Tsc1 Regulates the Proliferation Capacity of Bone-Marrow Derived Mesenchymal Stem Cells
Abstract
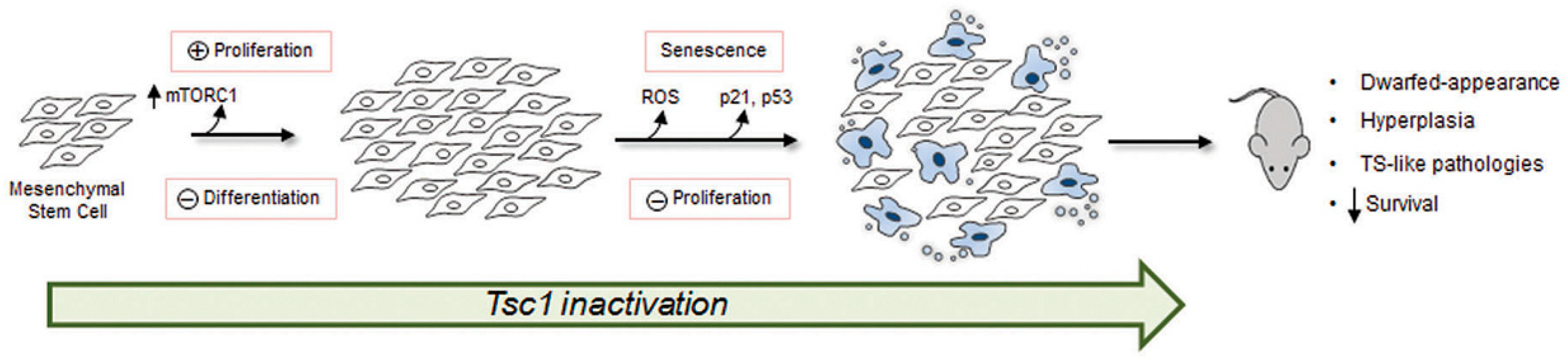
1. Introduction
2. Materials and Methods
2.1. Generation of the Mouse Model
2.2. Isolation and Expansion of Mouse Mesenchymal Stem Cells (mMSCs)
2.3. Flow Cytometric Analysis for Mmsc-Associated Cell Surface Markers
2.4. Bromodeoxyuridine (BrdU) Labeling
2.5. Colony Forming Assay (CFU-F)
2.6. Flow Cytometry Analysis of Reactive Oxygen Species (ROS)
2.7. Mouse Histopathology and Immunohistochemistry
2.8. Senescence-Associated β-Galactosidase (SA β-G) Staining
2.8.1. In Situ Whole Organ Staining
2.8.2. In Vitro Cell Culture
2.9. Lentiviral shRNA Vector Generation and Transduction
2.10. Western Blot Analyses
2.11. MSC Differentiation Assays
2.11.1. Adipogenesis
2.11.2. Smooth Muscle (SM) Myogenesis
2.12. RNA Extraction and RT-PCR
2.13. Everolimus Treatment
2.14. Statistical Analyses
3. Results
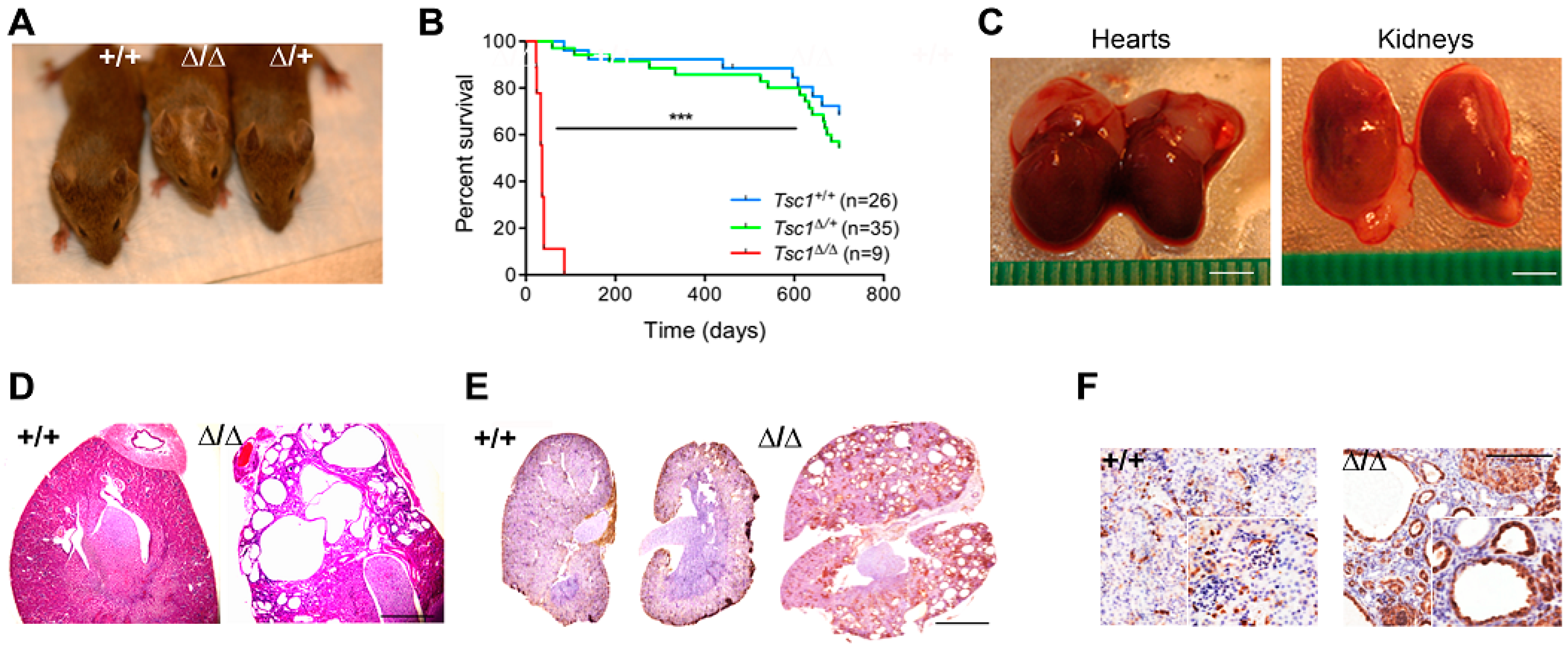
3.1. Tagln-Mediated Tsc1 Inactivation Targets SM and MSC Populations and Recapitulates Features of Human Tuberous Sclerosis
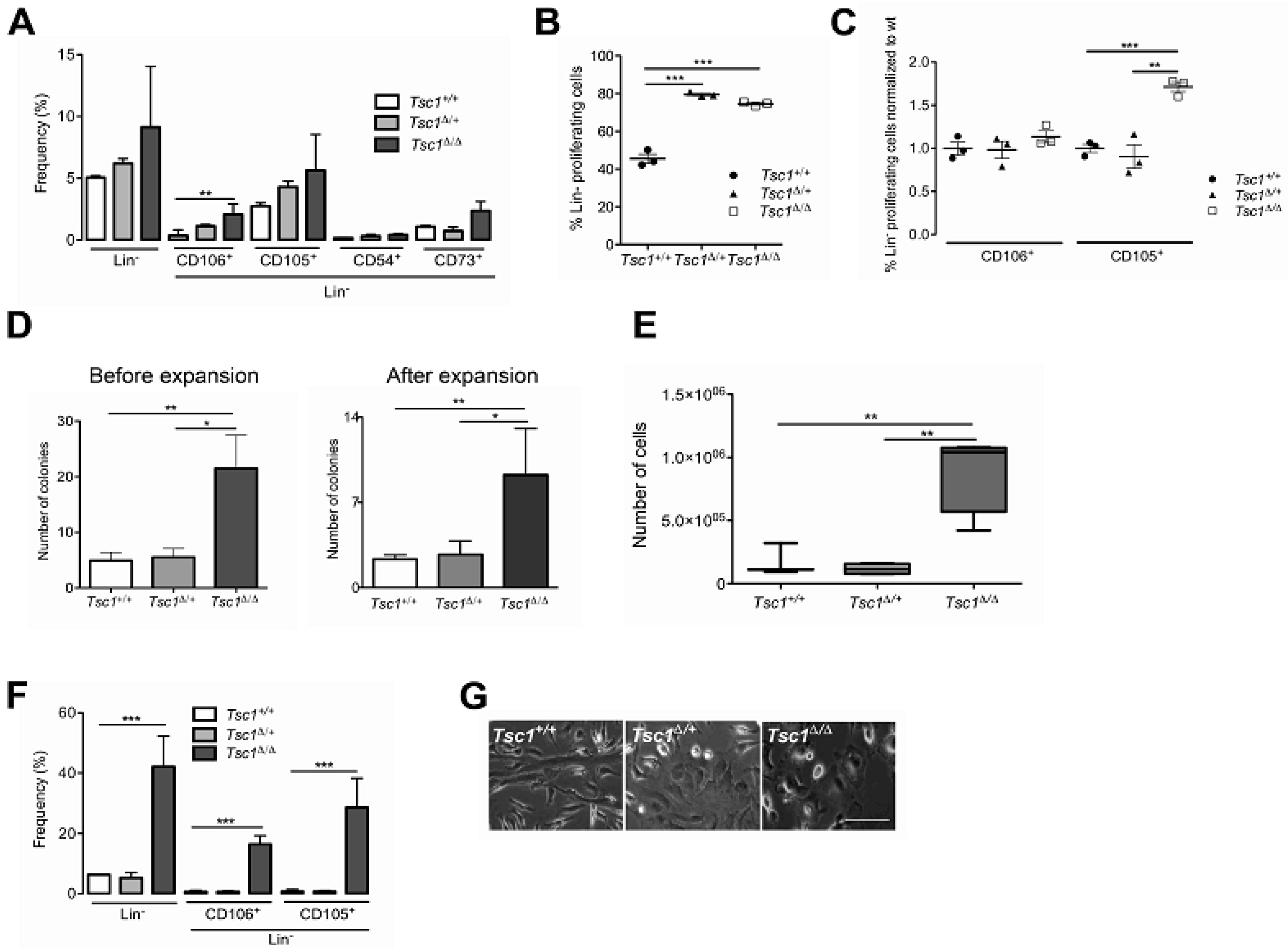
3.2. Tsc1 Deletion Leads to Expansion of the BM-MSC Pool in Young (28 Day Old) Mice
3.3. Aged (1.5 yr old) Mice Do Not Exhibit A Hyperproliferative BM-MSC Phenotype Following Tsc1 Loss
3.4. Tsc1 Inactivation Leads to ROS Production and Senescence
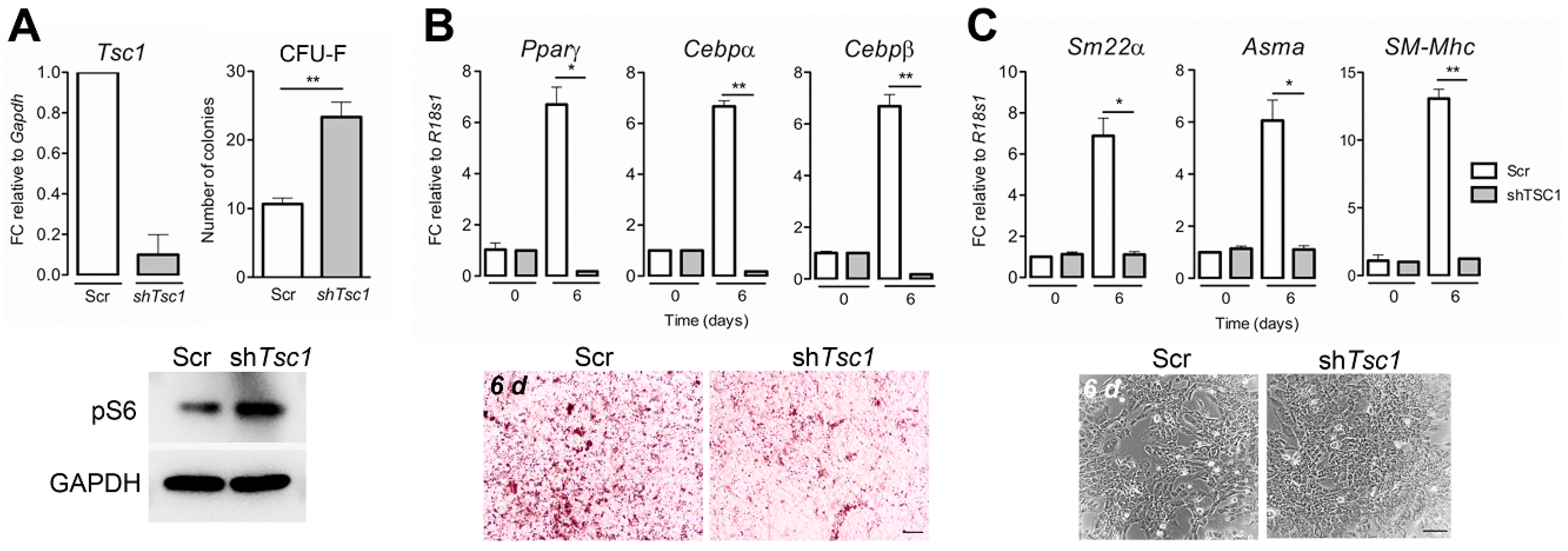
3.5. Tsc1 Knockdown in Wt BM-Mscs Increases Their Clonogenic Potential and Suppresses Adipocyte and Smooth Muscle Differentiation In Vitro
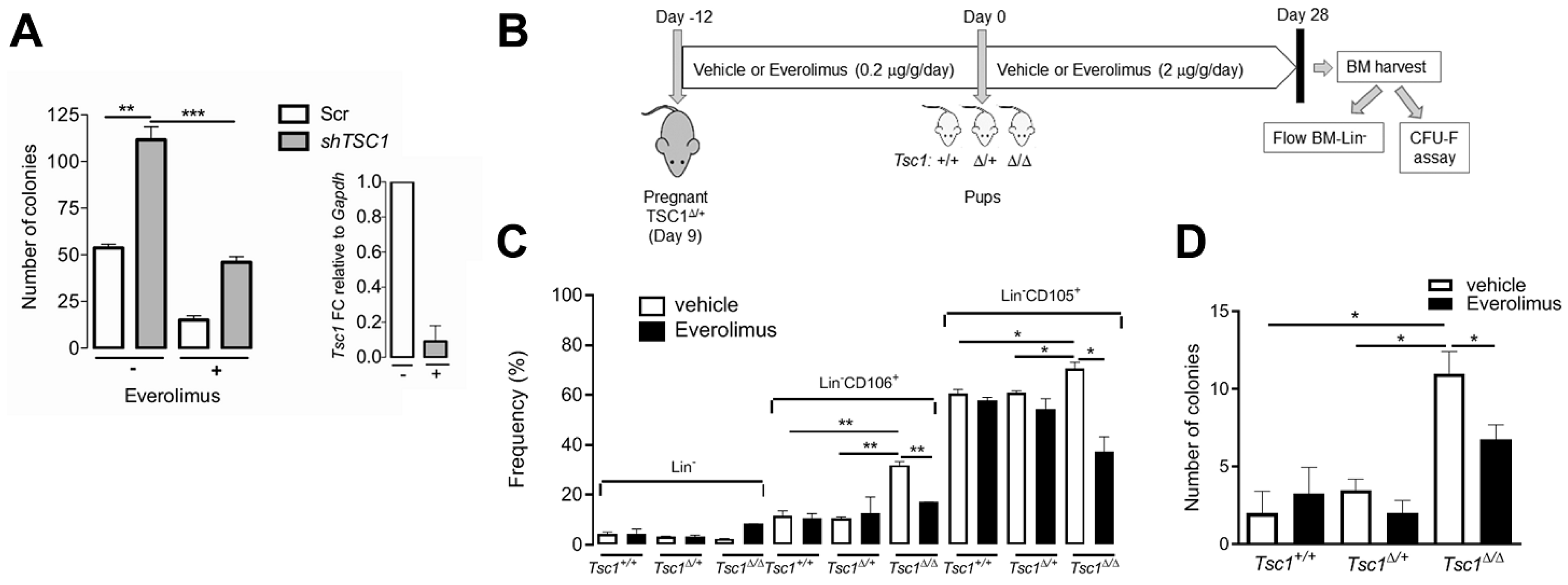
3.6. mTOR Activation is Required for BM-MSC Expansion Following Tsc1 Inactivation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Crino, P.B.; Nathanson, K.L.; Henske, E.P. The tuberous sclerosis complex. N. Engl. J. Med. 2006, 355, 1345–1356. [Google Scholar] [CrossRef] [PubMed]
- Curatolo, P.; Bombardieri, R.; Jozwiak, S. Tuberous sclerosis. Lancet 2008, 372, 657–668. [Google Scholar] [CrossRef]
- Ewalt, D.H.; Sheffield, E.; Sparagana, S.P.; Delgado, M.R.; Roach, E.S. Renal lesion growth in children with tuberous sclerosis complex. J. Urol. 1998, 160, 141–145. [Google Scholar] [CrossRef]
