An Evolutionary Cancer Epigenetic Approach Revealed DNA Hypermethylation of Ultra-Conserved Non-Coding Elements in Squamous Cell Carcinoma of Different Mammalian Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Study Population and Brushing Collection
2.3. Ultra-Conserved Non-Coding Element Selection and Primer Designing
2.4. DNA Methylation Analysis
3. Results
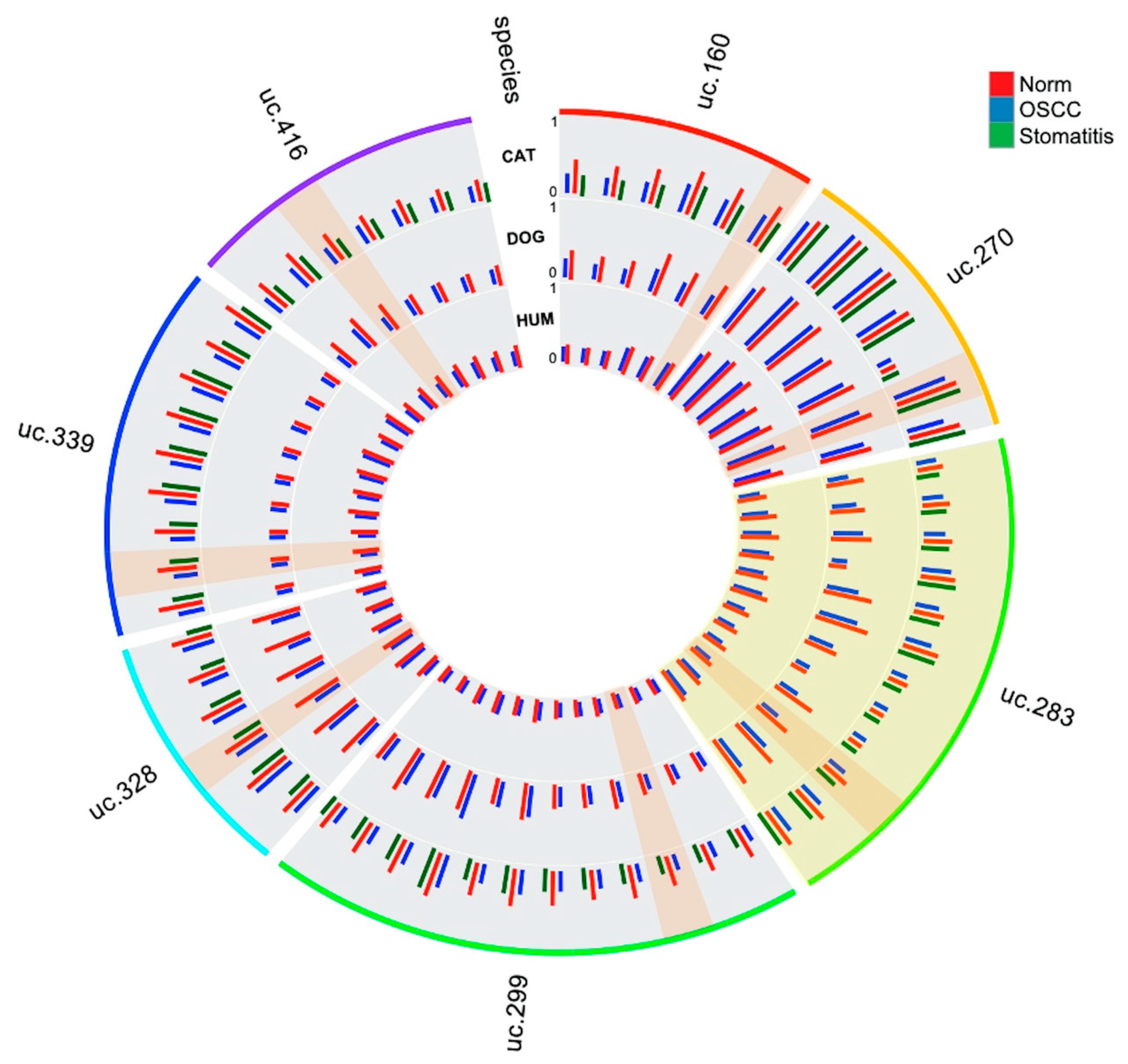
3.1. Methylation Plotter Analysis
3.2. Principal Component Analysis (PCA)
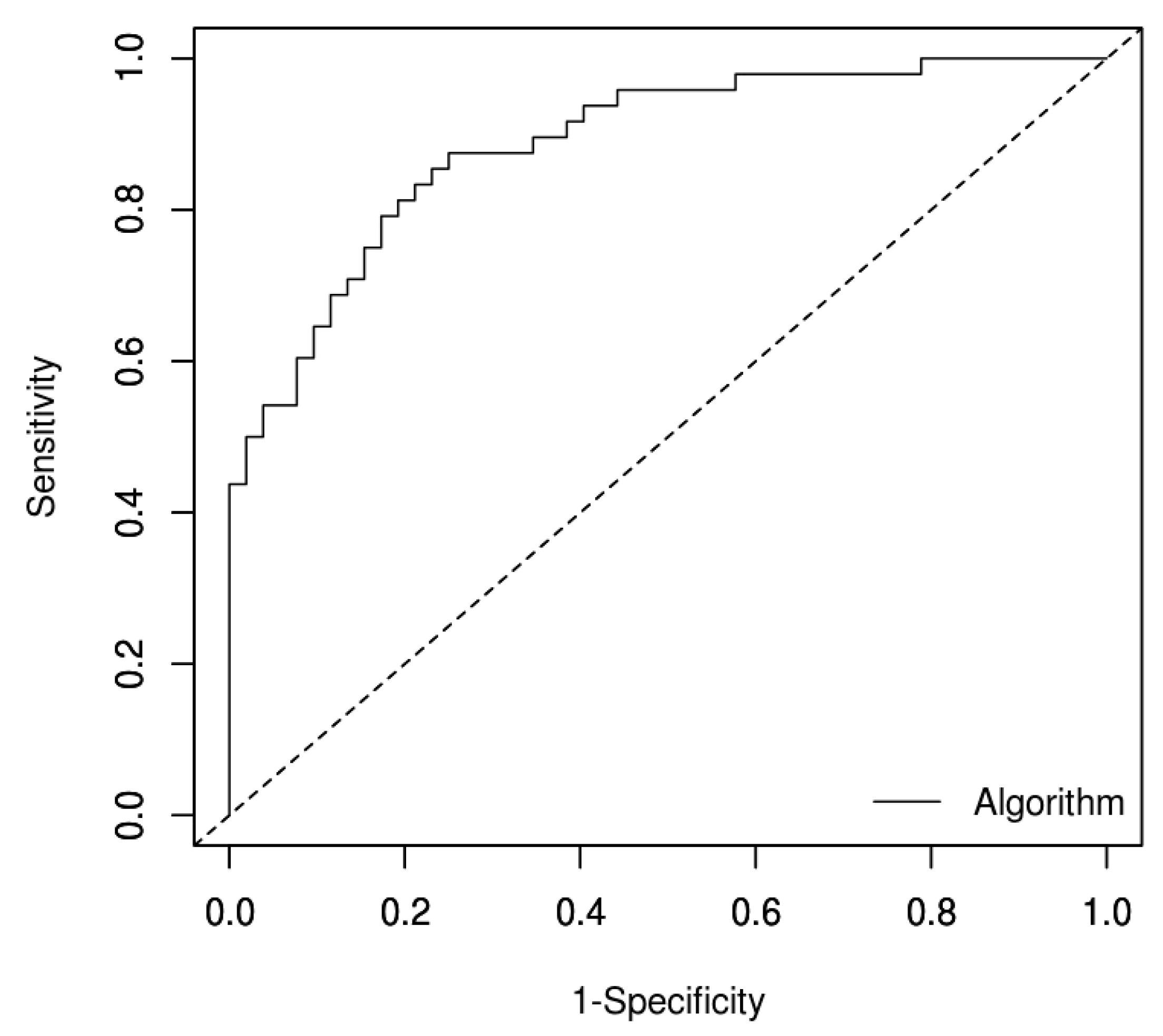
3.3. Receiver-Operating Characteristic (ROC) Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bejerano, G.; Pheasant, M.; Makunin, I.; Stephen, S.; Kent, W.J.; Mattick, J.S.; Haussler, D. Ultraconserved elements in the human genome. Science 2004, 304, 1321–1325. [Google Scholar] [CrossRef]
- Dimitrieva, S.; Bucher, P. UCNEbase—A database of ultraconserved non-coding elements and genomic regulatory blocks. Nucleic Acids Res. 2013, 41, D101–D109. [Google Scholar] [CrossRef]
- Habic, A.; Mattick, J.S.; Calin, G.A.; Krese, R.; Konc, J.; Kunej, T. Genetic Variations of Ultraconserved Elements in the Human Genome. OMICS 2019, 23, 549–559. [Google Scholar] [CrossRef]
- Dickel, D.E.; Ypsilanti, A.R.; Pla, R.; Zhu, Y.; Barozzi, I.; Mannion, B.J.; Khin, Y.S.; Fukuda-Yuzawa, Y.; Plajzer-Frick, I.; Pickle, C.S.; et al. Ultraconserved Enhancers Are Required for Normal Development. Cell 2018, 172, 491–499.e15. [Google Scholar] [CrossRef] [PubMed]
- Ryu, T.; Seridi, L.; Ravasi, T. The evolution of ultraconserved elements with different phylogenetic origins. BMC Evol. Biol. 2012, 12, 236. [Google Scholar] [CrossRef] [PubMed]
- Terracciano, D.; Terreri, S.; de Nigris, F.; Costa, V.; Calin, G.A.; Cimmino, A. The role of a new class of long noncoding RNAs transcribed from ultraconserved regions in cancer. Biochim. Biophys Acta Rev. Cancer 2017, 1868, 449–455. [Google Scholar] [CrossRef]
- Calin, G.A.; Liu, C.; Ferracin, M.; Hyslop, T.; Spizzo, R.; Sevignani, C.; Fabbri, M.; Cimmino, A.; Lee, E.J.; Wojcik, S.E.; et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell 2007, 12, 215–229. [Google Scholar] [CrossRef]
- Fassan, M.; Dall’Olmo, L.; Galasso, M.; Braconi, C.; Pizzi, M.; Realdon, S.; Volinia, S.; Valeri, N.; Gasparini, P.; Baffa, R.; et al. Transcribed ultraconserved noncoding RNAs (T-UCR) are involved in Barrett’s esophagus carcinogenesis. Oncotarget 2014, 5, 7162–7171. [Google Scholar] [CrossRef]
