FGF2 Inhibits Early Pancreatic Lineage Specification during Differentiation of Human Embryonic Stem Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Human ESC Culture and Differentiation
2.2. Gene and Expression Analysis
2.3. Immunofluorescence
2.4. SHH-ELISA
2.5. SHH Activity Assay
2.6. Transcriptomics
2.7. Proteomics
2.8. Western Blot
2.9. Statistics
3. Results
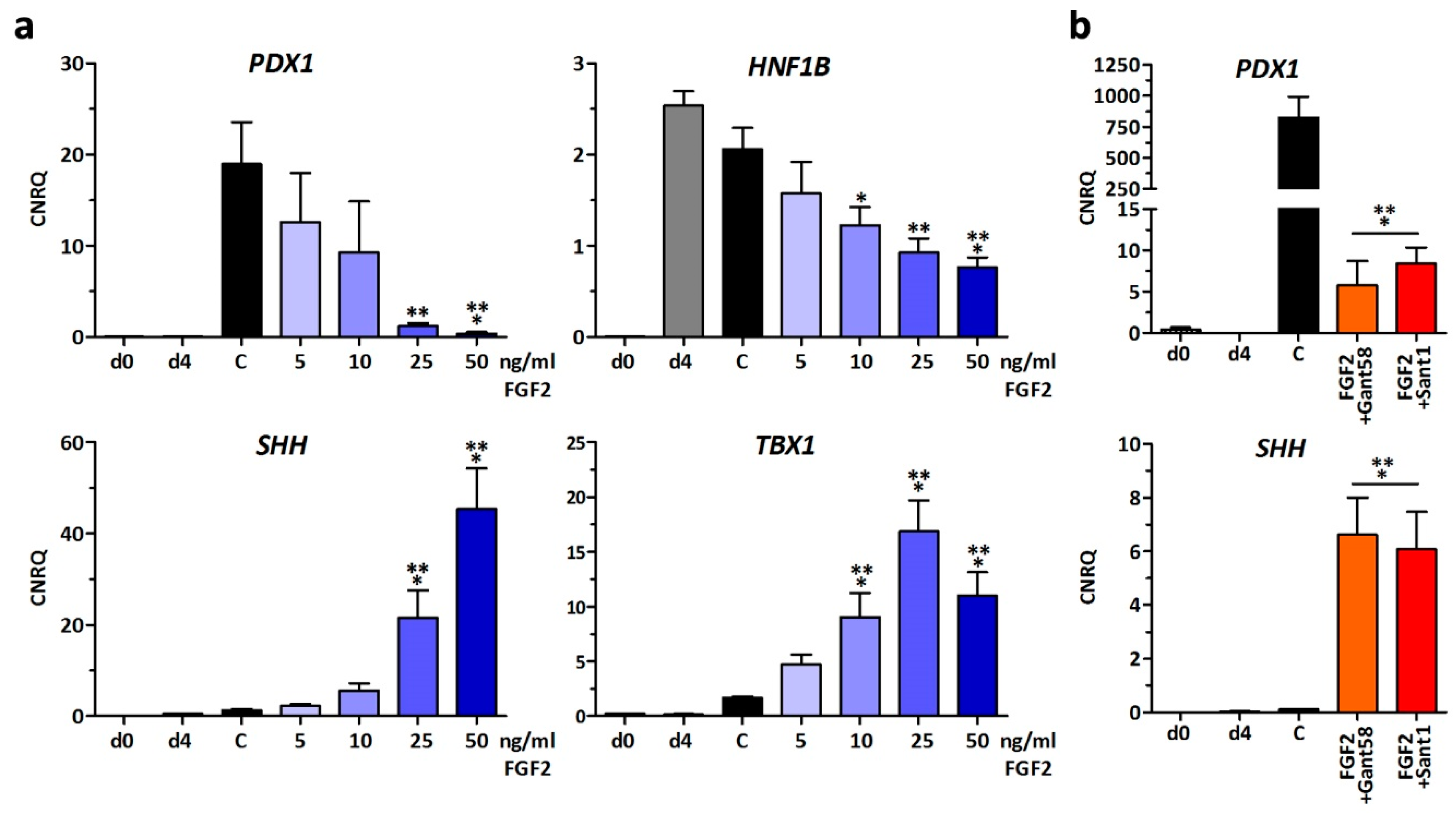
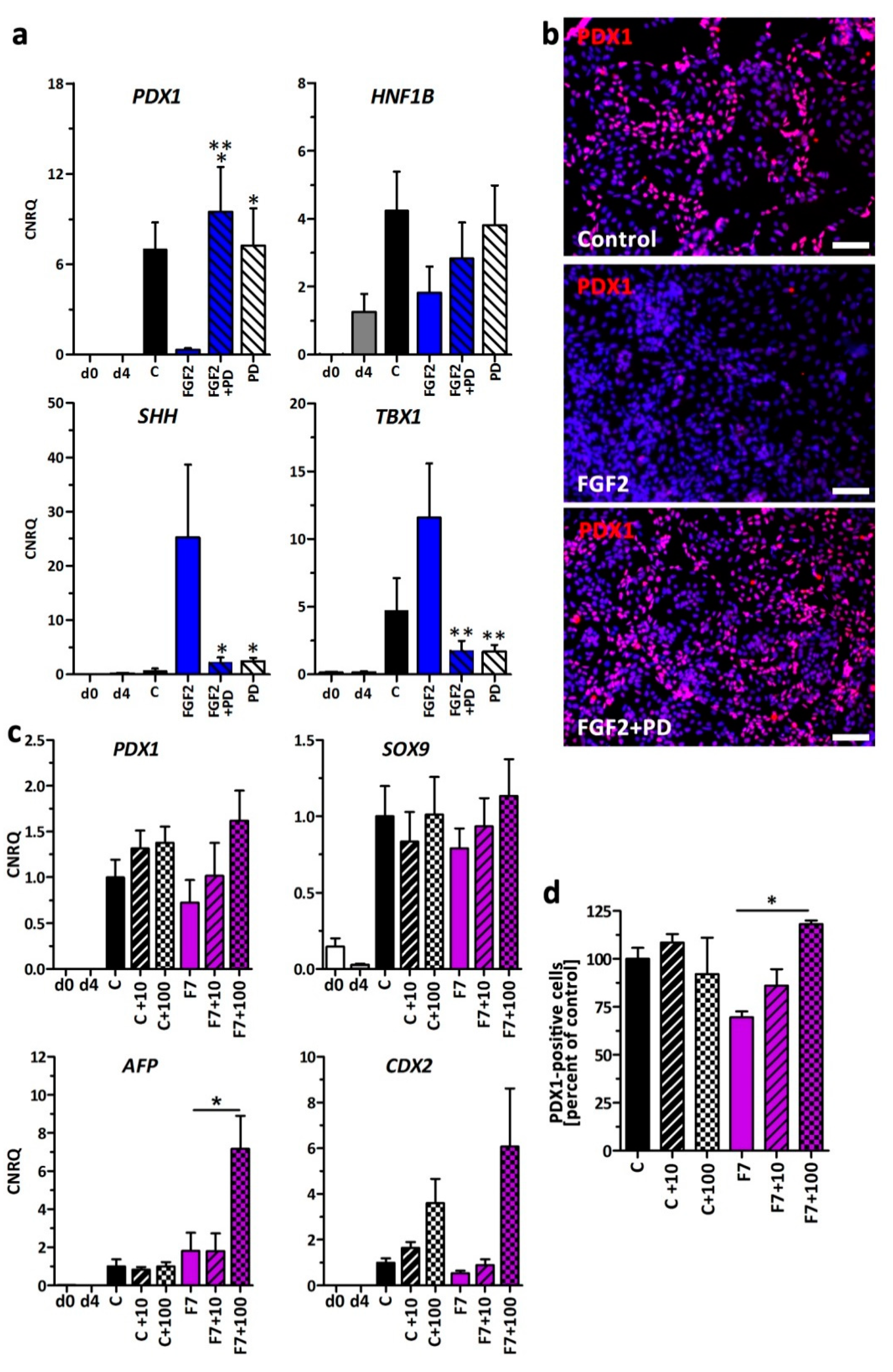
3.1. Effect of Growth Factors on Early Pancreatic Development
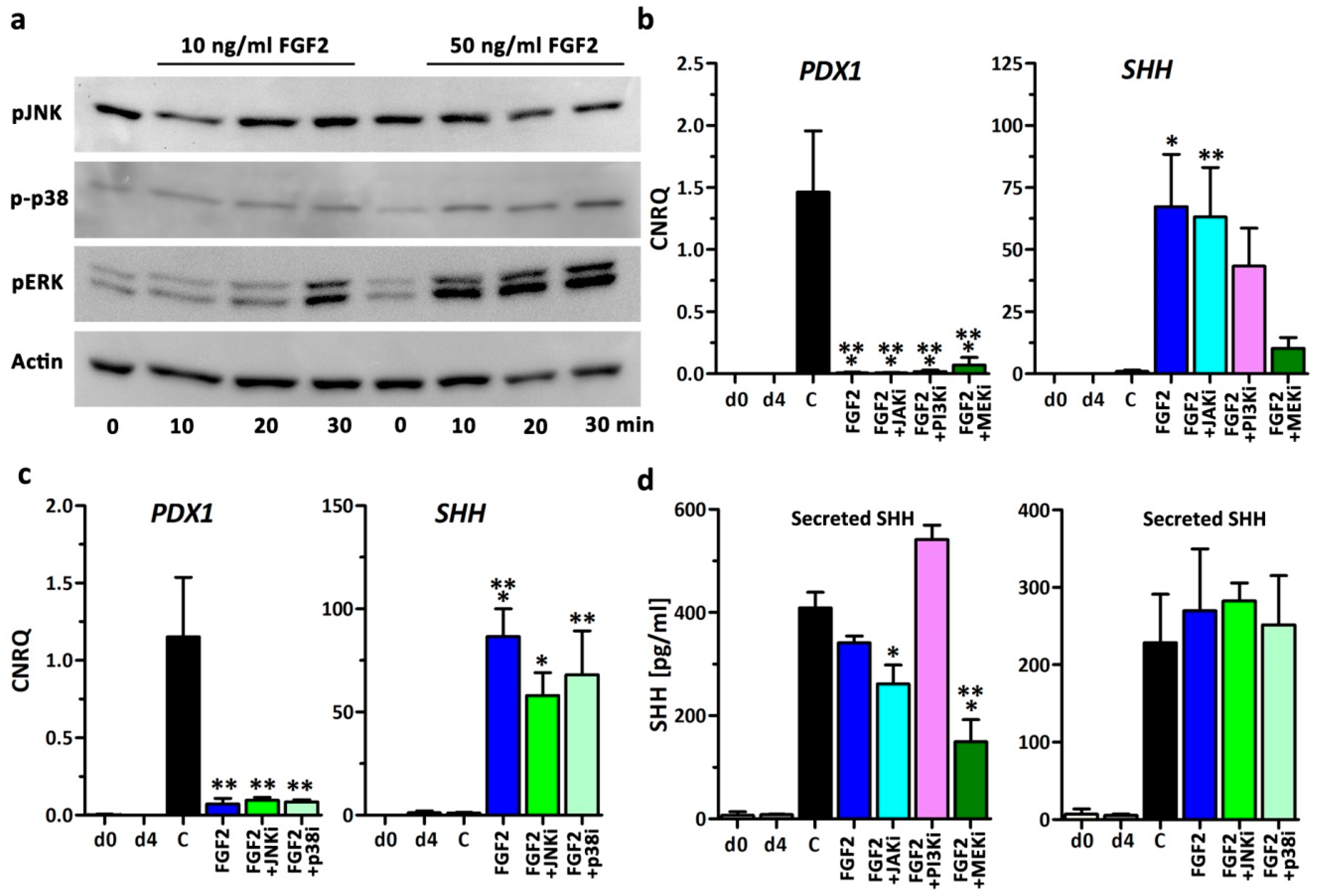
3.2. Small Molecule Inhibition of FGF-Signaling
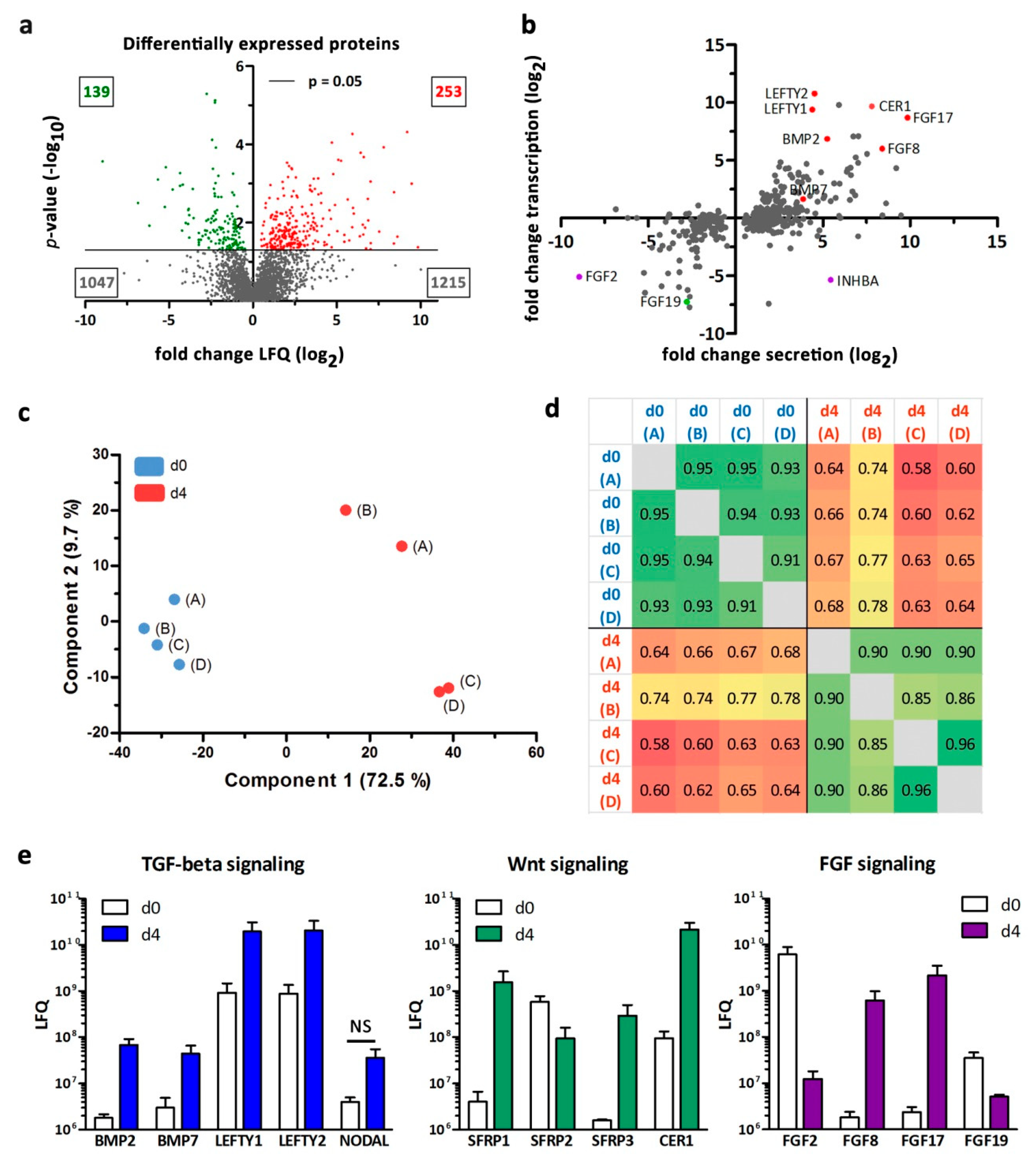
3.3. Secretome Analysis
3.4. FGF-Signaling through FGFR1c/3c
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bastidas-Ponce, A.; Scheibner, K.; Lickert, H.; Bakhti, M. Cellular and molecular mechanisms coordinating pancreas development. Development 2017, 144, 2873–2888. [Google Scholar] [CrossRef] [PubMed]