- Johnson, S.R.; Taveira-DaSilva, A.M.; Moss, J. Lymphangioleiomyomatosis. Clin. Chest Med. 2016, 37, 389–403. [Google Scholar] [CrossRef] [PubMed]
- Hinton, R.B.; Prakash, A.; Romp, R.L.; Krueger, D.A.; Knilans, T.K.; International Tuberous Sclerosis Consensus Group. Cardiovascular manifestations of tuberous sclerosis complex and summary of the revised diagnostic criteria and surveillance and management recommendations from the international tuberous sclerosis consensus group. J. Am. Heart Assoc. 2014, 3, e001493. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.C.; Chuang, H.C.; Chen, T.D.; Chi, C.L.; Ng, K.F.; Yeh, T.S.; Chen, T.C. Alterations of the mtor pathway in hepatic angiomyolipoma with emphasis on the epithelioid variant and loss of heterogeneity of tsc1/tsc2. Histopathology 2015, 66, 695–705. [Google Scholar] [CrossRef]
- Huang, J.; Manning, B.D. The tsc1-tsc2 complex: A molecular switchboard controlling cell growth. Biochem. J. 2008, 412, 179–190. [Google Scholar] [CrossRef]
- Chen, C.; Liu, Y.; Liu, R.; Ikenoue, T.; Guan, K.L.; Liu, Y.; Zheng, P. Tsc-mtor maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J. Exp. Med. 2008, 205, 2397–2408. [Google Scholar] [CrossRef]
- Gan, B.; Sahin, E.; Jiang, S.; Sanchez-Aguilera, A.; Scott, K.L.; Chin, L.; Williams, D.A.; Kwiatkowski, D.J.; DePinho, R.A. Mtorc1-dependent and -independent regulation of stem cell renewal, differentiation, and mobilization. Proc. Natl. Acad. Sci. USA 2008, 105, 19384–19389. [Google Scholar] [CrossRef]
- Yilmaz, O.H.; Valdez, R.; Theisen, B.K.; Guo, W.; Ferguson, D.O.; Wu, H.; Morrison, S.J. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature 2006, 441, 475–482. [Google Scholar] [CrossRef]
- Castilho, R.M.; Squarize, C.H.; Chodosh, L.A.; Williams, B.O.; Gutkind, J.S. Mtor mediates wnt-induced epidermal stem cell exhaustion and aging. Cell Stem Cell 2009, 5, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Bernardi, R.; Pandolfi, P.P. A novel signaling network as a critical rheostat for the biology and maintenance of the normal stem cell and the cancer-initiating cell. Curr. Opin. Genet. Dev. 2009, 19, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Nardella, C.; Chen, Z.; Salmena, L.; Carracedo, A.; Alimonti, A.; Egia, A.; Carver, B.; Gerald, W.; Cordon-Cardo, C.; Pandolfi, P.P. Aberrant rheb-mediated mtorc1 activation and pten haploinsufficiency are cooperative oncogenic events. Genes Dev. 2008, 22, 2172–2177. [Google Scholar] [CrossRef] [PubMed]
