cell. Physiol. Biochem. 2018, 48, 1355–1368. [Google Scholar] [CrossRef]
- Huang, Z.-G.; He, R.-Q.; Mo, Z.-N. Prognostic value and potential function of splicing events in prostate adenocarcinoma. Int. J. Oncol. 2018, 53, 2473–2487. [Google Scholar] [CrossRef]
- Lin, P.; He, R.-Q.; Ma, F.-C.; Liang, L.; He, Y.; Yang, H.; Dang, Y.-W.; Chen, G. Systematic Analysis of Survival-Associated Alternative Splicing Signatures in Gastrointestinal Pan-Adenocarcinomas. EBioMedicine 2018, 34, 46–60. [Google Scholar] [CrossRef]
- Zhu, J.; Chen, Z.; Yong, L. Systematic profiling of alternative splicing signature reveals prognostic predictor for ovarian cancer. Gynecol. Oncol. 2018, 148, 368–374. [Google Scholar] [CrossRef]
- Ryan, M.; Wong, W.C.; Brown, R.; Akbani, R.; Su, X.; Broom, B.; Melott, J.; Weinstein, J. TCGASpliceSeq a compendium of alternative mRNA splicing in cancer. Nucleic Acids Res. 2016, 44, D1018–D1022. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lichtenberg, T.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018, 173, 400–416. [Google Scholar] [CrossRef] [PubMed]
- Lex, A.; Gehlenborg, N.; Strobelt, H.; Vuillemot, R.; Pfister, H. UpSet: Visualization of Intersecting Sets. IEEE Trans. Vis. Comput. Gr. 2014, 20, 1983–1992. [Google Scholar] [CrossRef] [PubMed]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef] [PubMed]
- Tibshirani, R. The lasso method for variable selection in the Cox model. Stat. Med. 1997, 16, 385–395. [Google Scholar] [CrossRef]
- Yang, W.; Gao, Y.; Shi, Y.; Cao, L. MRM-Lasso: A Sparse Multiview Feature Selection Method via Low-Rank Analysis. IEEE Trans. Neural Netw. Learn. Syst. 2015, 26, 2801–2815. [Google Scholar] [CrossRef]
- Hanley, J.A.; McNeil, B.J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982, 143, 29–36. [Google Scholar] [CrossRef]