- Gittes, G.K. Developmental biology of the pancreas: A comprehensive review. Dev. Biol. 2009, 326, 4–35. [Google Scholar] [CrossRef] [PubMed]
- Hebrok, M.; Kim, S.K.; Melton, D.A. Notochord repression of endodermal Sonic hedgehog permits pancreas development. Genes Dev. 1998, 12, 1705–1713. [Google Scholar] [CrossRef] [PubMed]
- Hebrok, M.; Kim, S.K.; St Jacques, B.; McMahon, A.P.; Melton, D.A. Regulation of pancreas development by hedgehog signaling. Development 2000, 127, 4905–4913. [Google Scholar] [PubMed]
- Elghazi, L.; Cras-Meneur, C.; Czernichow, P.; Scharfmann, R. Role for FGFR2IIIb-mediated signals in controlling pancreatic endocrine progenitor cell proliferation. Proc. Natl. Acad. Sci. USA 2002, 99, 3884–3889. [Google Scholar] [CrossRef]
- Bhushan, A.; Itoh, N.; Kato, S.; Thiery, J.P.; Czernichow, P.; Bellusci, S.; Scharfmann, R. Fgf10 is essential for maintaining the proliferative capacity of epithelial progenitor cells during early pancreatic organogenesis. Development 2001, 128, 5109–5117. [Google Scholar]