- Colwell, M.; Drown, M.; Showel, K.; Drown, C.; Palowski, A.; Faulk, C. Evolutionary conservation of DNA methylation in CpG sites within ultraconserved noncoding elements. Epigenetics 2018, 13, 49–60. [Google Scholar] [CrossRef]
- Lujambio, A.; Portela, A.; Liz, J.; Melo, S.A.; Rossi, S.; Spizzo, R.; Croce, C.M.; Calin, G.A.; Esteller, M. CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene 2010, 29, 6390–6401. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Azevedo-Pouly, A.C.P.; Redis, R.S.; Lee, E.J.; Gusev, Y.; Allard, D.; Sutaria, D.S.; Badawi, M.; Elgamal, O.A.; Lerner, M.R.; et al. Globally increased ultraconserved noncoding RNA expression in pancreatic adenocarcinoma. Oncotarget 2016, 7, 53165–53177. [Google Scholar] [CrossRef]
- Galasso, M.; Dama, P.; Previati, M.; Sandhu, S.; Palatini, J.; Coppola, V.; Warner, S.; Sana, M.E.; Zanella, R.; Abujarour, R.; et al. A large scale expression study associates uc.283-plus lncRNA with pluripotent stem cells and human glioma. Genome Med. 2014, 6, 76. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vannini, I.; Wise, P.M.; Challagundla, K.B.; Plousiou, M.; Raffini, M.; Bandini, E.; Fanini, F.; Paliaga, G.; Crawford, M.; Ferracin, M.; et al. Transcribed ultraconserved region 339 promotes carcinogenesis by modulating tumor suppressor microRNAs. Nat Commun 2017, 8, 1801. [Google Scholar] [CrossRef]
- Guerrero-Preston, R.; Michailidi, C.; Marchionni, L.; Pickering, C.R.; Frederick, M.J.; Myers, J.N.; Yegnasubramanian, S.; Hadar, T.; Noordhuis, M.G.; Zizkova, V.; et al. Key tumor suppressor genes inactivated by “greater promoter” methylation and somatic mutations in head and neck cancer. Epigenetics 2014, 9, 1031–1046. [Google Scholar] [CrossRef] [PubMed]
- Sekino, Y.; Sakamoto, N.; Goto, K.; Honma, R.; Shigematsu, Y.; Quoc, T.P.; Sentani, K.; Oue, N.; Teishima, J.; Kawakami, F.; et al. Uc.416 + A promotes epithelial-to-mesenchymal transition through miR-153 in renal cell carcinoma. BMC Cancer 2018, 18, 952. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.H.; O’Sullivan, B. Overview of the 8th Edition TNM Classification for Head and Neck Cancer. Curr. Treat. Options Oncol. 2017, 18, 40. [Google Scholar] [CrossRef] [PubMed]