- Alimonti, A.; Nardella, C.; Chen, Z.; Clohessy, J.G.; Carracedo, A.; Trotman, L.C.; Cheng, K.; Varmeh, S.; Kozma, S.C.; Thomas, G.; et al. A novel type of cellular senescence that can be enhanced in mouse models and human tumor xenografts to suppress prostate tumorigenesis. J. Clin. Investig. 2010, 120, 681–693. [Google Scholar] [CrossRef]
- Harrison, D.E.; Strong, R.; Sharp, Z.D.; Nelson, J.F.; Astle, C.M.; Flurkey, K.; Nadon, N.L.; Wilkinson, J.E.; Frenkel, K.; Carter, C.S.; et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 2009, 460, 392–395. [Google Scholar] [CrossRef]
- Schieke, S.M.; Finkel, T. Mitochondrial signaling, tor, and life span. Biol. Chem. 2006, 387, 1357–1361. [Google Scholar] [CrossRef]
- Rossi, D.J.; Jamieson, C.H.; Weissman, I.L. Stems cells and the pathways to aging and cancer. Cell 2008, 132, 681–696. [Google Scholar] [CrossRef]
- Nombela-Arrieta, C.; Ritz, J.; Silberstein, L.E. The elusive nature and function of mesenchymal stem cells. Nat. Rev. Mol. Cell Biol. 2011, 12, 126–131. [Google Scholar] [CrossRef]
- Da Silva Meirelles, L.; Chagastelles, P.C.; Nardi, N.B. Mesenchymal stem cells reside in virtually all post-natal organs and tissues. J. Cell Sci. 2006, 119, 2204–2213. [Google Scholar] [CrossRef]
- Colter, D.C.; Class, R.; DiGirolamo, C.M.; Prockop, D.J. Rapid expansion of recycling stem cells in cultures of plastic-adherent cells from human bone marrow. Proc. Natl. Acad. Sci. USA 2000, 97, 3213–3218. [Google Scholar] [CrossRef]
- Pittenger, M.F.; Mackay, A.M.; Beck, S.C.; Jaiswal, R.K.; Douglas, R.; Mosca, J.D.; Moorman, M.A.; Simonetti, D.W.; Craig, S.; Marshak, D.R. Multilineage potential of adult human mesenchymal stem cells. Science 1999, 284, 143–147. [Google Scholar] [CrossRef] [PubMed]
- Holtwick, R.; Gotthardt, M.; Skryabin, B.; Steinmetz, M.; Potthast, R.; Zetsche, B.; Hammer, R.E.; Herz, J.; Kuhn, M. Smooth muscle-selective deletion of guanylyl cyclase-a prevents the acute but not chronic effects of anp on blood pressure. Proc. Natl. Acad. Sci. USA 2002, 99, 7142–7147. [Google Scholar] [CrossRef]