- Piva, F.; Giulietti, M.; Burini, A.B.; Principato, G. SpliceAid 2: A database of human splicing factors expression data and RNA target motifs. Hum. Mutat. 2012, 33, 81–85. [Google Scholar] [CrossRef]
- Koedoot, E.; Smid, M.; Foekens, J.A.; Martens, J.W.M.; Le Devedec, S.E.; van de Water, B. Co-regulated gene expression of splicing factors as drivers of cancer progression. Sci. Rep. 2019, 9, 5484. [Google Scholar] [CrossRef] [PubMed]
- Seiler, M.; Peng, S.; Agrawal, A.A.; Palacino, J.; Teng, T.; Zhu, P.; Smith, P.G.; Caesar-Johnson, S.J.; Demchok, J.A.; Felau, I.; et al. Somatic Mutational Landscape of Splicing Factor Genes and Their Functional Consequences across 33 Cancer Types. Cell Rep. 2018, 23, 282–296. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- McGlincy, N.J.; Smith, C.W.J. Alternative splicing resulting in nonsense-mediated mRNA decay: What is the meaning of nonsense? Trends Biochem. Sci. 2008, 33, 385–393. [Google Scholar] [CrossRef]
- Matlin, A.J.; Clark, F.; Smith, C.W.J. Understanding alternative splicing: Towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005, 6, 386–398. [Google Scholar] [CrossRef]
- Chen, J.; Weiss, W.A. Alternative splicing in cancer: Implications for biology and therapy. Oncogene 2015, 34, 1–14. [Google Scholar] [CrossRef]
- Pajares, M.A.J.; Ezponda, T.; Catena, R.L.; Calvo, A.; Pio, R.; Montuenga, L.M. Alternative splicing: An emerging topic in molecular and clinical oncology. Lancet Oncol. 2007, 8, 349–357. [Google Scholar] [CrossRef]
- Correa, B.R.; de Araujo, P.R.; Qiao, M.; Burns, S.C.; Chen, C.; Schlegel, R.; Agarwal, S.; Galante, P.A.F.; Penalva, L.O.F. Functional genomics analyses of RNA-binding proteins reveal the splicing regulator SNRPB as an oncogenic candidate in glioblastoma. Genome Biol. 2016, 17, 125. [Google Scholar] [CrossRef]