- Kademani, D.; Bell, R.B.; Bagheri, S.; Holmgren, E.; Dierks, E.; Potter, B.; Homer, L. Prognostic factors in intraoral squamous cell carcinoma: The influence of histologic grade. J. Oral. Maxillofac. Surg. 2005, 63, 1599–1605. [Google Scholar] [CrossRef] [PubMed]
- Morandi, L.; Gissi, D.; Tarsitano, A.; Asioli, S.; Gabusi, A.; Marchetti, C.; Montebugnoli, L.; Foschini, M.P. CpG location and methylation level are crucial factors for the early detection of oral squamous cell carcinoma in brushing samples using bisulfite sequencing of a 13-gene panel. Clin. Epigenet. 2017, 9, 85. [Google Scholar] [CrossRef] [PubMed]
- Gissi, D.B.; Tarsitano, A.; Gabusi, A.; Rossi, R.; Attardo, G.; Lenzi, J.; Marchetti, C.; Montebugnoli, L.; Foschini, M.P.; Morandi, L. 13-gene DNA Methylation Analysis from Oral Brushing: A Promising Non Invasive Tool in the Follow-up of Oral Cancer Patients. J. Clin. Med. 2019, 8, 2107. [Google Scholar] [CrossRef] [PubMed]
- Li, L.-C.; Dahiya, R. MethPrimer: Designing primers for methylation PCRs. Bioinformatics 2002, 18, 1427–1431. [Google Scholar] [CrossRef]
- Gabusi, A.; Gissi, D.B.; Montebugnoli, L.; Asioli, S.; Tarsitano, A.; Marchetti, C.; Balbi, T.; Helliwell, T.R.; Foschini, M.P.; Morandi, L. Prognostic impact of intra-field heterogeneity in oral squamous cell carcinoma. Virchows Arch. 2019, 476, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Ricci, C.; Morandi, L.; Righi, A.; Gibertoni, D.; Maletta, F.; Ambrosi, F.; Agostinelli, C.; Uccella, S.; Asioli, S.; Sessa, F.; et al. PD-1 (PDCD1) promoter methylation in Merkel cell carcinoma: Prognostic relevance and relationship with clinico-pathological parameters. Mod. Pathol. 2019, 32, 1359–1372. [Google Scholar] [CrossRef] [PubMed]
- Gabusi, A.; Gissi, D.B.; Tarsitano, A.; Asioli, S.; Marchetti, C.; Montebugnoli, L.; Foschini, M.P.; Morandi, L. Intratumoral Heterogeneity in Recurrent Metastatic Squamous Cell Carcinoma of the Oral Cavity: New Perspectives Afforded by Multiregion DNA Sequencing and mtDNA Analysis. J. Oral Maxillofac. Surg. 2019, 77, 440–455. [Google Scholar] [CrossRef]
- Renzi, A.; De Bonis, P.; Morandi, L.; Lenzi, J.; Tinto, D.; Rigillo, A.; Bettini, G.; Bellei, E.; Sabattini, S. Prevalence of p53 dysregulations in feline oral squamous cell carcinoma and non-neoplastic oral mucosa. PLoS ONE 2019, 14, e0215621. [Google Scholar] [CrossRef] [PubMed]
