Dim Light at Night Induced Neurodegeneration and Ameliorative Effect of Curcumin
Abstract
1. Introduction
2. Materials and Methods
2.1. Biochemical Reagents and Instruments
2.2. Animals and Experimental Conditions
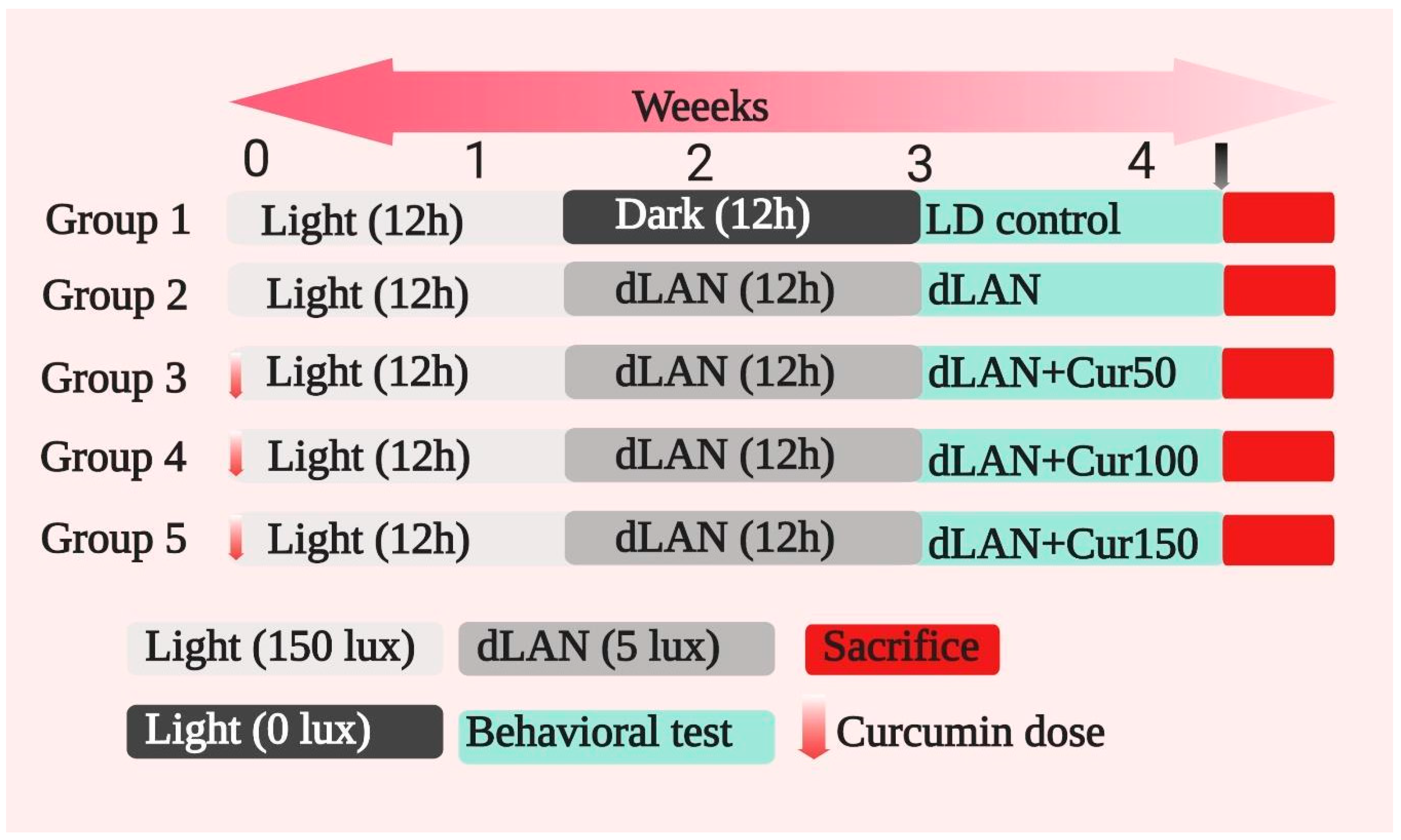
2.3. Experimental Design and Treatment
- LD control group (12:12 light (~150 lux)/dark (~0 lux), 1% CMC);
- dLAN control group (12:12 light (~150 lux)/dim light (~5 lux), 1% CMC);
- dLAN + 50 mg/kg curcumin (dLAN + Cur50);
- dLAN + 100 mg/kg curcumin (dLAN + Cur100);
- dLAN + 150 mg/kg curcumin (dLAN + Cur150).
- LD control group (12:12 light (~150 lux)/dark (~0 lux), 1% CMC) (n = 6);
- dLAN exposed group (12:12 light (~150 lux)/dim light (~5 lux), 1% CMC) (n = 6);
- dLAN + 50 mg/kg curcumin (dLAN + Cur50) (n = 6);
- dLAN + 100 mg/kg curcumin (dLAN + Cur100) (n = 6);
- dLAN + 150 mg/kg curcumin (dLAN + Cur150) (n = 6).
2.4. Behaviour Studies
2.4.1. Open-Field Test
- Total number of crossings from one square to another (locomotor activity)—NLC;
- Crossing in the centre square (stress and anxiety level)—CSE;
- Time spent in the centre square (stress and anxiety level)—DCS;
- Rearing frequency (number of times the animal stood on their hind paws) (anxiety-like behaviour)—RR.