- Anczukow, O.; Akerman, M.; Clery, A.; Wu, J.; Shen, C.; Shirole, N.H.; Raimer, A.; Sun, S.; Jensen, M.A.; Hua, Y.; et al. SRSF1-Regulated Alternative Splicing in Breast Cancer. Mol. Cell 2015, 60, 105–117. [Google Scholar] [CrossRef]
- Akcakanat, A.; Zhang, L.; Tsavachidis, S.; Meric-Bernstam, F. The rapamycin-regulated gene expression signature determines prognosis for breast cancer. Mol. Cancer 2009, 8, 75. [Google Scholar] [CrossRef]
- Gu, Y.; Li, P.; Peng, F.; Zhang, M.; Zhang, Y.; Liang, H.; Zhao, W.; Qi, L.; Wang, H.; Wang, C.; et al. Autophagy-related prognostic signature for breast cancer. Mol. Carcinog. 2016, 55, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Moriya, T.; Ishida, T.; Shibata, H.; Sasano, H.; Ohuchi, N.; Ishioka, C. Prediction of breast cancer prognosis by gene expression profile of TP53 status. Cancer Sci. 2008, 99, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lai, Y.; Ma, J.; Liu, Y.; Bi, J.; Zhang, L.; Chen, L.; Yao, C.; Lv, W.; Chang, G.; et al. miR-17-5p suppresses cell proliferation and invasion by targeting ETV1 in triple-negative breast cancer. BMC Cancer 2017, 17, 745. [Google Scholar] [CrossRef] [PubMed]
- Ploeger, C.; Waldburger, N.; Fraas, A.; Goeppert, B.; Pusch, S.; Breuhahn, K.; Wang, X.W.; Schirmacher, P.; Roessler, S. Chromosome 8p tumor suppressor genes SH2D4A and SORBS3 cooperate to inhibit interleukin-6 signaling in hepatocellular carcinoma. Hepatology 2016, 64, 828–842. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Li, X.; Cheng, Y.; Wu, W.; Xie, Z.; Xi, Q.; Han, J.; Wu, G.; Fang, J.; Feng, Y. BCLAF1 and its splicing regulator SRSF10 regulate the tumorigenic potential of colon cancer cells. Nat. Commun. 2014, 5, 4581. [Google Scholar] [CrossRef] [PubMed]
- Gaur, S.; Shively, J.E.; Yen, Y.; Gaur, R.K. Altered splicing of CEACAM1 in breast cancer: Identification of regulatory sequences that control splicing of CEACAM1 into long or short cytoplasmic domain isoforms. Mol. Cancer 2008, 7, 46. [Google Scholar] [CrossRef]