- Zhang, J.; Link, D.C. Targeting of mesenchymal stromal cells by cre-recombinase transgenes commonly used to target osteoblast lineage cells. J. Bone Min. Res. 2016, 31, 2001–2007. [Google Scholar] [CrossRef] [PubMed]
- Kwiatkowski, D.J.; Zhang, H.; Bandura, J.L.; Heiberger, K.M.; Glogauer, M.; el-Hashemite, N.; Onda, H. A mouse model of tsc1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70s6 kinase activity in tsc1 null cells. Hum. Mol. Genet. 2002, 11, 525–534. [Google Scholar] [CrossRef] [PubMed]
- Soriano, P. Generalized lacz expression with the rosa26 cre reporter strain. Nat. Genet. 1999, 21, 70–71. [Google Scholar] [CrossRef] [PubMed]
- Meikle, L.; McMullen, J.R.; Sherwood, M.C.; Lader, A.S.; Walker, V.; Chan, J.A.; Kwiatkowski, D.J. A mouse model of cardiac rhabdomyoma generated by loss of tsc1 in ventricular myocytes. Hum. Mol. Genet. 2005, 14, 429–435. [Google Scholar] [CrossRef] [PubMed]
- Uezumi, A.; Fukada, S.; Yamamoto, N.; Takeda, S.; Tsuchida, K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat. Cell Biol. 2010, 12, 143–152. [Google Scholar] [CrossRef]
- Lepore, J.J.; Cheng, L.; Min Lu, M.; Mericko, P.A.; Morrisey, E.E.; Parmacek, M.S. High-efficiency somatic mutagenesis in smooth muscle cells and cardiac myocytes in sm22alpha-cre transgenic mice. Genesis 2005, 41, 179–184. [Google Scholar] [CrossRef]
- Kobayashi, T.; Minowa, O.; Sugitani, Y.; Takai, S.; Mitani, H.; Kobayashi, E.; Noda, T.; Hino, O. A germ-line tsc1 mutation causes tumor development and embryonic lethality that are similar, but not identical to, those caused by tsc2 mutation in mice. Proc. Natl. Acad. Sci. USA 2001, 98, 8762–8767. [Google Scholar] [CrossRef]
- Malhowski, A.J.; Hira, H.; Bashiruddin, S.; Warburton, R.; Goto, J.; Robert, B.; Kwiatkowski, D.J.; Finlay, G.A. Smooth muscle protein-22-mediated deletion of tsc1 results in cardiac hypertrophy that is mtorc1-mediated and reversed by rapamycin. Hum. Mol. Genet. 2011, 20, 1290–1305. [Google Scholar] [CrossRef]
