Synthetic Notch-Receptor-Mediated Transmission of a Transient Signal into Permanent Information via CRISPR/Cas9-Based Genome Editing
Abstract
:1. Introduction
2. Materials and Methods
2.1. SynNotch Receptor and Response Element
2.2. Cell Culture Conditions
2.3. Lentiviral Preparation
2.4. Generation of HEK293 Receiver Cells
2.5. Generation of Clonal Receiver Cell Lines
2.6. Activation of HEK293 Receiver Cells
2.7. Flow Cytometry
2.8. GFP Sender Cells
2.9. SynNotch-Mediated Gene Editing Approaches
2.10. Gene Editing Experiments
2.11. Detection of Gene Editing Events
2.12. Statistical Analyses
3. Results
3.1. Generation of synNotch Receiver Cell Lines
3.2. Evaluation of Most Suitable Conditions for SynNotch Receptor Activation
3.3. SynNotch-Mediated Gene Editing
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wang, H.; Zang, C.; Liu, X.S.; Aster, J.C. The role of Notch receptors in transcriptional regulation. J. Cell Physiol. 2015, 230, 982–988. [Google Scholar] [CrossRef]
- Esensten, J.H.; Bluestone, J.A.; Lim, W.A. Engineering Therapeutic T Cells: From Synthetic Biology to Clinical Trials. Annu. Rev. Pathol. 2017, 12, 305–330. [Google Scholar] [CrossRef] [Green Version]
- Morsut, L.; Roybal, K.T.; Xiong, X.; Gordley, R.M.; Coyle, S.M.; Thomson, M.; Lim, W.A. Engineering Customized Cell Sensing and Response Behaviors Using Synthetic Notch Receptors. Cell 2016, 164, 780–791. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Wang, F.; Zhong, J.; Zhu, T.; Zheng, Y.; Zhao, T.; Xie, Q.; Ma, F.; Li, R.; Tang, Q.; et al. Using apelin-based synthetic Notch receptors to detect angiogenesis and treat solid tumors. Nat. Commun. 2020, 11, 2163. [Google Scholar] [CrossRef]
- Luo, H.; Wu, X.; Sun, R.; Su, J.; Wang, Y.; Dong, Y.; Shi, B.; Sun, Y.; Jiang, H.; Li, Z. Target-Dependent Expression of IL12 by synNotch Receptor-Engineered NK92 Cells Increases the Antitumor Activities of CAR-T Cells. Front. Oncol. 2019, 9, 1448. [Google Scholar] [CrossRef]