2.4.2. The Novel Object Recognition Test
2.4.3. Morris Water Maze Test
2.4.4. Tissue Sample Collection
2.5. Estimation of Oxidative Stress
2.5.1. Lipid Peroxidation
2.5.2. Superoxide Dismutase Activity
2.5.3. Catalase Activity
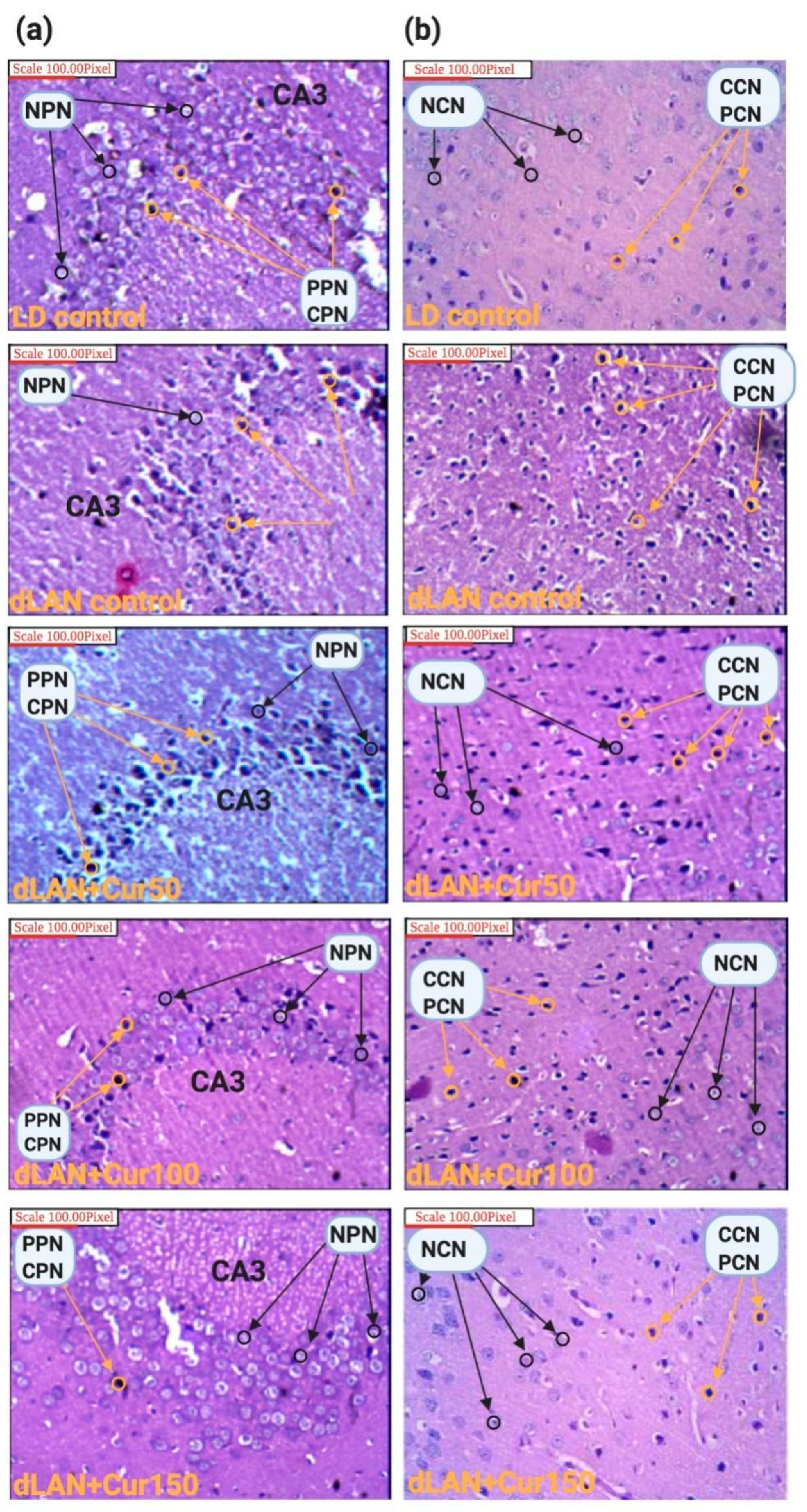
2.6. Morphometric and Histopathological Analyses
2.7. Hippocampus Protein Estimation
2.8. RNA Extraction and cDNA Synthesis
2.8.1. Quantitative RT-PCR
2.8.2. miRNA Profiling in Brain Tissue Using SYBR Green
2.9. Statistical Analyses
3. Results
3.1. Curcumin Improved the Locomotor Activity and Anxiety-Like Behaviour in Mice Exposed to dLAN
3.2. Curcumin Improved the dLAN Induced Deterioration of Spatial and Retention Memory
3.3. Curcumin Reduced dLAN Induced Abrasion of Recognition Memory
3.4. Curcumin Reduced dLAN Induced Lipid Peroxidation and Enhanced Superoxide Dismutase and Catalase Activity
3.5. Curcumin Reduced dLAN Induced Neuronal Abnormality
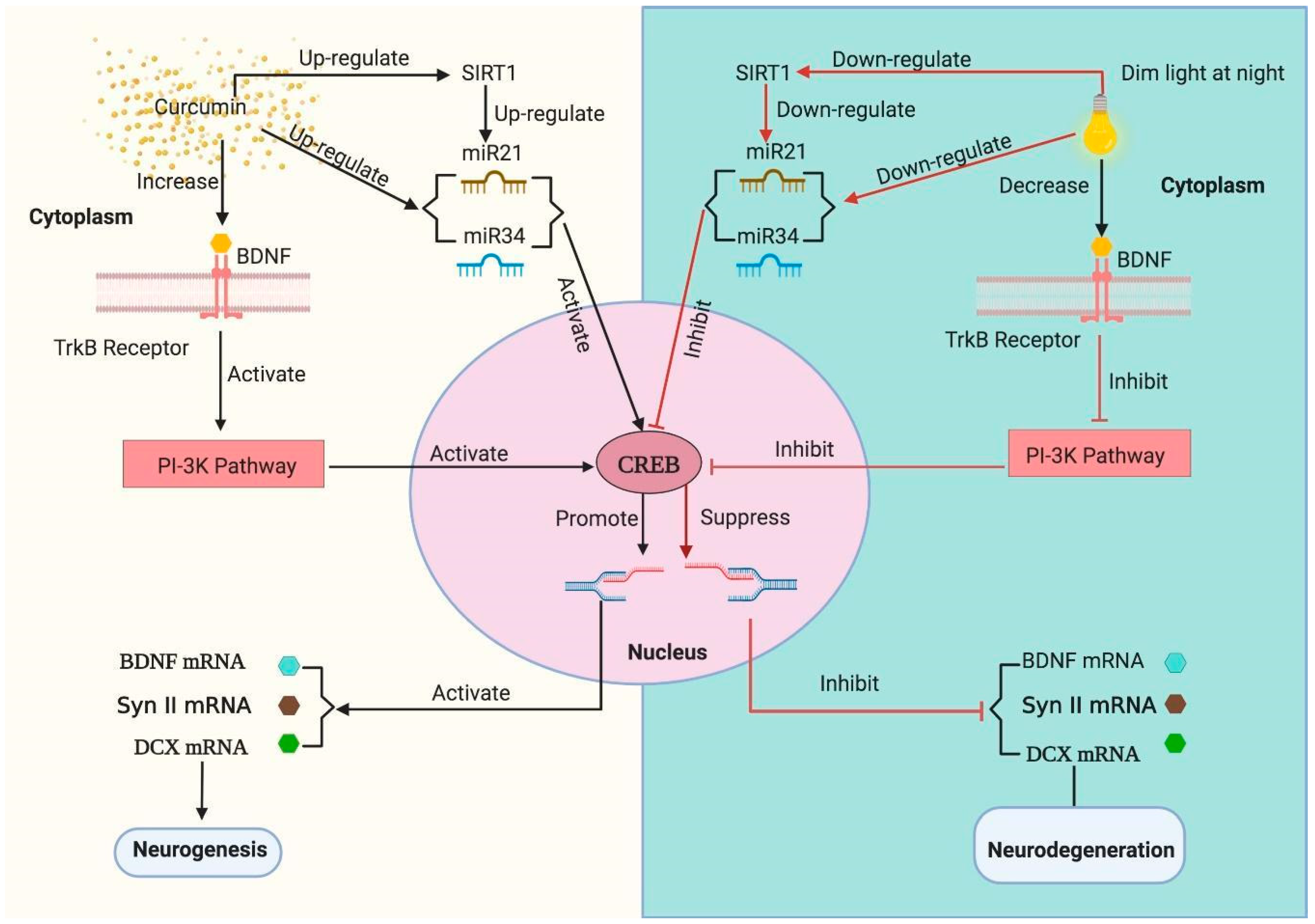
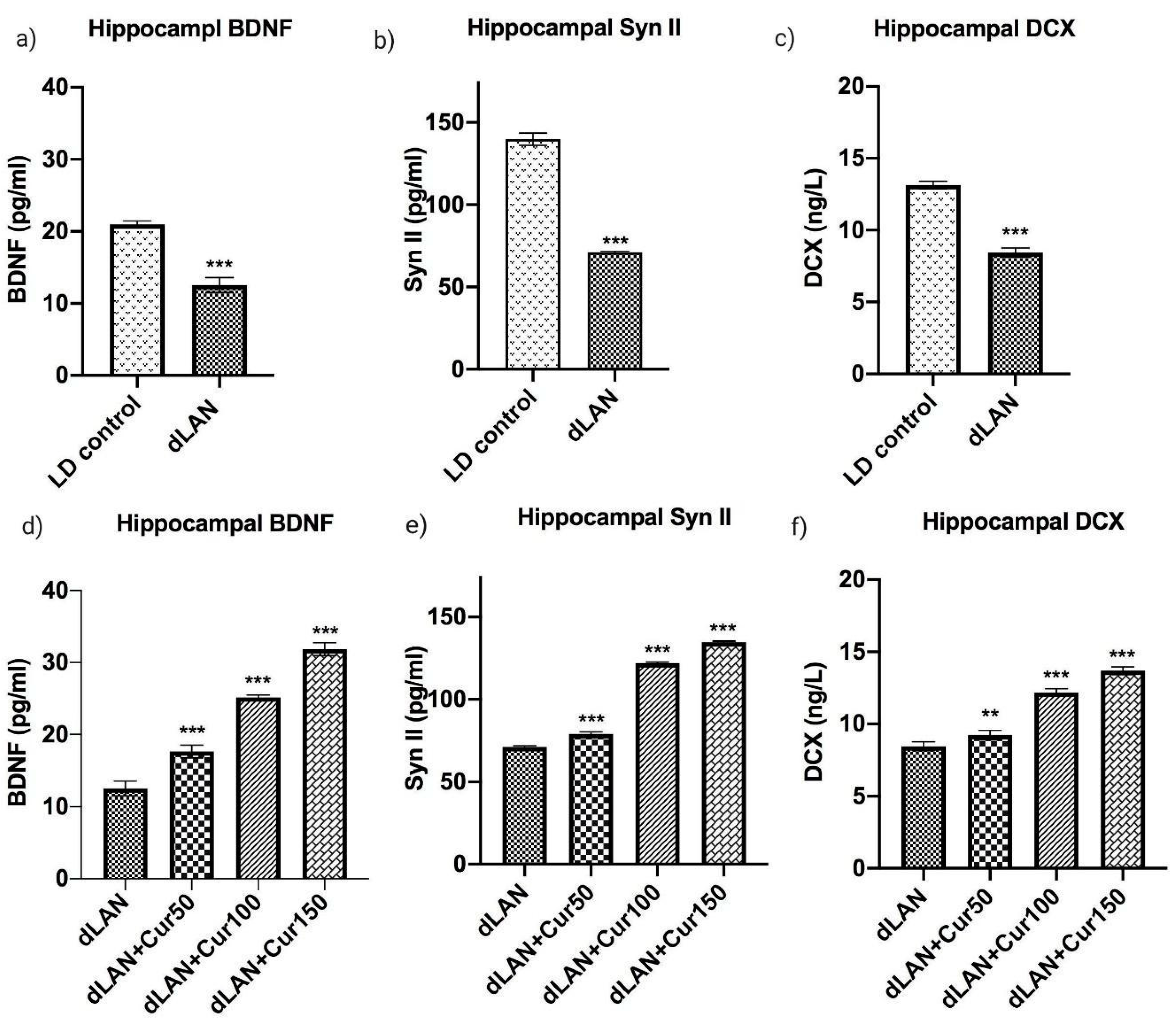
3.6. Curcumin Up-Regulates the dLAN Induced Down-Regulation of Hippocampal BDNF, Synapsin II, and DCX Proteins in Dose Dependent Manner
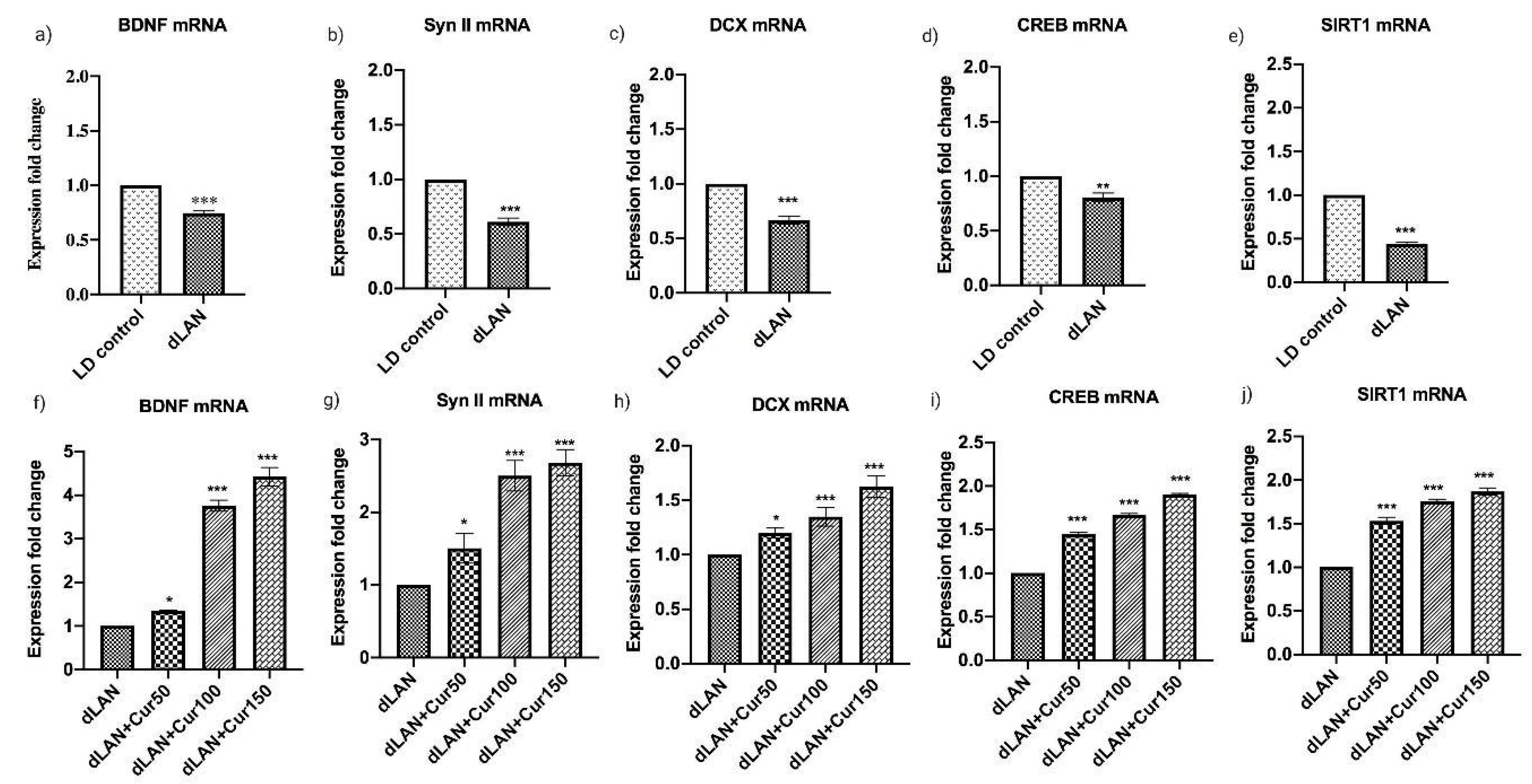
3.7. Curcumin Enhanced the mRNA Levels of Hippocampal BDNF, Synapsin II, DCX, CREB and SIRT1 in Mice Exposed to dLAN
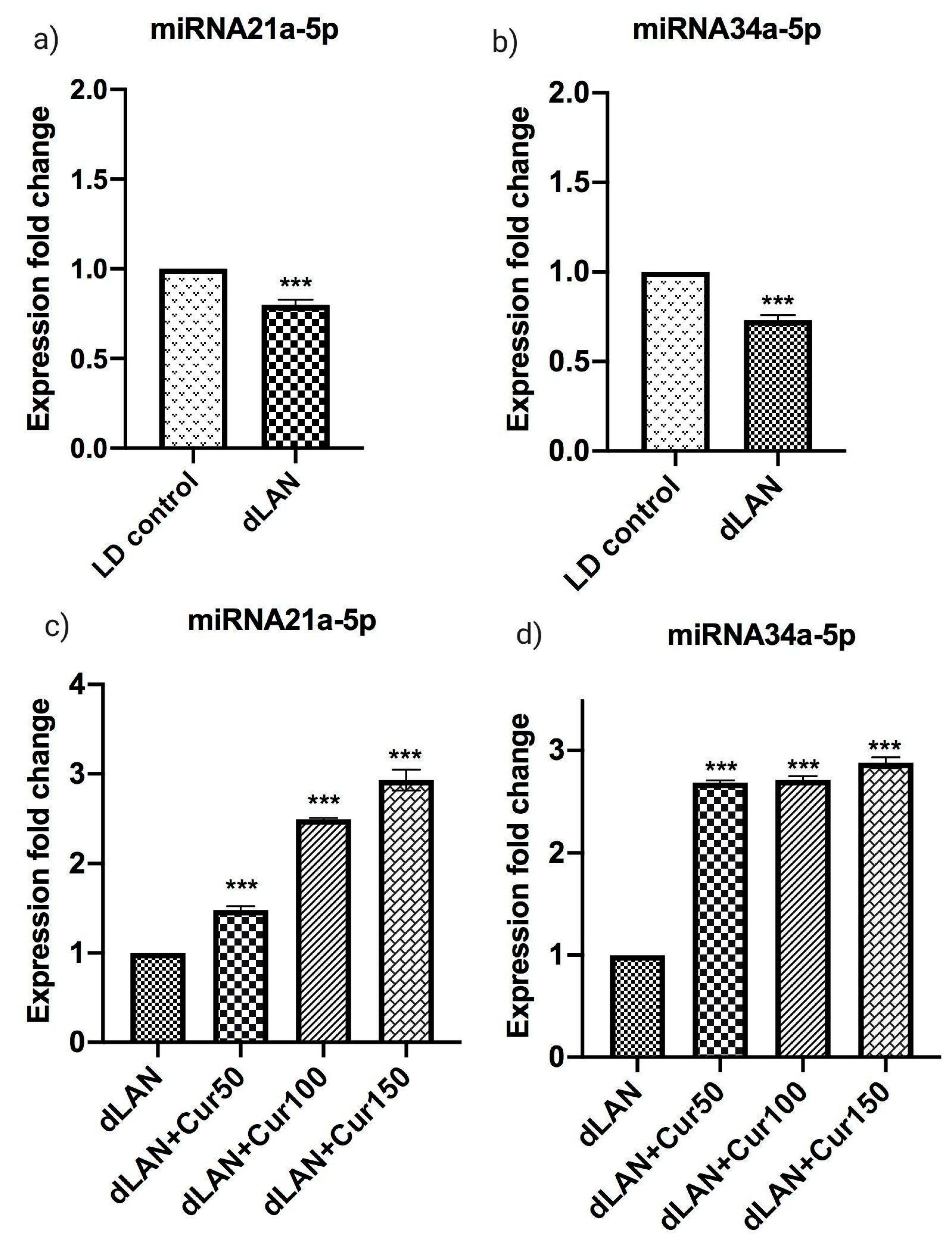
3.8. Influence of Curcumin on Altered Expression of Hippocampal miRNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Roberts, M. Modernity, mental illness and the crisis of meaning. J. Psychiatr. Ment. Health Nurs. 2007, 14, 277–281. [Google Scholar] [CrossRef] [PubMed]
- Bedrosian, T.A.; Nelson, R.J. Timing of light exposure affects mood and brain circuits. Transl. Psychiatry 2017, 7, e1017. [Google Scholar] [CrossRef]
- Bumgarner, J.R.; Walker, W.H., II; Liu, J.A.; Walton, J.C.; Nelson, R.J. Dim Light at Night Exposure Induces Cold Hyperalgesia and Mechanical Allodynia in Male Mice. Neuroscience 2020, 434, 111–119. [Google Scholar] [CrossRef]
- Castro, J.P.M.V.; Frussa-Filho, R.; Fukushiro, D.F.; Chinen, C.C.; Abílio, V.C.; Silva, R.H. Effects of long-term continuous exposure to light on memory and anxiety in mice. Physiol. Behav. 2005, 86, 218–223. [Google Scholar] [CrossRef]
- Fonken, L.K.; Nelson, R.J. Illuminating the deleterious effects of light at night. F1000 Med. Rep. 2011, 3, 18. [Google Scholar] [CrossRef]
- Gerstner, J.R. On the evolution of memory: A time for clocks. Front. Mol. Neurosci. 2012, 5, 23. [Google Scholar] [CrossRef]
- Fonken, L.K.; Bedrosian, T.A.; Zhang, N.; Weil, Z.M.; DeVries, A.C.; Nelson, R.J. Dim light at night impairs recovery from global cerebral ischemia. Exp. Neurol. 2019, 317, 100–109. [Google Scholar] [CrossRef]
- Morris, C.J.; Purvis, T.E.; Hu, K.; Scheer, F.A.J.L. Circadian misalignment increases cardiovascular disease risk factors in humans. Proc. Natl. Acad. Sci. USA 2016, 113, E1402–E1411. [Google Scholar] [CrossRef]
- Maguire, C.A.; León, S.; Carroll, R.S.; Kaiser, U.B.; Navarro, V.M. Altered circadian feeding behavior and improvement of metabolic syndrome in obese Tac1-deficient mice. Int. J. Obes. (Lond.) 2017, 41, 1798–1804. [Google Scholar] [CrossRef][Green Version]