- Guijarro, M.V.; Dahiya, S.; Danielson, L.S.; Segura, M.F.; Vales-Lara, F.M.; Menendez, S.; Popiolek, D.; Mittal, K.; Wei, J.J.; Zavadil, J.; et al. Dual pten/tp53 suppression promotes sarcoma progression by activating notch signaling. Am. J. Pathol. 2013, 182, 2015–2027. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Fujiwara, Y.; Chapdelaine, A.; Yang, H.; Orkin, S.H. Activation of egfp expression by cre-mediated excision in a new rosa26 reporter mouse strain. Blood 2001, 97, 324–326. [Google Scholar] [CrossRef] [PubMed]
- Houlihan, D.D.; Mabuchi, Y.; Morikawa, S.; Niibe, K.; Araki, D.; Suzuki, S.; Okano, H.; Matsuzaki, Y. Isolation of mouse mesenchymal stem cells on the basis of expression of sca-1 and pdgfr-alpha. Nat. Protoc. 2012, 7, 2103–2111. [Google Scholar] [CrossRef]
- Peister, A.; Mellad, J.A.; Larson, B.L.; Hall, B.M.; Gibson, L.F.; Prockop, D.J. Adult stem cells from bone marrow (mscs) isolated from different strains of inbred mice vary in surface epitopes, rates of proliferation, and differentiation potential. Blood 2004, 103, 1662–1668. [Google Scholar] [CrossRef] [PubMed]
- Moravcikova, E.; Meyer, E.M.; Corselli, M.; Donnenberg, V.S.; Donnenberg, A.D. Proteomic profiling of native unpassaged and culture-expanded mesenchymal stromal cells (msc). Cytom. Part A 2018, 93, 894–904. [Google Scholar] [CrossRef]
- Jeon, Y.J.; Kim, J.; Cho, J.H.; Chung, H.M.; Chae, J.I. Comparative analysis of human mesenchymal stem cells derived from bone marrow, placenta, and adipose tissue as sources of cell therapy. J. Cell Biochem. 2016, 117, 1112–1125. [Google Scholar] [CrossRef] [PubMed]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The international society for cellular therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Gan, B.; DePinho, R.A. Mtorc1 signaling governs hematopoietic stem cell quiescence. Cell Cycle 2009, 8, 1003–1006. [Google Scholar] [CrossRef]
- Wullschleger, S.; Loewith, R.; Hall, M.N. Tor signaling in growth and metabolism. Cell 2006, 124, 471–484. [Google Scholar] [CrossRef]
- Zhou, J.; Brugarolas, J.; Parada, L.F. Loss of tsc1, but not pten, in renal tubular cells causes polycystic kidney disease by activating mtorc1. Hum. Mol. Genet. 2009, 18, 4428–4441. [Google Scholar] [CrossRef]