- Toda, S.; Blauch, L.R.; Tang, S.K.Y.; Morsut, L.; Lim, W.A. Programming self-organizing multicellular structures with synthetic cell-cell signaling. Science 2018, 361, 156–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.; Zhang, X.; Lv, J.; Yang, H.; Wang, X.; Ma, S.; Shao, R.; Peng, X.; Lin, Y.; Rong, Z. Cell-cell contact-induced gene editing/activation in mammalian cells using a synNotch-CRISPR/Cas9 system. Protein Cell 2020, 11, 299–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.J.; Yu, Z.Y.; Cai, Y.M.; Du, R.R.; Cai, L. Engineering of an enhanced synthetic Notch receptor by reducing ligand-independent activation. Commun. Biol. 2020, 3, 116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roybal, K.T.; Rupp, L.J.; Morsut, L.; Walker, W.J.; McNally, K.A.; Park, J.S.; Lim, W.A. Precision Tumor Recognition by T Cells With Combinatorial Antigen-Sensing Circuits. Cell 2016, 164, 770–779. [Google Scholar] [CrossRef] [Green Version]
- McKenna, A.; Findlay, G.M.; Gagnon, J.A.; Horwitz, M.S.; Schier, A.F.; Shendure, J. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 2016, 353, aaf7907. [Google Scholar] [CrossRef] [Green Version]
- Perli, S.D.; Cui, C.H.; Lu, T.K. Continuous genetic recording with self-targeting CRISPR-Cas in human cells. Science 2016, 353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farzadfard, F.; Gharaei, N.; Higashikuni, Y.; Jung, G.; Cao, J.; Lu, T.K. Single-Nucleotide-Resolution Computing and Memory in Living Cells. Mol. Cell 2019, 75, 769–780 e764. [Google Scholar] [CrossRef] [PubMed]
- Knopp, Y.; Geis, F.K.; Heckl, D.; Horn, S.; Neumann, T.; Kuehle, J.; Meyer, J.; Fehse, B.; Baum, C.; Morgan, M.; et al. Transient Retrovirus-Based CRISPR/Cas9 All-in-One Particles for Efficient, Targeted Gene Knockout. Mol. Ther. Nucleic Acids 2018, 13, 256–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fridy, P.C.; Li, Y.; Keegan, S.; Thompson, M.K.; Nudelman, I.; Scheid, J.F.; Oeffinger, M.; Nussenzweig, M.C.; Fenyo, D.; Chait, B.T.; et al. A robust pipeline for rapid production of versatile nanobody repertoires. Nat. Methods 2014, 11, 1253–1260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicholson, I.C.; Lenton, K.A.; Little, D.J.; Decorso, T.; Lee, F.T.; Scott, A.M.; Zola, H.; Hohmann, A.W. Construction and characterisation of a functional CD19 specific single chain Fv fragment for immunotherapy of B lineage leukaemia and lymphoma. Mol. Immunol. 1997, 34, 1157–1165. [Google Scholar] [CrossRef]