- Zanini, I.M.Y.; Soneson, C.; Lorenzi, L.E.; Azzalin, C.M. Human cactin interacts with DHX8 and SRRM2 to assure efficient pre-mRNA splicing and sister chromatid cohesion. J. Cell. Sci. 2017, 130, 767–778. [Google Scholar] [CrossRef]
- Tomsic, J.; He, H.; Akagi, K.; Liyanarachchi, S.; Pan, Q.; Bertani, B.; Nagy, R.; Symer, D.E.; Blencowe, B.J.; de la Chapelle, A. A germline mutation in SRRM2, a splicing factor gene, is implicated in papillary thyroid carcinoma predisposition. Sci. Rep. 2015, 5, 1–13. [Google Scholar] [CrossRef]
- Awasthi, S.; Chakrapani, B.; Mahesh, A.; Chavali, P.L.; Chavali, S.; Dhayalan, A. DDX39B promotes translation through regulation of pre-ribosomal RNA levels. RNA Biol. 2018, 15, 1157–1166. [Google Scholar] [CrossRef]
- Chanarat, S.; Mishra, S.K. Emerging Roles of Ubiquitin-like Proteins in Pre-mRNA Splicing. Trends Biochem. Sci. 2018, 43, 896–907. [Google Scholar] [CrossRef]
- Zou, L.; Jaramillo, M.; Whaley, D.; Wells, A.; Panchapakesa, V.; Das, T.; Roy, P. Profilin-1 is a negative regulator of mammary carcinoma aggressiveness. Br. J. Cancer 2007, 97, 1361–1371. [Google Scholar] [CrossRef] [PubMed]
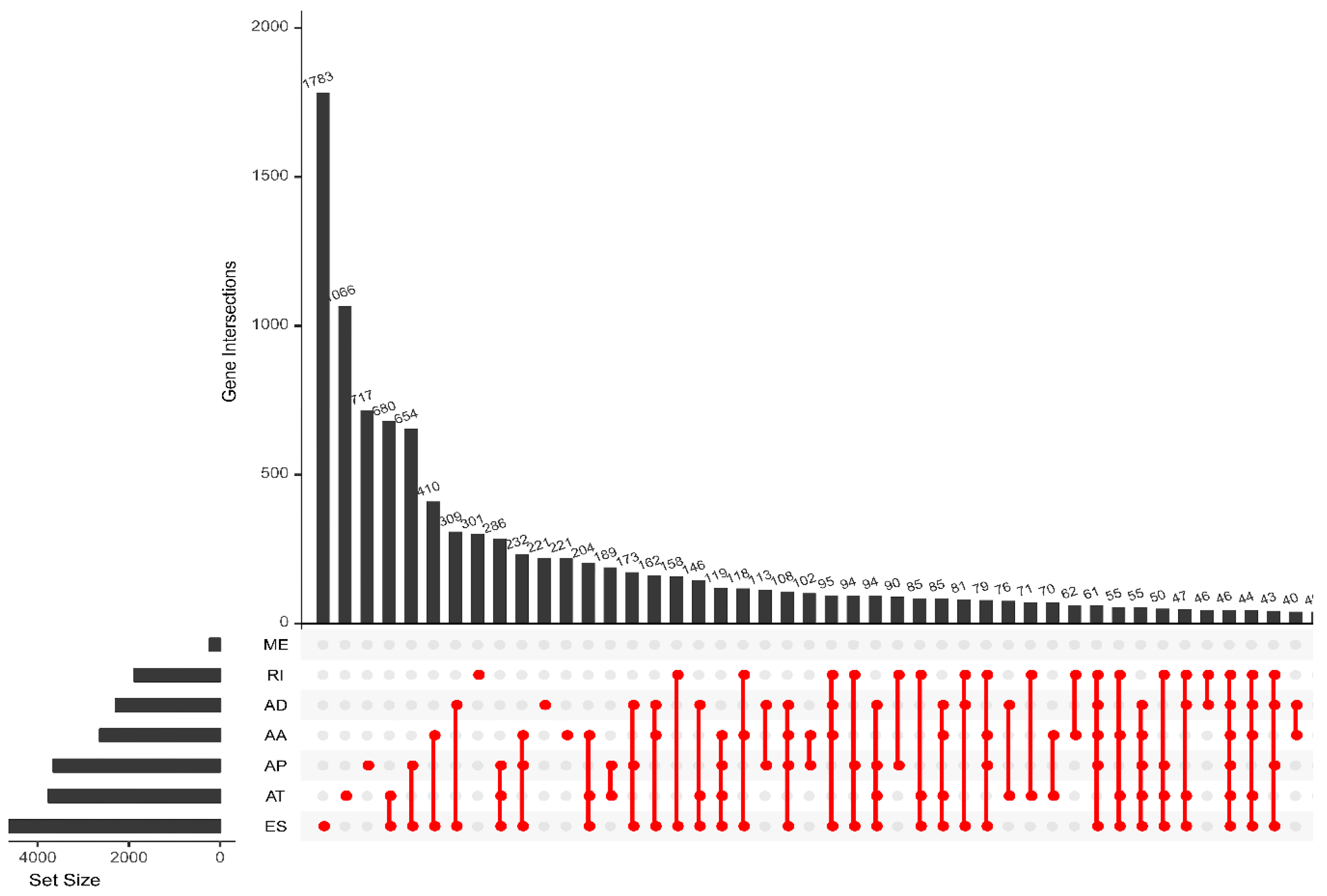
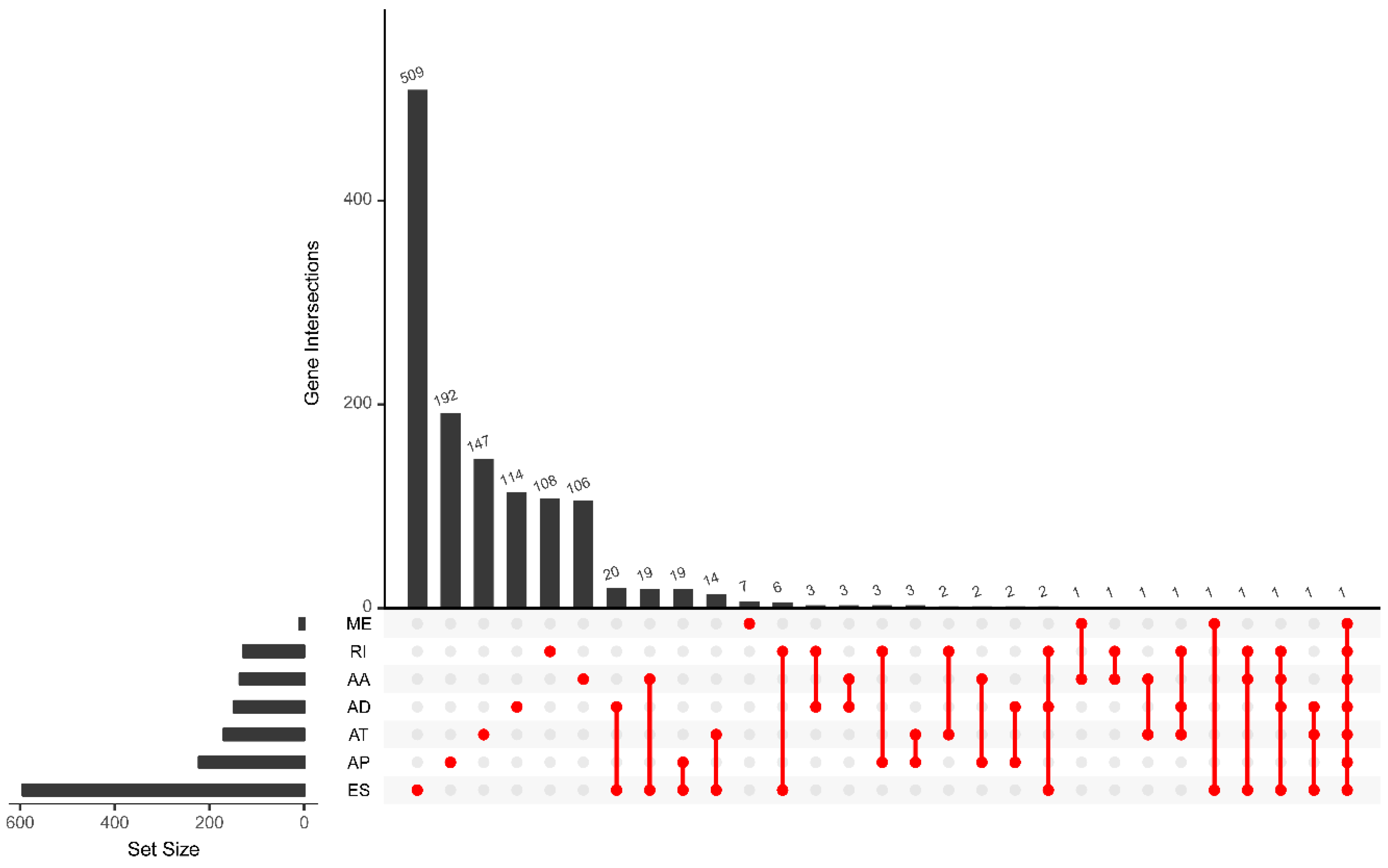


| Type | Total Splicing Events | SSEs | ||
|---|---|---|---|---|
| Splicing Events | Genes | Splicing Events | Genes | |
| AA | 3731 | 2628 | 141 | 136 |
| AD | 3246 | 2278 | 158 | 147 |
| AP | 9112 | 3654 | 308 | 221 |
| AT | 8595 | 3755 | 247 | 169 |
| ES | 17,702 | 6811 | 695 | 593 |
| ME | 233 | 227 | 9 | 9 |
| RI | 2802 | 1878 | 140 | 128 |
| Total | 45,421 | 21,231 | 1698 | 1403 |
| Clinical Variable | Univariate | Multivariate | ||
|---|---|---|---|---|
| HR (95% CI) | p-Value | HR (95% CI) | p-Value | |
| Age | 1.064 (1.030–1.101) | 0.0002 | 1.079 (1.042–1.117) | 1.65 × 10−5 |