- Gissi, D.B.; Gabusi, A.; Tarsitano, A.; Asioli, S.; Rossi, R.; Marchetti, C.; Montebugnoli, L.; Foschini, M.P.; Morandi, L. Application of a non-invasive oral brushing procedure based on bisulfite sequencing of a 13-gene panel to study high-risk OSCC patients. Cancer Biomark 2020, 28, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Foschini, M.P.; Morandi, L.; Sanchez, A.M.; Santoro, A.; Mulè, A.; Zannoni, G.F.; Varga, Z.; Moskovszky, L.; Cucchi, M.C.; Moelans, C.B.; et al. Methylation Profile of X-Chromosome-Related Genes in Male Breast Cancer. Front. Oncol. 2020, 10, 784. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Cech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef]
- Krainer, J.; Weinhäusel, A.; Hanak, K.; Pulverer, W.; Özen, S.; Vierlinger, K.; Pabinger, S. EPIC-TABSAT: Analysis tool for targeted bisulfite sequencing experiments and array-based methylation studies. Nucleic Acids Res. 2019, 47, W166–W170. [Google Scholar] [CrossRef]
- Mallona, I.; Díez-Villanueva, A.; Peinado, M.A. Methylation plotter: A web tool for dynamic visualization of DNA methylation data. Source Code Biol. Med. 2014, 9, 11. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef]
- Renzi, A.; Morandi, L.; Lenzi, J.; Rigillo, A.; Bettini, G.; Bellei, E.; Giacomini, A.; Tinto, D.; Sabattini, S. Analysis of DNA methylation and TP53 mutational status for differentiating feline oral squamous cell carcinoma from non-neoplastic mucosa: A preliminary study. Vet. Comp. Oncol. 2020, 18, 12624. [Google Scholar] [CrossRef] [PubMed]
- Planello, A.C.; Singhania, R.; Kron, K.J.; Bailey, S.D.; Roulois, D.; Lupien, M.; Line, S.R.P.; de Souza, A.P.; De Carvalho, D.D. Pre-neoplastic epigenetic disruption of transcriptional enhancers in chronic inflammation. Oncotarget 2016, 7, 15772–15786. [Google Scholar] [CrossRef]
- Shin, Y.J.; Choung, H.W.; Lee, J.H.; Rhyu, I.C.; Kim, H.D. Association of Periodontitis with Oral Cancer: A Case-Control Study. J. Dent. Res. 2019, 98, 526–533. [Google Scholar] [CrossRef] [PubMed]
- van Vlodrop, I.J.H.; Niessen, H.E.C.; Derks, S.; Baldewijns, M.M.L.L.; van Criekinge, W.; Herman, J.G.; van Engeland, M. Analysis of promoter CpG island hypermethylation in cancer: Location, location, location! Clin. Cancer Res. 2011, 17, 4225–4231. [Google Scholar] [CrossRef] [PubMed]
- Dotto, G.P.; Rustgi, A.K. Squamous Cell Cancers: A Unified Perspective on Biology and Genetics. Cancer Cell 2016, 29, 622–637. [Google Scholar] [CrossRef]