- Mao, Z.; Bozzella, M.; Seluanov, A.; Gorbunova, V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle 2008, 7, 2902–2906. [Google Scholar] [CrossRef]
- Frankenberg-Schwager, M.; Becker, M.; Garg, I.; Pralle, E.; Wolf, H.; Frankenberg, D. The role of nonhomologous DNA end joining, conservative homologous recombination, and single-strand annealing in the cell cycle-dependent repair of DNA double-strand breaks induced by H2O2 in mammalian cells. Radiat Res. 2008, 170, 784–793. [Google Scholar] [CrossRef]
- Baeumler, T.A.; Ahmed, A.A.; Fulga, T.A. Engineering Synthetic Signaling Pathways with Programmable dCas9-Based Chimeric Receptors. Cell Rep. 2017, 20, 2639–2653. [Google Scholar] [CrossRef] [Green Version]
- Lienert, F.; Lohmueller, J.J.; Garg, A.; Silver, P.A. Synthetic biology in mammalian cells: Next generation research tools and therapeutics. Nat. Rev. Mol. Cell Biol. 2014, 15, 95–107. [Google Scholar] [CrossRef]
- Lim, W.A. Designing customized cell signalling circuits. Nat. Rev. Mol. Cell Biol. 2010, 11, 393–403. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, J.; Schussler-Lenz, M.; Bondanza, A.; Buchholz, C.J. Clinical development of CAR T cells-challenges and opportunities in translating innovative treatment concepts. EMBO Mol. Med. 2017, 9, 1183–1197. [Google Scholar] [CrossRef] [PubMed]
- Hoepfner, J.; Kleinsorge, M.; Papp, O.; Ackermann, M.; Alfken, S.; Rinas, U.; Solodenko, W.; Kirschning, A.; Sgodda, M.; Cantz, T. Biphasic modulation of Wnt signaling supports efficient foregut endoderm formation from human pluripotent stem cells. Cell Biol. Int. 2016, 40, 534–548. [Google Scholar] [CrossRef] [PubMed]