- Russart, K.L.G.; Chbeir, S.A.; Nelson, R.J.; Magalang, U.J. Light at night exacerbates metabolic dysfunction in a polygenic mouse model of type 2 diabetes mellitus. Life Sci. 2019, 231, 116574. [Google Scholar] [CrossRef]
- Rijo-Ferreira, F.; Carvalho, T.; Afonso, C.; Sanches-Vaz, M.; Costa, R.M.; Figueiredo, L.M.; Takahashi, J.S. Sleeping sickness is a circadian disorder. Nat. Commun. 2018, 9, 62. [Google Scholar] [CrossRef] [PubMed]
- Ohkubo, R.; Chen, D. Aging: Rewiring the circadian clock. Nat. Struct. Mol. Biol. 2017, 24, 687–688. [Google Scholar] [CrossRef] [PubMed]
- Grace, A.A. Dysregulation of the dopamine system in the pathophysiology of schizophrenia and depression. Nat. Rev. Neurosci. 2016, 17, 524–532. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Jang, S.; Choe, H.K.; Chung, S.; Son, G.H.; Kim, K. Implications of Circadian Rhythm in Dopamine and Mood Regulation. Mol. Cells 2017, 40, 450–456. [Google Scholar] [CrossRef]
- Cissé, Y.M.; Peng, J.; Nelson, R.J. Effects of Dim Light at Night on Food Intake and Body Mass in Developing Mice. Front. Neurosci. 2017, 11, 294. [Google Scholar] [CrossRef]
- Cleary-Gaffney, M.; Coogan, A.N. Limited evidence for affective and diurnal rhythm responses to dim light-at-night in male and female C57Bl/6 mice. Physiol. Behav. 2018, 189, 78–85. [Google Scholar] [CrossRef]
- Gaston, K.J.; Holt, L.A. Nature, extent and ecological implications of night-time light from road vehicles. J. Appl. Ecol. 2018, 55, 2296–2307. [Google Scholar] [CrossRef]
- Rajaratnam, S.M.W.; Arendt, J. Health in a 24-h society. Lancet 2001, 358, 999–1005. [Google Scholar] [CrossRef]
- Walbeek, T.J.; Gorman, M.R. Simple Lighting Manipulations Facilitate Behavioral Entrainment of Mice to 18-h Days. J. Biol. Rhythms. 2017, 32, 309–322. [Google Scholar] [CrossRef]
- González, M.M.C. Dim Light at Night and Constant Darkness: Two Frequently Used Lighting Conditions That Jeopardize the Health and Well-being of Laboratory Rodents. Front. Neurol. 2018, 9, 609. [Google Scholar] [CrossRef]
- Bedrosian, T.A.; Weil, Z.M.; Nelson, R.J. Chronic dim light at night provokes reversible depression-like phenotype: Possible role for TNF. Mol. Psychiatry 2012, 18, 930–936. [Google Scholar] [CrossRef]
- Bedrosian, T.A.; Fonken, L.K.; Walton, J.C.; Haim, A.; Nelson, R.J. Dim light at night provokes depression-like behaviors and reduces CA1 dendritic spine density in female hamsters. Psychoneuroendocrinology 2011, 36, 1062–1069. [Google Scholar] [CrossRef]
- Hacioglu, G.; Senturk, A.; Ince, I.; Alver, A. Assessment of oxidative stress parameters of brain-derived neurotrophic factor heterozygous mice in acute stress model. Iran. J. Basic Med Sci. 2016, 19, 388–393. [Google Scholar]