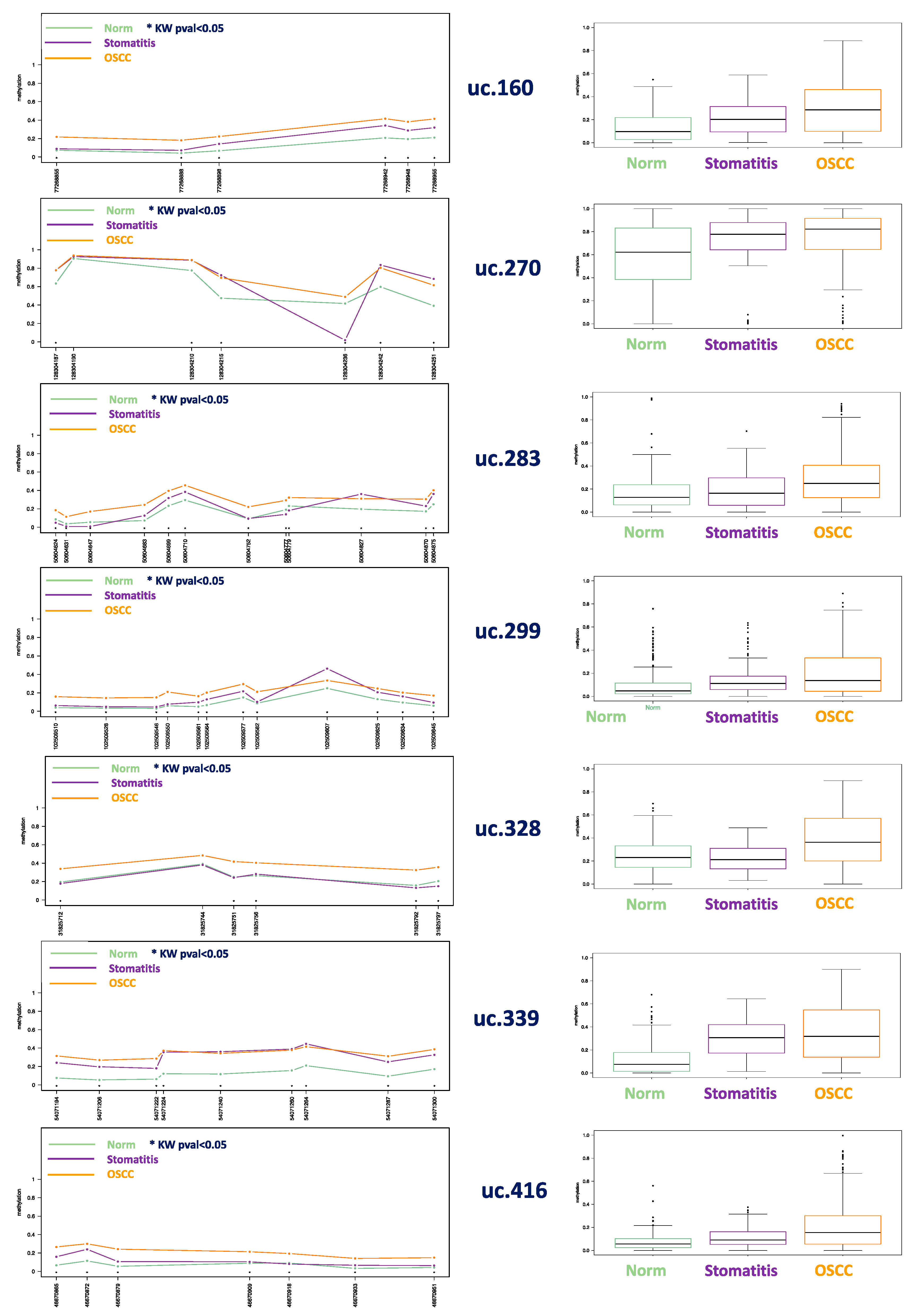

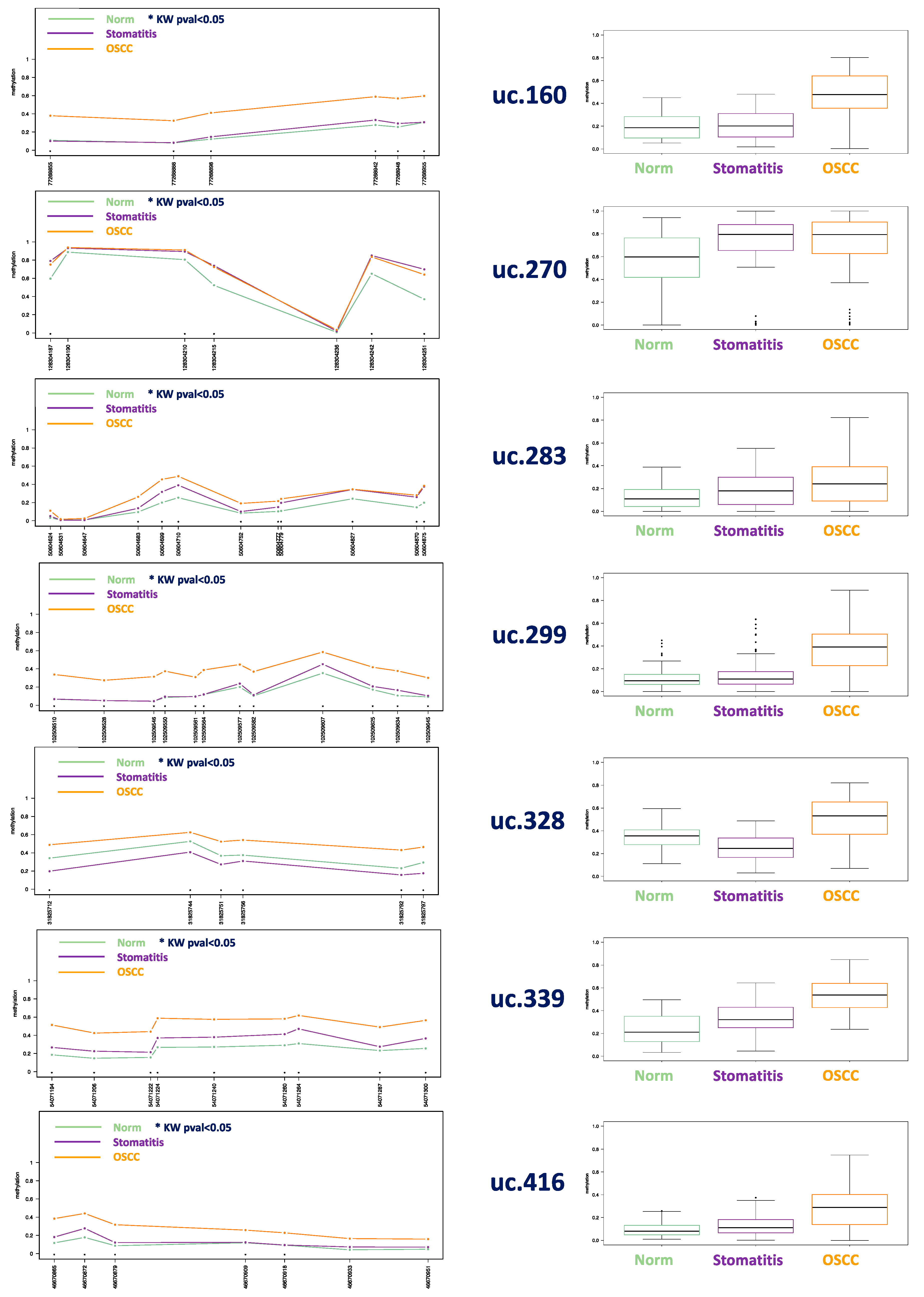
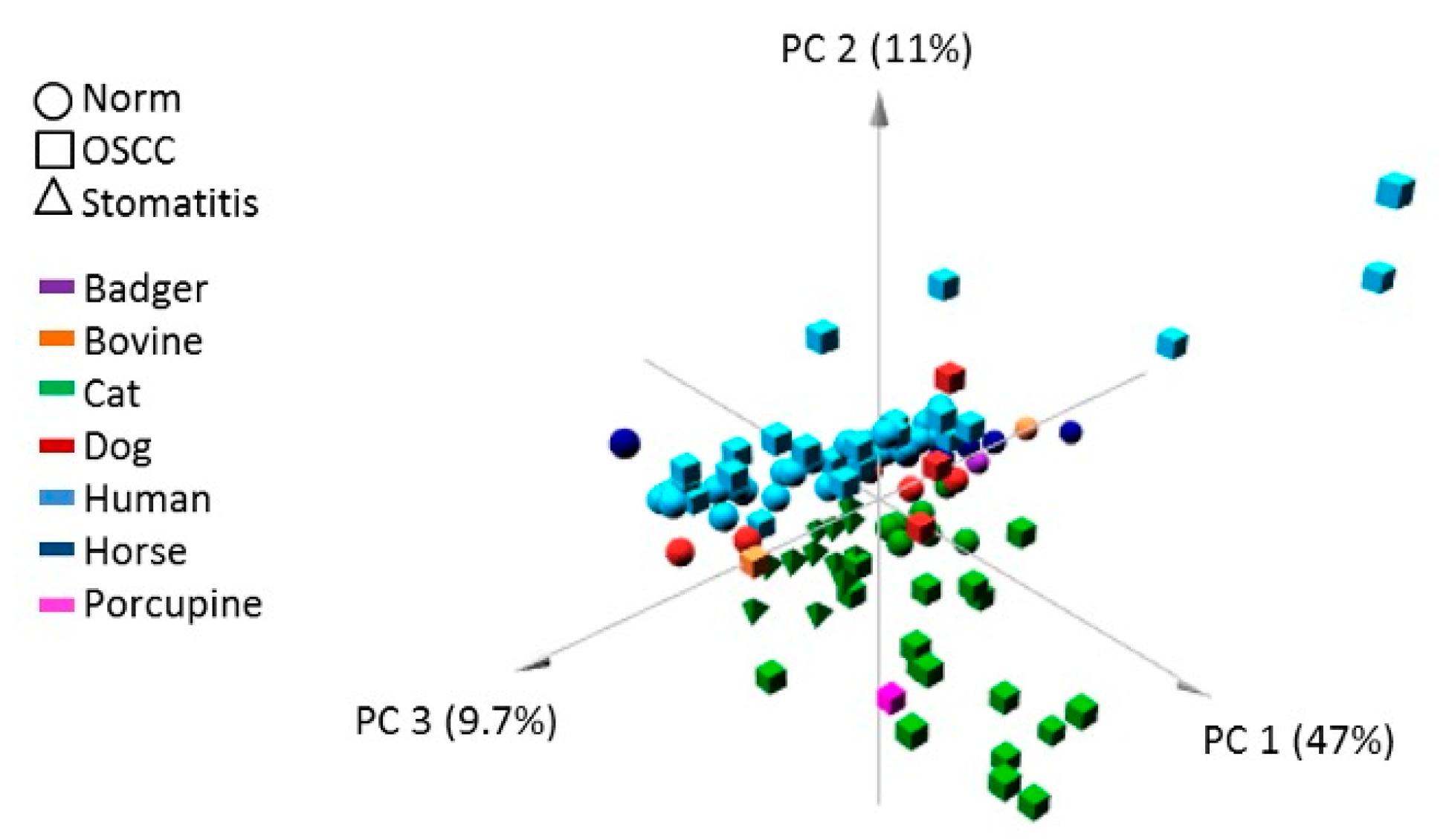
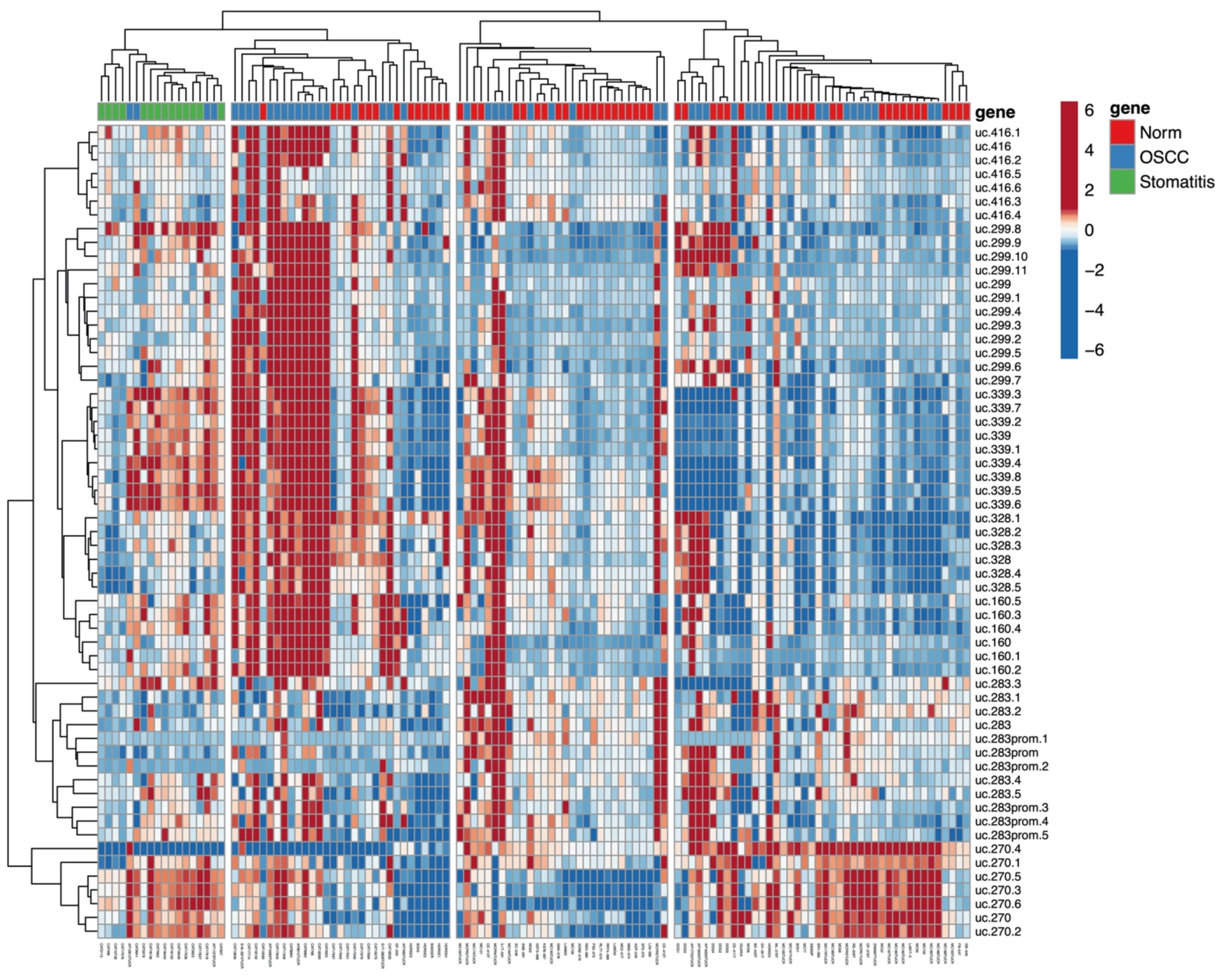
| ID | Sample | Diagnosis | Species | Breed | Location | Sex | Age |
|---|---|---|---|---|---|---|---|
| AP15289 | FFPE | SCC | Felis catus (Cat) | Domestic short-haired | Skin (head) | Fs | 9 y |
| AP8012 | FFPE | SCC | Felis catus (Cat) | Domestic short-haired | Skin (chin) | Mc | 10 y |
| AP11378 | FFPE | SCC | Felis catus (Cat) | Domestic short-haired | Skin (ear) | Fs | 12 y |
| AP11306 | FFPE | SCC | Felis catus (Cat) | Domestic short-haired | Oral cavity | Fs | 12 y |
| AP18329 | FFPE | SCC | Felis catus (Cat) | Domestic short-haired | Oral cavity | Mc | 11 y |
| AP18369 | FFPE | SCC | Felis catus (Cat) | Domestic short-haired | Oral cavity | Fs | 13 y |
| AP4925 | FFPE | SCC | Felis catus (Cat) | Domestic short-haired | Oral cavity | Fs | 19 y |
| AP9641 | FFPE | SCC | Felis catus (Cat) | Domestic short-haired | Oral cavity | Mc | 16 y |
| AP9960 | FFPE | SCC | Felis catus (Cat) | Persian | Oral cavity | Mc | 11 y |
| AP17845 | FFPE | SCC | Felis catus (Cat) | Domestic short-haired | Oral cavity | Mc | 11 y |
| AP17919 | FFPE | SCC | Felis catus (Cat) | Domestic short-haired | Oral cavity | Fs | 10 y |
| AP13806 | FFPE | SCC | Felis catus (Cat) | Domestic short-haired | Oral cavity | Mc | 15 y |
| AP17595 | FFPE | SCC | Felis catus (Cat) | Thai | Oral cavity | Fs | 12 y |
| AP17714 | FFPE | SCC | Felis catus (Cat) | Domestic short-haired | Oral cavity | Mc | 9 y |
| AP18102 | FFPE | SCC | Felis catus (Cat) | Domestic short-haired | Oral cavity | Mc | 12 y |
| AP768 | FFPE | SCC | Felis catus (Cat) | Domestic short-haired | Oral cavity | Fs | 15 y |
| AP8694 | FFPE | SCC | Felis catus (Cat) | Domestic short-haired | Oral cavity | Fs | 12 y |
| AP16359 | FFPE | SCC | Canis lupus familiaris (Dog) | Beagle | Prepuce | M | 12 y |
| AP17735 | FFPE | SCC | Canis lupus familiaris (Dog) | Labrador retriever | Skin | F | 11 y |
| AP16030 | FFPE | SCC | Canis lupus familiaris (Dog) | Labrador retriever | Oral cavity | Fs | 14 y |
| AP7483 | FFPE | SCC | Equus ferus caballus (Horse) | Paint horse | Vulva | F | 15 y |
| AP12812 | FFPE | SCC | Bos taurus (Bovine) | Holstein fresian | Eyelid | Mc | 8 y |