- Sgodda, M.; Dai, Z.; Zweigerdt, R.; Sharma, A.D.; Ott, M.; Cantz, T. A Scalable Approach for the Generation of Human Pluripotent Stem Cell-Derived Hepatic Organoids with Sensitive Hepatotoxicity Features. Stem Cells Dev. 2017, 26, 1490–1504. [Google Scholar] [CrossRef] [PubMed]
- Koike, H.; Iwasawa, K.; Ouchi, R.; Maezawa, M.; Giesbrecht, K.; Saiki, N.; Ferguson, A.; Kimura, M.; Thompson, W.L.; Wells, J.M.; et al. Modelling human hepato-biliary-pancreatic organogenesis from the foregut-midgut boundary. Nature 2019, 574, 112–116. [Google Scholar] [CrossRef] [PubMed]







| Construct | % dsRed-Express-Positive Cells | % α-myc-APC-Positive Cells |
|---|---|---|
| GFP-tTA | 10.3 | 80.2 |
| CD19-Gal4 | 8.4 | 76.5 |
| HER2.3-Gal4 | 7.3 | 57.3 |
| HER2.5-Gal4 | 6.8 | 45.6 |
| Construct | % Activation in Bulk Population | % Activation in Clonal Cell Lines |
|---|---|---|
| GFP-tTA | 31.3 ± (−1.7) | 73.5 ± 2.1 |
| CD19-Gal4 | 18.9 ± 0.8 | 56.1 ± 5.9 |
| HER2.3-Gal4 | 20.4 ± 0.9 | 53.1 ± 2.0 |
| HER2.5-Gal4 | 25.3 ± 1.7 | 47.3 ± 6.5 |
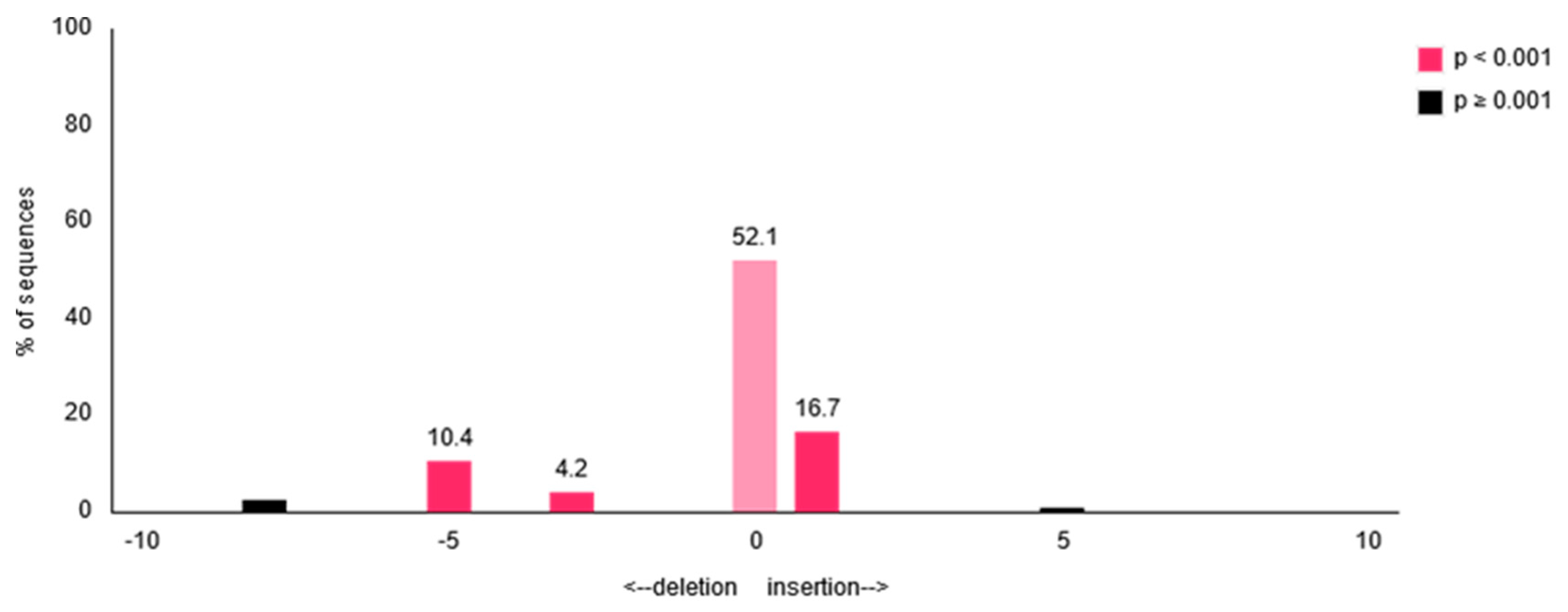
| Experiment | Overall Efficiency | Insertion | Deletion |
|---|---|---|---|
| Experiment 1 | 34.5% (p < 0.001)< | 17.5% | 17.0% |
| Experiment 2 | 41.3% (p < 0.001)< | 25.9% | 15.4% |
| Experiment 3 | 45.4% (p < 0.001)< | 30.3% | 15.1% |
| HEK293 cells | 0.9% (p ≥ 0.01) | 0.2% | 0.5% |
| Scramble control | 2.4% (p ≥ 0.01) | 0.1% | 2.3% |
| Positive control | 47.6% (p < 0.001)< | 30.9% | 16.7% |
| Not activated receiver cells | 2.7% (p < 0.001) | 1.3% | 1.4% |
| HEKempty activated receiver cells | 3.7% (p ≥ 0.001) | 1.6% | 2.1% |
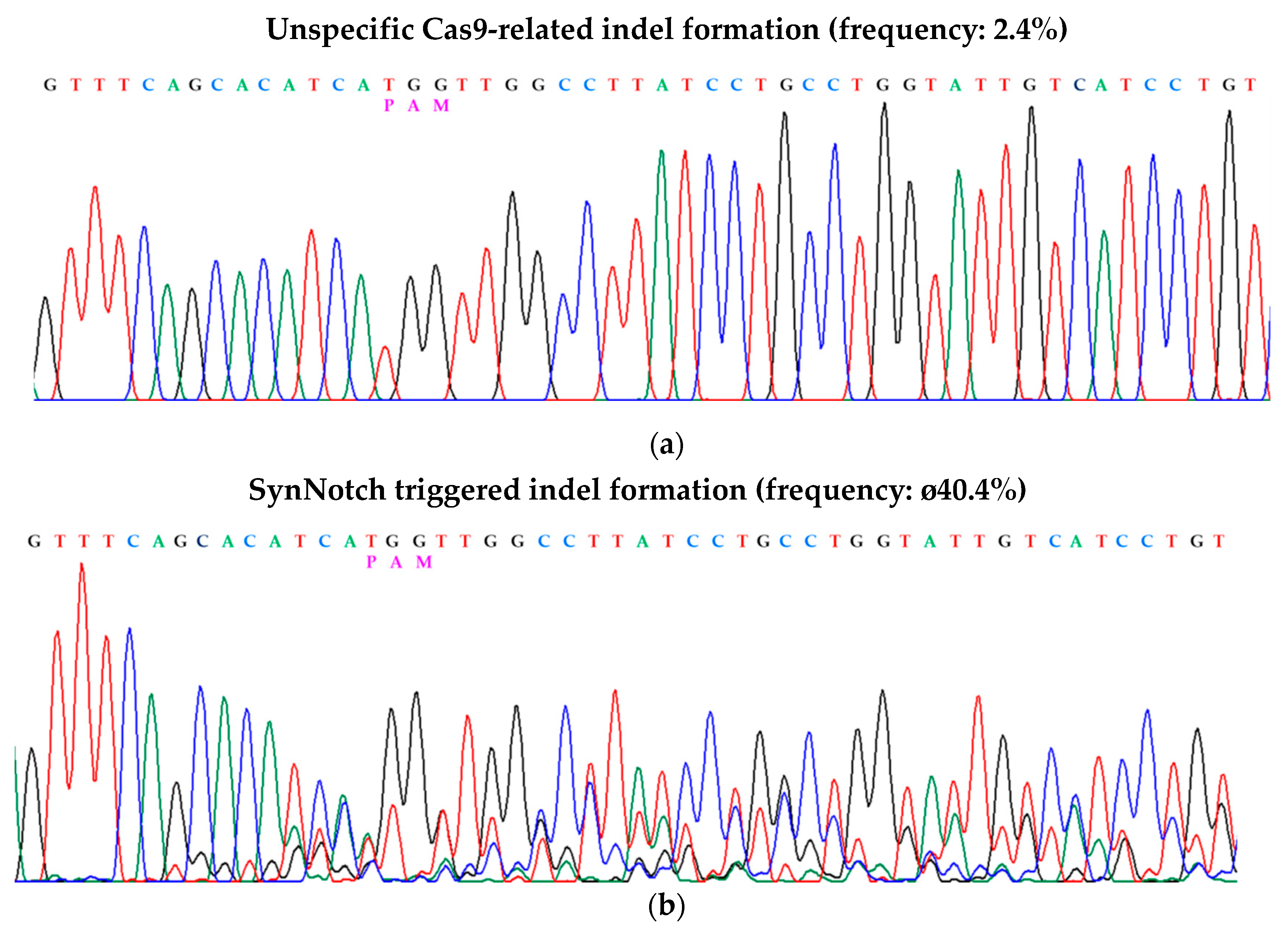
| PAM | indel | |
|---|---|---|
| CXCR4 | GTGTTCCAGTTTCAGCAC-ATCATGGTTGGCCTTATCCTGCCTGGTATT | |
| Clone 2 | GTGTTCCAGTTTCAGCAC-ATCATGGTTGGCCTTATCCTGCCTGGTATT | |
| Clone 5 | GTGTTCCAGTTTCAGCAC-ATCATGGTTGGCCTTATCCTGCCTGGTATT | |
| Clone 6 | GTGTTCCAGTTTCAGCAC-ATCATGGTTGGCCTTATCCTGCCTGGTATT | |
| Clone 9 | GTGTTCCAGTTTCAGCAC-ATCATGGTTGGCCTTATCCTGCCTGGTATT | |
| Clone 11 | GTGTTCCAGTTTCAGCAC-ATCATGGTTGGCCTTATCCTGCCTGGTATT | |
| Clone 12 | GTGTTCCAGTTTCAGCACAATCATGGTTGGCCTTATCCTGCCTGGTATT | +1 |