| Stage | 1.875 (1.413–2.487) | 1.31 × 10−5 | 1.690 (0.809–3.529) | 0.1627 |
| T | 1.484 (1.120–1.966) | 0.006 | 0.887 (0.582–1.351) | 0.557 |
| M | 3.116 (1.412–6.874) | 0.0049 | 0.621 (0.192–2.009) | 0.426 |
| N | 1.775 (1.413–2.229) | 8.00 × 10−7 | 1.427 (0.946–2.153) | 0.09 |
| PS–AA | 1.085 (1.065–1.105) | 2.88 × 10−18 | 1.088 (1.065–1.111) | 6.15 × 10−15 |
| PS–AD | 1.132 (1.102–1.1164) | 3.28 × 10−19 | 1.133 (1.098–1.169) | 6.92 × 10−15 |
| PS–AP | 1.043 (1.032–1.055) | 3.54 × 10−14 | 1.041 (1.029–1.054) | 3.41 × 10−11 |
| PS–AT | 1.011 (1.006–1.016) | 1.13 × 10−5 | 1.012 (1.007–1.017) | 4.79 × 10−6 |
| PS–ES | 1.025 (1.018–1.032) | 1.87 × 10−12 | 1.023 (1.015–1.031) | 2.87 × 10−8 |
| PS–ME | 1.502 (1.309–1.724) | 6.79 × 10−9 | 1.546 (1.343–1.778) | 1.08 × 10−9 |
| PS–RI | 1.011 (1.007–1.015) | 9.69 × 10−8 | 1.011 (1.007–1.015) | 5.22 × 10−8 |
| PS–ALL | 1.006 (1.003–1.009) | 0.0002 | 1.006 (1.003–1.009) | 4.87 × 10−5 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Wang, Y.; Li, B.; Ma, W. Prognostic Value and Potential Regulatory Mechanism of Alternative Splicing in Geriatric Breast Cancer. Genes 2020, 11, 200. https://doi.org/10.3390/genes11020200
Li X, Wang Y, Li B, Ma W. Prognostic Value and Potential Regulatory Mechanism of Alternative Splicing in Geriatric Breast Cancer. Genes. 2020; 11(2):200. https://doi.org/10.3390/genes11020200
Chicago/Turabian StyleLi, Xin, Yaxuan Wang, Bingjie Li, and Wang Ma. 2020. "Prognostic Value and Potential Regulatory Mechanism of Alternative Splicing in Geriatric Breast Cancer" Genes 11, no. 2: 200. https://doi.org/10.3390/genes11020200
APA StyleLi, X., Wang, Y., Li, B., & Ma, W. (2020). Prognostic Value and Potential Regulatory Mechanism of Alternative Splicing in Geriatric Breast Cancer. Genes, 11(2), 200. https://doi.org/10.3390/genes11020200