- Shepherd, C.W.; Gomez, M.R.; Lie, J.T.; Crowson, C.S. Causes of death in patients with tuberous sclerosis. Mayo Clin. Proc. 1991, 66, 792–796. [Google Scholar] [CrossRef]
- Amin, S.; Lux, A.; Calder, N.; Laugharne, M.; Osborne, J.; O’Callaghan, F. Causes of mortality in individuals with tuberous sclerosis complex. Dev. Med. Child. Neurol 2017, 59, 612–617. [Google Scholar] [CrossRef]
- Rosu-Myles, M.; Fair, J.; Pearce, N.; Mehic, J. Non-multipotent stroma inhibit the proliferation and differentiation of mesenchymal stromal cells in vitro. Cytotherapy 2010, 12, 818–830. [Google Scholar] [CrossRef] [PubMed]
- Suto, E.G.; Mabuchi, Y.; Suzuki, N.; Suzuki, K.; Ogata, Y.; Taguchi, M.; Muneta, T.; Sekiya, I.; Akazawa, C. Prospectively isolated mesenchymal stem/stromal cells are enriched in the cd73+ population and exhibit efficacy after transplantation. Sci. Rep. 2017, 7, 4838. [Google Scholar] [CrossRef] [PubMed]
- Halfon, S.; Abramov, N.; Grinblat, B.; Ginis, I. Markers distinguishing mesenchymal stem cells from fibroblasts are downregulated with passaging. Stem Cells Dev. 2011, 20, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Friedenstein, A.J.; Chailakhjan, R.K.; Lalykina, K.S. The development of fibroblast colonies in monolayer cultures of guinea-pig bone marrow and spleen cells. Cell Tissue Kinet 1970, 3, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Friedenstein, A.J.; Deriglasova, U.F.; Kulagina, N.N.; Panasuk, A.F.; Rudakowa, S.F.; Luria, E.A.; Ruadkow, I.A. Precursors for fibroblasts in different populations of hematopoietic cells as detected by the in vitro colony assay method. Exp. Hematol. 1974, 2, 83–92. [Google Scholar] [PubMed]
- Digirolamo, C.M.; Stokes, D.; Colter, D.; Phinney, D.G.; Class, R.; Prockop, D.J. Propagation and senescence of human marrow stromal cells in culture: A simple colony-forming assay identifies samples with the greatest potential to propagate and differentiate. Br. J. Haematol. 1999, 107, 275–281. [Google Scholar] [CrossRef]
- Ma, J.; Meng, Y.; Kwiatkowski, D.J.; Chen, X.; Peng, H.; Sun, Q.; Zha, X.; Wang, F.; Wang, Y.; Jing, Y.; et al. Mammalian target of rapamycin regulates murine and human cell differentiation through stat3/p63/jagged/notch cascade. J. Clin. Investig. 2010, 120, 103–114. [Google Scholar] [CrossRef]