- Fonken, L.K.; Kitsmiller, E.; Smale, L.; Nelson, R.J. Dim Nighttime Light Impairs Cognition and Provokes Depressive-Like Responses in a Diurnal Rodent. J. Biol. Rhythms. 2012, 27, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Fujioka, A.; Fujioka, T.; Tsuruta, R.; Izumi, T.; Kasaoka, S.; Maekawa, T. Effects of a constant light environment on hippocampal neurogenesis and memory in mice. Neurosci. Lett. 2011, 488, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.K.; Agarwal, S.; Tripathi, A.; Chaturvedi, R.K. Bisphenol-A Mediated Inhibition of Hippocampal Neurogenesis Attenuated by Curcumin via Canonical Wnt Pathway. Mol. Neurobiol. 2015, 53, 3010–3029. [Google Scholar] [CrossRef] [PubMed]
- Ułamek-Kozioł, M.; Czuczwar, S.J.; Januszewski, S.; Pluta, R. Substantiation for the Use of Curcumin during the Development of Neurodegeneration after Brain Ischemia. Int. J. Mol. Sci. 2020, 21, 517. [Google Scholar] [CrossRef]
- Anand, P.; Kunnumakkara, A.B.; Newman, R.A.; Aggarwal, B.B. Bioavailability of Curcumin: Problems and Promises. Mol. Pharm. 2007, 4, 807–818. [Google Scholar] [CrossRef]
- Perrone, L.; Squillaro, T.; Napolitano, F.; Terracciano, C.; Sampaolo, S.; Melone, M.A.B. The Autophagy Signaling Pathway: A Potential Multifunctional Therapeutic Target of Curcumin in Neurological and Neuromuscular Diseases. Nutrients 2019, 11, 1881. [Google Scholar] [CrossRef]
- Popa-Wagner, A.; Dumitrascu, D.I.; Capitanescu, B.; Petcu, E.B.; Surugiu, R.; Fang, W.-H.; Dumbrava, D.-A. Dietary habits, lifestyle factors and neurodegenerative diseases. Neural Regen. Res. 2020, 15, 394–400. [Google Scholar] [CrossRef]
- Sharma, R.A.; Gescher, A.J.; Steward, W.P. Curcumin: The story so far. Eur. J. Cancer 2005, 41, 1955–1968. [Google Scholar] [CrossRef]
- Rakotoarisoa, M.; Angelov, B.; Espinoza, S.; Khakurel, K.; Bizien, T.; Angelova, A. Cubic liquid crystalline nanostructures involving catalase and curcumin: BioSAXS study and catalase peroxidatic function after cubosomal nanoparticle treatment of differentiated SH-SY5Y cells. Molecules 2019, 24, 3058. [Google Scholar] [CrossRef] [PubMed]
- Rakotoarisoa, M.; Angelova, A. Amphiphilic nanocarrier systems for curcumin delivery in neurodegenerative disorders. Medicines 2018, 5, 126. [Google Scholar] [CrossRef] [PubMed]
- Guerzoni, L.P.; Nicolas, V.; Angelova, A. In vitro modulation of TrkB receptor signaling upon sequential delivery of curcumin-DHA loaded carriers towards promoting neuronal survival. Pharm. Res. 2017, 34, 492–505. [Google Scholar] [CrossRef] [PubMed]
- Rakotoarisoa, M.; Angelov, B.; Garamus, V.M.; Angelova, A. Curcumin-and fish oil-loaded spongosome and cubosome nanoparticles with neuroprotective potential against H2O2-induced oxidative stress in differentiated human SH-SY5Y cells. ACS Omega 2019, 4, 3061–3073. [Google Scholar] [CrossRef]
- Conboy, L.; Foley, A.G.; O’Boyle, N.M.; Lawlor, M.; Gallagher, H.C.; Murphy, K.J.; Regan, C.M. Curcumin-induced degradation of PKCδ is associated with enhanced dentate NCAM PSA expression and spatial learning in adult and aged Wistar rats. Biochem. Pharmacol. 2009, 77, 1254–1265. [Google Scholar] [CrossRef]
- Gupta, S.K.; Kumar, B.; Nag, T.C.; Agrawal, S.S.; Agrawal, R.; Agrawal, P.; Saxena, R.; Srivastava, S. Curcumin Prevents Experimental Diabetic Retinopathy in Rats Through Its Hypoglycemic, Antioxidant, and Anti-Inflammatory Mechanisms. J. Ocul. Pharmacol. Ther. 2011, 27, 123–130. [Google Scholar] [CrossRef]