| S17_3828 | FFPE | SCC | Meles meles (European badger) | Lung | F | 14 y | |
| S18_5874 | FFPE | SCC | Hystrix cristata (Porcupine) | Mandible | F | 14 y | |
| AP10728 | FFPE | Stomatitis | Felis catus (Cat) | Domestic short-haired | Oral cavity | Mc | 9 y |
| AP2633 | FFPE | Stomatitis | Felis catus (Cat) | Domestic short-haired | Oral cavity | Fs | 10 y |
| AP5978 | FFPE | Stomatitis | Felis catus (Cat) | Domestic short-haired | Oral cavity | Fs | 7 y |
| AP847 | FFPE | Stomatitis | Felis catus (Cat) | Domestic short-haired | Oral cavity | Fs | 12 y |
| AP18349 | FFPE | Stomatitis | Felis catus (Cat) | Domestic short-haired | Oral cavity | Mc | 8 y |
| AP16102 | FFPE | Stomatitis | Felis catus (Cat) | Domestic short-haired | Oral cavity | Mc | 17 y |
| AP17827 | FFPE | Stomatitis | Felis catus (Cat) | Domestic short-haired | Oral cavity | Mc | 9 y |
| AP17936 | FFPE | Stomatitis | Felis catus (Cat) | Domestic short-haired | Oral cavity | Fs | 4 y |
| AP18136 | FFPE | Stomatitis | Felis catus (Cat) | Domestic short-haired | Oral cavity | Mc | 5 y |
| AP18404 | FFPE | Stomatitis | Felis catus (Cat) | Domestic short-haired | Oral cavity | Fs | 8 y |
| AP2115 | FFPE | Stomatitis | Felis catus (Cat) | Domestic short-haired | Oral cavity | Fs | 12 y |
| AP498 | FFPE | Stomatitis | Felis catus (Cat) | Domestic short-haired | Oral cavity | Mc | 1 y |
| AP7692 | FFPE | Stomatitis | Felis catus (Cat) | Domestic short-haired | Oral cavity | Fs | 13 y |
| AP17076 | FFPE | Stomatitis | Felis catus (Cat) | Domestic short-haired | Oral cavity | Mc | 10 y |
| CAT-1 | Brushing | Norm | Felis catus (Cat) | Domestic short-haired | Oral cavity | Mc | 15 y |
| CAT-2 | Brushing | Norm | Felis catus (Cat) | Domestic short-haired | Oral cavity | Fs | 10 y |
| CAT-3 | Brushing | Norm | Felis catus (Cat) | Domestic short-haired | Oral cavity | Fs | 9 y |
| CAT-4 | Brushing | Norm | Felis catus (Cat) | Domestic short-haired | Oral cavity | Mc | 10 y |
| CAT-5 | Brushing | Norm | Felis catus (Cat) | Domestic short-haired | Oral cavity | Fs | 9 y |
| CAT-6 | Brushing | Norm | Felis catus (Cat) | Persian | Oral cavity | Fs | 11 y |
| CAT-7 | Brushing | Norm | Felis catus (Cat) | Domestic short-haired | Oral cavity | Mc | 12 y |
| DOG-1 | Brushing | Norm | Canis lupus familiaris (Dog) | Border Collie | Oral cavity | Mc | 5 y |