- Martin, K.A.; Merenick, B.L.; Ding, M.; Fetalvero, K.M.; Rzucidlo, E.M.; Kozul, C.D.; Brown, D.J.; Chiu, H.Y.; Shyu, M.; Drapeau, B.L.; et al. Rapamycin promotes vascular smooth muscle cell differentiation through insulin receptor substrate-1/phosphatidylinositol 3-kinase/akt2 feedback signaling. J. Biol. Chem. 2007, 282, 36112–36120. [Google Scholar] [CrossRef]
- Hegner, B.; Lange, M.; Kusch, A.; Essin, K.; Sezer, O.; Schulze-Lohoff, E.; Luft, F.C.; Gollasch, M.; Dragun, D. Mtor regulates vascular smooth muscle cell differentiation from human bone marrow-derived mesenchymal progenitors. Arter. Thromb. Vasc. Biol. 2009, 29, 232–238. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Wu, Z.; Li, P.; Cong, Q.; Chen, R.; Xu, W.; Biswas, S.; Liu, H.; Xia, X.; Li, S.; et al. Bone size and quality regulation: Concerted actions of mtor in mesenchymal stromal cells and osteoclasts. Stem Cell Rep. 2017, 8, 1600–1616. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Zhang, Z.; Jin, D.; Cai, C.; Jia, C.; Liu, W.; Wang, T.; Li, S.; Zhang, H.; Huang, B.; et al. Mtorc1 regulates pthrp to coordinate chondrocyte growth, proliferation and differentiation. Nat. Commun. 2016, 7, 11151. [Google Scholar] [CrossRef] [PubMed]
- Roforth, M.M.; Farr, J.N.; Fujita, K.; McCready, L.K.; Atkinson, E.J.; Therneau, T.M.; Cunningham, J.M.; Drake, M.T.; Monroe, D.G.; Khosla, S. Global transcriptional profiling using rna sequencing and DNA methylation patterns in highly enriched mesenchymal cells from young versus elderly women. Bone 2015, 76, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Gharibi, B.; Farzadi, S.; Ghuman, M.; Hughes, F.J. Inhibition of akt/mtor attenuates age-related changes in mesenchymal stem cells. Stem Cells 2014, 32, 2256–2266. [Google Scholar] [CrossRef] [PubMed]
- Davalli, P.; Mitic, T.; Caporali, A.; Lauriola, A.; D’Arca, D. Ros, cell senescence, and novel molecular mechanisms in aging and age-related diseases. Oxid Med. Cell Longev. 2016, 2016, 3565127. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Liu, Y.; Liu, Y.; Zheng, P. Mtor regulation and therapeutic rejuvenation of aging hematopoietic stem cells. Sci. Signal. 