- Ikram, M.; Saeed, K.; Khan, A.; Muhammad, T.; Khan, M.S.; Jo, M.G.; Rehman, S.U.; Kim, M.O. Natural Dietary Supplementation of Curcumin Protects Mice Brains against Ethanol-Induced Oxidative Stress-Mediated Neurodegeneration and Memory Impairment via Nrf2/TLR4/RAGE Signaling. Nutrients 2019, 11, 1082. [Google Scholar] [CrossRef]
- Khan, M.S.; Muhammad, T.; Ikram, M.; Kim, M.O. Dietary Supplementation of the Antioxidant Curcumin Halts Systemic LPS-Induced Neuroinflammation-Associated Neurodegeneration and Memory/Synaptic Impairment via the JNK/NF-κB/Akt Signaling Pathway in Adult Rats. Oxid. Med. Cell. Longev. 2019, 2019, 7860650. [Google Scholar] [CrossRef]
- Pyrzanowska, J.; Piechal, A.; Blecharz-Klin, K.; Lehner, M.; Skórzewska, A.; Turzyńska, D.; Sobolewska, A.; Plaznik, A.; Widy-Tyszkiewicz, E. The influence of the long-term administration of Curcuma longa extract on learning and spatial memory as well as the concentration of brain neurotransmitters and level of plasma corticosterone in aged rats. Pharmacol. Biochem. Behav. 2010, 95, 351–358. [Google Scholar] [CrossRef]
- Rinwa, P.; Kumar, A. Piperine potentiates the protective effects of curcumin against chronic unpredictable stress-induced cognitive impairment and oxidative damage in mice. Brain Res. 2012, 1488, 38–50. [Google Scholar] [CrossRef]
- Namgyal, D.; Ali, S.; Mehta, R.; Sarwat, M. The Neuroprotective effect of curcumin against Cd-induced neurotoxicity and hippocampal neurogenesis promotion through CREB-BDNF signaling pathway. Toxicology 2020, 442, 152542. [Google Scholar] [CrossRef] [PubMed]
- Bailey, K.R.; Crawley, J.N. Anxiety-related behaviors in mice. In Methods of Behavior Analysis in Neuroscience, 2nd ed.; Buccafusco, J.J., Ed.; CRC Press: Boca Raton, FL, USA, 2009; pp. 77–101. [Google Scholar]
- Lueptow, L.M. Novel Object Recognition Test for the Investigation of Learning and Memory in Mice. J. Vis. Exp. JoVE 2017. [Google Scholar] [CrossRef] [PubMed]
- Vorhees, C.V.; Williams, M.T. Morris water maze: Procedures for assessing spatial and related forms of learning and memory. Nat. Protoc. 2006, 1, 848–858. [Google Scholar] [CrossRef] [PubMed]
- Wills, E.D. Mechanisms of lipid peroxide formation in animal tissues. Biochem. J. 1966, 99, 667–676. [Google Scholar] [CrossRef]
- Weydert, C.J.; Cullen, J.J. Measurement of superoxide dismutase, catalase and glutathione peroxidase in cultured cells and tissue. Nat. Protoc. 2010, 5, 51–66. [Google Scholar] [CrossRef]
- Gupta, S.; Singh, P.; Sharma, B.; Sharma, B. Neuroprotective effects of agomelatine and vinpocetine against chronic cerebral hypoperfusion induced vascular dementia. Curr. Neurovasc. Res. 2015, 12, 240–252. [Google Scholar] [CrossRef]
- Aggarwal, M.; Burnsed, J.; Martin, L.J.; Northington, F.J.; Zhang, J. Imaging neurodegeneration in the mouse hippocampus after neonatal hypoxia-ischemia using oscillating gradient diffusion MRI. Magn. Reson. Med. 2014, 72, 829–840. [Google Scholar] [CrossRef]