| Clone 3 | GTGTTCCAGTTTCAGCACAATCATGGTTGGCCTTATCCTGCCTGGTATT | +1 |
| Clone 8 | GTGTTCCAGTTTCAGCACAATCATGGTTGGCCTTATCCTGCCTGGTATT | +1 |
| Clone 7 | GTGTTCCAGTTTCAGCACAATCATGGTTGGCCTTATCCTGCCTGGTATT | +1 |
| Clone 4 | GTGTTCCAGTTTCAGCAC----ATGGTTGGCCTTATCCTGCCTGGTATT | −3 |
| Clone 10 | GTGTTCCAGTTTCAGC------ATGGTTGGCCTTATCCTGCCTGGTATT | −5 |
| Clone 1 | GTGTTCCAGTTTCAGC------ATGGTTGGCCTTATCCTGCCTGGTATT | −5 |
| Experiment | Indel Formation Frequency | Insertion | Deletion |
|---|---|---|---|
| Low FBS | 37.4% (p < 0.001) | 23.3% | 14.1% |
| No FBS | 31.4% (p < 0.001) | 19.0% | 12.4% |
| Nocodazole | 34.4% (p < 0.001) | 26.5% | 7.9% |
| Low FBS; HEKempty stimulated | 3.7% (p < 0.001) | 1.6% | 2.1% |
| No FBS; HEKempty stimulated | 3.3% (p < 0.001) | 1.5% | 1.8% |
| Nocodazole; HEKempty stimulated | 3.1% (p < 0.001) | 2.2% | 0.9% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sgodda, M.; Alfken, S.; Schambach, A.; Eggenschwiler, R.; Fidzinski, P.; Hummel, M.; Cantz, T. Synthetic Notch-Receptor-Mediated Transmission of a Transient Signal into Permanent Information via CRISPR/Cas9-Based Genome Editing. Cells 2020, 9, 1929. https://doi.org/10.3390/cells9091929
Sgodda M, Alfken S, Schambach A, Eggenschwiler R, Fidzinski P, Hummel M, Cantz T. Synthetic Notch-Receptor-Mediated Transmission of a Transient Signal into Permanent Information via CRISPR/Cas9-Based Genome Editing. Cells. 2020; 9(9):1929. https://doi.org/10.3390/cells9091929
Chicago/Turabian StyleSgodda, Malte, Susanne Alfken, Axel Schambach, Reto Eggenschwiler, Pawel Fidzinski, Michael Hummel, and Tobias Cantz. 2020. "Synthetic Notch-Receptor-Mediated Transmission of a Transient Signal into Permanent Information via CRISPR/Cas9-Based Genome Editing" Cells 9, no. 9: 1929. https://doi.org/10.3390/cells9091929
APA StyleSgodda, M., Alfken, S., Schambach, A., Eggenschwiler, R., Fidzinski, P., Hummel, M., & Cantz, T. (2020). Synthetic Notch-Receptor-Mediated Transmission of a Transient Signal into Permanent Information via CRISPR/Cas9-Based Genome Editing. Cells, 9(9), 1929. https://doi.org/10.3390/cells9091929