2009, 2, ra75. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Inoki, K.; Karbowniczek, M.; Petroulakis, E.; Sonenberg, N.; Henske, E.P.; Guan, K.L. Constitutive mtor activation in tsc mutants sensitizes cells to energy starvation and genomic damage via p53. EMBO J. 2007, 26, 4812–4823. [Google Scholar] [CrossRef] [PubMed]


| Gene | Primer Sequences |
|---|---|
| Tsc1 | F: 5′-ATGGCCCAGTTAGCCAACATT-3′ |
| R: 5′-CAGAATTGAGGGACTCCTTGAAG-3′ | |
| Gapdh | F: 5′-CCTGGAGAAACCTGCCAAGTATG-3′ |
| R: 5′-AGAGTGGGAGTTGCTGTTGAAGTC-3′ | |
| 18S rRNA | F: 5′-TTGTACACACCGCCCGTCGC-3′ |
| R: 5′-CTTCTCAGCGCTCCGCCAGG-3′ | |
| Asma | F: 5′-GAGAAGCCCAGCCAGTCG-3′ |
| R: 5′-CTCTTGCTCTGGGCTTCA-3′ | |
| Tagln (SM22α) | F: 5′-TAATGGCTTTGGGCAGTTTG-3′ |
| R: 5′-TGCAGTTGGCTGTCTGTGAA -3′ | |
| Myh11 (SM-Mhc) | F: 5′-GCAGAAGGCTCAGACCAAAG-3′ |
| R: 5′-TATCCAGAATGCCCAGGAAG-3′ | |
| Cebpα | F: 5′-GCCGAGATAAAGCCAAACAAC-3′ |
| R: 5′-GACCCGAAACCATCCTCTG-3′ | |
| Cebpβ | F: 5′-GCCAAGAAGACGGTGGACA-3′ |
| F: 5′-ACAAGTTCCGCAGGGTGCT-3′ | |
| Pparγ | F: 5′-TTGCTGAACGTGAAGCCCATCGAGG-3′ |
| R: 5′-GTCCTTGTAGATCTCCTGGAGCAG-3′ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guijarro, M.V.; Danielson, L.S.; Cañamero, M.; Nawab, A.; Abrahan, C.; Hernando, E.; Palmer, G.D. Tsc1 Regulates the Proliferation Capacity of Bone-Marrow Derived Mesenchymal Stem Cells. Cells 2020, 9, 2072. https://doi.org/10.3390/cells9092072
Guijarro MV, Danielson LS, Cañamero M, Nawab A, Abrahan C, Hernando E, Palmer GD. Tsc1 Regulates the Proliferation Capacity of Bone-Marrow Derived Mesenchymal Stem Cells. Cells. 2020; 9(9):2072. https://doi.org/10.3390/cells9092072
Chicago/Turabian StyleGuijarro, Maria V., Laura S. Danielson, Marta Cañamero, Akbar Nawab, Carolina Abrahan, Eva Hernando, and Glyn D. Palmer. 2020. "Tsc1 Regulates the Proliferation Capacity of Bone-Marrow Derived Mesenchymal Stem Cells" Cells 9, no. 9: 2072. https://doi.org/10.3390/cells9092072
APA StyleGuijarro, M. V., Danielson, L. S., Cañamero, M., Nawab, A., Abrahan, C., Hernando, E., & Palmer, G. D. (2020). Tsc1 Regulates the Proliferation Capacity of Bone-Marrow Derived Mesenchymal Stem Cells. Cells, 9(9), 2072. https://doi.org/10.3390/cells9092072