- Sarwat, M.; Naqvi, A.R. Heterologous expression of rice calnexin (OsCNX) confers drought tolerance in Nicotiana tabacum. Mol. Biol. Rep. 2013, 40, 5451–5464. [Google Scholar] [CrossRef]
- Hewlings, S.J.; Kalman, D.S. Curcumin: A review of its’ effects on human health. Foods 2017, 6, 92. [Google Scholar] [CrossRef]
- Namgyal, D.; Kumari, C.; Ali, S.; Mehta, R.; Sarwat, M. Curcumin improves the behaviour and memory in mice by modulating the core circadian genes and their associated micro-RNAs. JPP 2020, in press. [Google Scholar]
- Stenvers, D.J.; van Dorp, R.; Foppen, E.; Mendoza, J.; Opperhuizen, A.-L.; Fliers, E.; Bisschop, P.H.; Meijer, J.H.; Kalsbeek, A.; Deboer, T. Dim light at night disturbs the daily sleep-wake cycle in the rat. Sci. Rep. 2016, 6, 35662. [Google Scholar] [CrossRef] [PubMed]
- Cissé, Y.M.; Peng, J.; Nelson, R.J. Dim light at night prior to adolescence increases adult anxiety-like behaviors. Chronobiol. Int. 2016, 33, 1473–1480. [Google Scholar] [CrossRef] [PubMed]
- Fonken, L.K.; Aubrecht, T.G.; Meléndez-Fernández, O.H.; Weil, Z.M.; Nelson, R.J. Dim light at night disrupts molecular circadian rhythms and increases body weight. J. Biol. Rhythms. 2013, 28, 262–271. [Google Scholar] [CrossRef] [PubMed]
- Taufique, S.T.; Prabhat, A.; Kumar, V. Light at night affects hippocampal and nidopallial cytoarchitecture: Implication for impairment of brain function in diurnal corvids. J. Exp. Zool. Part A Ecol. Integr. Physiol. 2018, 331, 149–156. [Google Scholar] [CrossRef]
- Toda, T.; Parylak, S.L.; Linker, S.B.; Gage, F.H. The role of adult hippocampal neurogenesis in brain health and disease. Mol. Psychiatry 2019, 24, 67–87. [Google Scholar] [CrossRef]
- Taufique, S.K.T.; Prabhat, A.; Kumar, V. Illuminated night alters hippocampal gene expressions and induces depressive-like responses in diurnal corvids. Eur. J. Neurosci. 2018, 48, 3005–3018. [Google Scholar] [CrossRef]
- Michán, S.; Li, Y.; Chou, M.M.-H.; Parrella, E.; Ge, H.; Long, J.M.; Allard, J.S.; Lewis, K.; Miller, M.; Xu, W.; et al. SIRT1 is essential for normal cognitive function and synaptic plasticity. J. Neurosci. 2010, 30, 9695–9707. [Google Scholar] [CrossRef]
- Jeong, H.; Cohen, D.E.; Cui, L.; Supinski, A.; Savas, J.N.; Mazzulli, J.R.; Yates, J.R., 3rd; Bordone, L.; Guarente, L.; Krainc, D. Sirt1 mediates neuroprotection from mutant huntingtin by activation of the TORC1 and CREB transcriptional pathway. Nat. Med. 2011, 18, 159–165. [Google Scholar] [CrossRef]
- Nam, S.M.; Choi, J.H.; Yoo, D.Y.; Kim, W.; Jung, H.Y.; Kim, J.W.; Yoo, M.; Lee, S.; Kim, C.J.; Yoon, Y.S.; et al. Effects of curcumin (Curcuma longa) on learning and spatial memory as well as cell proliferation and neuroblast differentiation in adult and aged mice by upregulating brain-derived neurotrophic factor and CREB signaling. J. Med. Food 2014, 17, 641–649. [Google Scholar] [CrossRef]
- Xu, Y.; Ku, B.; Cui, L.; Li, X.; Barish, P.A.; Foster, T.C.; Ogle, W.O. Curcumin reverses impaired hippocampal neurogenesis and increases serotonin receptor 1A mRNA and brain-derived neurotrophic factor expression in chronically stressed rats. Brain Res. 2007, 1162, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, A.R.; Sarwat, M.; Hasan, S.; Roychodhury, N. Biogenesis, functions and fate of plant microRNAs. J. Cell. Physiol. 2012, 227, 3163–3168. [Google Scholar] [CrossRef] [PubMed]