- Cras-Meneur, C.; Elghazi, L.; Czernichow, P.; Scharfmann, R. Epidermal growth factor increases undifferentiated pancreatic embryonic cells in vitro: A balance between proliferation and differentiation. Diabetes 2001, 50, 1571–1579. [Google Scholar] [CrossRef]
- Pagliuca, F.W.; Millman, J.R.; Gurtler, M.; Segel, M.; Van Dervort, A.; Ryu, J.H.; Peterson, Q.P.; Greiner, D.; Melton, D.A. Generation of functional human pancreatic beta cells in vitro. Cell 2014, 159, 428–439. [Google Scholar] [CrossRef]
- Rezania, A.; Bruin, J.E.; Arora, P.; Rubin, A.; Batushansky, I.; Asadi, A.; O’Dwyer, S.; Quiskamp, N.; Mojibian, M.; Albrecht, T.; et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat. Biotechnol. 2014, 32, 1121–1133. [Google Scholar] [CrossRef]
- Ameri, J.; Borup, R.; Prawiro, C.; Ramond, C.; Schachter, K.A.; Scharfmann, R.; Semb, H. Efficient Generation of Glucose-Responsive Beta Cells from Isolated GP2(+) Human Pancreatic Progenitors. Cell Rep. 2017, 19, 36–49. [Google Scholar] [CrossRef]