| DOG-2 | Brushing | Norm | Canis lupus familiaris (Dog) | Golden Retriever | Oral cavity | Mc | 10 y |
| DOG-3 | Brushing | Norm | Canis lupus familiaris (Dog) | Newfoundland | Oral cavity | Fs | 9 y |
| DOG-4 | Brushing | Norm | Canis lupus familiaris (Dog) | Golden Retriever | Oral cavity | F | 7 y |
| DOG-5 | Brushing | Norm | Canis lupus familiaris (Dog) | Golden Retriever | Oral cavity | F | 5 y |
| HORSE-1 | Brushing | Norm | Equus ferus caballus (Horse) | Italian Saddle | Oral cavity | Mc | 16 y |
| HORSE-2 | Brushing | Norm | Equus ferus caballus (Horse) | Italian Saddle | Oral cavity | Mc | 9 y |
| HORSE-3 | Brushing | Norm | Equus ferus caballus (Horse) | Italian Saddle | Oral cavity | Mc | 8 y |
| HORSE-4 | Brushing | Norm | Equus ferus caballus (Horse) | Italian Saddle | Oral cavity | F | 10 y |
| HORSE-5 | Brushing | Norm | Equus ferus caballus (Horse) | Italian Saddle | Oral cavity | F | 9 y |
| BOV-1 | Brushing | Norm | Bos taurus (Bovine) | Holstein fresian | Oral cavity | F | 2 y |
| BOV-2 | Brushing | Norm | Bos taurus (Bovine) | Holstein fresian | Oral cavity | F | 1 y 6 m |
| BADGER-1 | Brushing | Norm | Meles meles (European badger) | Oral cavity | F | Unknown (adult) |
| UCNE Bejerano | UCNE ID | UCNE Name | Upstream Gene | Within Gene | Downstream Gene | Primer For | Primer Rev | Genome Coordinates (hg38) | Number of CpG | Amplicon Length |
|---|---|---|---|---|---|---|---|---|---|---|
| uc.160+ | 27423 | OTP_Lukas | AK128395 | NA | AP3B1 | TTTTTGATTTTAGGTGGAATTAGGAG | TACATTAAAACCTCAATTAATTAACCCTTA | Chr5+: 77972981−77973184 | 6 | 204 |
| uc.283+ | 6661 | DRGX_Toru | DRGX | NA | ERCC6 | TTGGGTGAAAATTAAATTTTAAAGTAT | AAAAAACACTTAATAATACAAAAAACC | Chr10+: 49396679−49396856 | 6 | 178 |
| uc.283 Prom | 6661 | DRGX_Toru | DRGX | NA | ERCC6 | TGTTTTTAGTAAGGGGTTTTTGAATT | AAAATTTAATTTTCACCCAAACAAA | Chr10+: 49396546−49396698 | 6 | 153 |
| uc.416+ | NA | NA | HOXB4 | HOXB5 | HOXB6 | GTTTTAAAGTGGTTGGAGGAGGT | CTATCAATTACTAAATTATAACAATAACAA | Chr17+: 48593480−48593629 | 7 | 150 |
| uc.339+ | NA | NA | ATP5G2 | NA | KIAA1536 | GTTATTATAGGAATGTAATTTTGTT | CCTTTCAAAAACTCAAAACCATAAA | Chr12+: 53677385−53677558 | 9 | 174 |
| uc.270+ | 35244 | NA | AB040954 | MAPKAP1 | LOC51145 | GTGATGGGTATTTAGAGAGTTGGTTAT | CTTACCAAAAATCATTATTCACCCT | Chr9+: 125541877−125541998 | 7 | 122 |