- Sarwat, M.; Naqvi, A.R.; Ahmad, P.; Ashraf, M.; Akram, N.A. Phytohormones and microRNAs as sensors and regulators of leaf senescence: Assigning macro roles to small molecules. Biotechnol. Adv. 2013, 31, 1153–1171. [Google Scholar] [CrossRef] [PubMed]
- Hébert, S.S.; De Strooper, B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009, 32, 199–206. [Google Scholar] [CrossRef]
- Yelamanchili, S.V.; Fox, H.S. Defining larger roles for "tiny" RNA molecules: Role of miRNAs in neurodegeneration research. J. Neuroimmune Pharmacol. 2010, 5, 63–69. [Google Scholar] [CrossRef]
- Alvarez-Erviti, L.; Seow, Y.; Schapira, A.H.V.; Rodriguez-Oroz, M.C.; Obeso, J.A.; Cooper, J.M. Influence of microRNA deregulation on chaperone-mediated autophagy and α-synuclein pathology in Parkinson’s disease. Cell. Death Dis. 2013, 4, e545. [Google Scholar] [CrossRef]
- Montalban, E.; Mattugini, N.; Ciarapica, R.; Provenzano, C.; Savino, M.; Scagnoli, F.; Prosperini, G.; Carissimi, C.; Fulci, V.; Matrone, C.; et al. MiR-21 is an Ngf-modulated microRNA that supports Ngf signaling and regulates neuronal degeneration in PC12 cells. Neuromol. Med. 2014, 16, 415–430. [Google Scholar] [CrossRef]
- Shi, J. Regulatory networks between neurotrophins and miRNAs in brain diseases and cancers. Acta Pharmacol. Sin. 2015, 36, 149–157. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, F.; Pan, C.; Lou, Y.; Zhang, P.; Guo, S.; Yin, J.; Deng, Z. Effects of low temperatures on proliferation-related signaling pathways in the hippocampus after traumatic brain injury. Exp. Biol. Med. 2012, 237, 1424–1432. [Google Scholar] [CrossRef]
- Yamakuchi, M.; Lowenstein, C.J. MiR-34, SIRT1, and p53: The feedback loop. Cell Cycle 2009, 8, 712–715. [Google Scholar] [CrossRef]
- Miñones-Moyano, E.; Porta, S.; Escaramís, G.; Rabionet, R.; Iraola, S.; Kagerbauer, B.; Espinosa-Parrilla, Y.; Ferrer, I.; Estivill, X.; Martí, E. MicroRNA profiling of Parkinson’s disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum. Mol. Genet. 2011, 20, 3067–3078. [Google Scholar] [CrossRef] [PubMed]
- Lou, G.; Liu, Y.; Wu, S.; Xue, J.; Yang, F.; Fu, H.; Zheng, M.; Chen, Z. The p53/miR-34a/SIRT1 Positive Feedback Loop in Quercetin-Induced Apoptosis. Cell. Physiol. Biochem. 2015, 35, 2192–2202. [Google Scholar] [CrossRef] [PubMed]
- Wurzelmann, M.; Romeika, J.; Sun, D. Therapeutic potential of brain-derived neurotrophic factor (BDNF) and a small molecular mimics of BDNF for traumatic brain injury. Neural. Regen. Res. 2017, 12, 7–12. [Google Scholar] [CrossRef] [PubMed]
| Behavioural Parameters | |||||||
|---|---|---|---|---|---|---|---|
| S.No. | Test | Parameters | Group | t-Test | |||
| 1 | LD control | dLAN exposed | t | df | p | ||
| OFT | NLC | 152.75 ± 5.50 a | 97.25 ± 1.47 b | 19.27 | 6 | p < 0.001 | |
| CSE (n) | 06.00 ± 0.81 a | 3.45 ± 0.42 b | 5.549 | 6 | p < 0.01 | ||
| CSD (s) | 12.48 ± 0.55 a | 7.91 ± 0.58 b | 11.45 | 6 | p < 0.001 | ||
| RR (n) | 45.25 ± 1.70 a | 23.25 ± 1.50 b | 19.36 | 6 | p < 0.001 | ||
| 2 | MWM | TSFP (s) | 42.25 ± 0.95 a | 64.25 ± 1.70 b | 22.47 | 6 | p < 0.001 |
| TSPQ (s) | 49.50 ± 1.29 a | 23.25 ± 1.70 b | 24.52 | 6 | p < 0.001 | ||
| 3 | NOR | T2/T1 | 1.24 ± 0.01 a | 0.48 ± 0.05 b | 29.04 | 6 | p < 0.001 |
| Biochemical Parameters | |||||||
| 4 | MDA (nM/mg) | 2.10 ± 0.01 a | 3.17 ± 0.05 b | 17.45 | 6 | p < 0.001 | |
| 5 | SOD (unit/mg) | 6.46 ± 0.31 a | 2.44 ± 0.22 b | 24.32 | 6 | p < 0.001 | |
| 6 | CAT (unit/mg) | 3.37 ± 0.08 a | 2.33 ± 0.08 b | 10.55 | 6 | p < 0.001 | |
| Behavioural Parameters | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| S.No. | Test | Parameters | Group | One-Way ANOVA | |||||
| 1 | OFT | dLAN | dLAN + Cur50 | dLAN + Cur100 | dLAN + Cur150 | F | df | p | |
| NLC (n) | 97.25 ± 1.47 | 107.75 ± 1.70 a | 120.00 ± 4.39 b | 124.25 ± 2.75 c | 73.17 | 12 | p < 0.001 | ||
| CSE (n) | 3.45 ± 0.42 | 4.50 ± 0.57 a | 5.50 ± 0.57 b | 6.25 ± 0.5 c | 21.65 | 12 | p < 0.001 | ||
| CSD (s) | 7.91 ± 0.58 | 8.11 ± 0.43 | 9.56 ± 0.45 | 14.37 ± 1.81 a | 36.22 | 12 | p < 0.001 | ||
| RR (n) | 23.25 ± 1.50 | 30.00 ± 1.82 a | 39.00 ± 1.41 b | 40.25 ± 0.95 c | 120.80 | 12 | p < 0.001 | ||
| 2 | MWM | TSFP (s) | 64.25 ± 1.70 | 58.39 ± 0.74 a | 55.87 ± 1.39 b | 42.76 ± 0.50 c | 232.80 | 12 | p < 0.001 |
| TSPQ (s) | 23.25 ± 1.70 | 26.41 ± 0.72 a | 32.50 ± 1.53 b | 44.82 ± 1.27 c | 194.70 | 12 | p < 0.001 | ||
| 3 | NOR | T2/T1 | 0.48 ± 0.05 | 0.63 ± 0.018 a | 0.94 ± 0.04 b | 1.19 ± 0.04 c | 227.30 | 12 | p < 0.001 |
| Biochemical Parameters | |||||||||
| 4 | MDA (nM/mg) | 3.17 ± 0.0476 | 2.66 ± 0.09 a | 1.89 ± 0.14 b | 1.52 ± 0.09 c | 167.40 | 12 | p < 0.001 | |
| 5 | SOD (unit/mg) | 2.44 ± 0.219 | 3.56 ± 0.10 a | 4.74 ± 0.18 b | 5.55 ± 0.21 c | 302.70 | 12 | p < 0.001 | |
| 6 | CAT (unit/mg) | 2.33 ± 0.08 | 2.64 ± 0.06 | 3.04 ± 0.11 a | 3.82 ± 0.13 b | 101.00 | 12 | p < 0.001 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Namgyal, D.; Chandan, K.; Sultan, A.; Aftab, M.; Ali, S.; Mehta, R.; El-Serehy, H.A.; Al-Misned, F.A.; Sarwat, M. Dim Light at Night Induced Neurodegeneration and Ameliorative Effect of Curcumin. Cells 2020, 9, 2093. https://doi.org/10.3390/cells9092093
Namgyal D, Chandan K, Sultan A, Aftab M, Ali S, Mehta R, El-Serehy HA, Al-Misned FA, Sarwat M. Dim Light at Night Induced Neurodegeneration and Ameliorative Effect of Curcumin. Cells. 2020; 9(9):2093. https://doi.org/10.3390/cells9092093
Chicago/Turabian StyleNamgyal, Dhondup, Kumari Chandan, Armiya Sultan, Mehreen Aftab, Sher Ali, Rachna Mehta, Hamed A. El-Serehy, Fahad A. Al-Misned, and Maryam Sarwat. 2020. "Dim Light at Night Induced Neurodegeneration and Ameliorative Effect of Curcumin" Cells 9, no. 9: 2093. https://doi.org/10.3390/cells9092093
APA StyleNamgyal, D., Chandan, K., Sultan, A., Aftab, M., Ali, S., Mehta, R., El-Serehy, H. A., Al-Misned, F. A., & Sarwat, M. (2020). Dim Light at Night Induced Neurodegeneration and Ameliorative Effect of Curcumin. Cells, 9(9), 2093. https://doi.org/10.3390/cells9092093