| uc.299+ | NA | NA | HIF1AN | PAX2 | C10orf6 | TTTTGTTGTGTGTGGGGTGT | CCTCACCTACCCAAAATTTTACTAAC | Chr10+: 100749700−100749920 | 12 | 221 |
| uc.328+ | NA | NA | ELP4 | PAX6 | RCN1 | TTTGATATTTGTTTTTAAAATAAAATTAGT | ACAAAATAACAAAACTACCACAAAC | Chr11+: 31804130−31804274 | 6 | 145 |
| UCNE | Best Position | Best AUC Value | Worst Position | Worst AUC Value | Best to Worst AUC Gap |
|---|---|---|---|---|---|
| uc.160 | 77268955 | 0.783 | 77268898 | 0.725 | 0.058 |
| uc.270 | 128304242 | 0.814 | 128304236 | 0.589 | 0.225 |
| uc.283 | 50604683 | 0.824 | 50604779 | 0.653 | 0.171 |
| uc.299 | 102509546 | 0.752 | 102509564 | 0.686 | 0.066 |
| uc.328 | 31825751 | 0.713 | 31825744 | 0.624 | 0.089 |
| uc.339 | 54071206 | 0.826 | 54071264 | 0.742 | 0.084 |
| uc.416 | 46670879 | 0.843 | 46670918 | 0.679 | 0.164 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morandi, L.; Sabattini, S.; Renzi, A.; Rigillo, A.; Bettini, G.; Dervas, E.; Schauer, A.; Morandi, M.; Gissi, D.B.; Tarsitano, A.; et al. An Evolutionary Cancer Epigenetic Approach Revealed DNA Hypermethylation of Ultra-Conserved Non-Coding Elements in Squamous Cell Carcinoma of Different Mammalian Species. Cells 2020, 9, 2092. https://doi.org/10.3390/cells9092092
Morandi L, Sabattini S, Renzi A, Rigillo A, Bettini G, Dervas E, Schauer A, Morandi M, Gissi DB, Tarsitano A, et al. An Evolutionary Cancer Epigenetic Approach Revealed DNA Hypermethylation of Ultra-Conserved Non-Coding Elements in Squamous Cell Carcinoma of Different Mammalian Species. Cells. 2020; 9(9):2092. https://doi.org/10.3390/cells9092092
Chicago/Turabian StyleMorandi, Luca, Silvia Sabattini, Andrea Renzi, Antonella Rigillo, Giuliano Bettini, Eva Dervas, Alexandria Schauer, Marco Morandi, Davide B. Gissi, Achille Tarsitano, and et al. 2020. "An Evolutionary Cancer Epigenetic Approach Revealed DNA Hypermethylation of Ultra-Conserved Non-Coding Elements in Squamous Cell Carcinoma of Different Mammalian Species" Cells 9, no. 9: 2092. https://doi.org/10.3390/cells9092092
APA StyleMorandi, L., Sabattini, S., Renzi, A., Rigillo, A., Bettini, G., Dervas, E., Schauer, A., Morandi, M., Gissi, D. B., Tarsitano, A., Evangelisti, S., & Tonon, C. (2020). An Evolutionary Cancer Epigenetic Approach Revealed DNA Hypermethylation of Ultra-Conserved Non-Coding Elements in Squamous Cell Carcinoma of Different Mammalian Species. Cells, 9(9), 2092. https://doi.org/10.3390/cells9092092