- Nostro, M.C.; Sarangi, F.; Yang, C.; Holland, A.; Elefanty, A.G.; Stanley, E.G.; Greiner, D.L.; Keller, G. Efficient generation of NKX6-1+ pancreatic progenitors from multiple human pluripotent stem cell lines. Stem Cell Rep. 2015, 4, 591–604. [Google Scholar] [CrossRef] [PubMed]
- Davenport, C.; Diekmann, U.; Budde, I.; Detering, N.; Naujok, O. Anterior-Posterior Patterning of Definitive Endoderm Generated from Human Embryonic Stem Cells Depends on the Differential Signaling of Retinoic Acid, Wnt-, and BMP-Signaling. Stem Cells 2016. [Google Scholar] [CrossRef] [PubMed]
- Diekmann, U.; Wolling, H.; Dettmer, R.; Niwolik, I.; Naujok, O.; Buettner, F.F.R. Chemically defined and xenogeneic-free differentiation of human pluripotent stem cells into definitive endoderm in 3D culture. Sci. Rep. 2019, 9, 996. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, D.; Diekmann, U.; Fiedler, J.; Just, A.; Thum, T.; Lenzen, S.; Naujok, O. miRNome Profiling of Purified Endoderm and Mesoderm Differentiated from hESCs Reveals Functions of miR-483-3p and miR-1263 for Cell-Fate Decisions. Stem Cell Rep. 2017, 9, 1588–1603. [Google Scholar] [CrossRef]
- Diekmann, U.; Lenzen, S.; Naujok, O. A reliable and efficient protocol for human pluripotent stem cell differentiation into the definitive endoderm based on dispersed single cells. Stem Cells Dev. 2015, 24, 190–204. [Google Scholar] [CrossRef]
- Sasaki, H.; Hui, C.; Nakafuku, M.; Kondoh, H. A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development 1997, 124, 1313–1322. [Google Scholar]
- Wolling, H.; Konze, S.A.; Hofer, A.; Erdmann, J.; Pich, A.; Zweigerdt, R.; Buettner, F.F.R. Quantitative Secretomics Reveals Extrinsic Signals Involved in Human Pluripotent Stem Cell Cardiomyogenesis. Proteomics 2018, 18, e1800102. [Google Scholar] [CrossRef]
- Rabilloud, T.; Triboulet, S. Two-dimensional SDS-PAGE fractionation of biological samples for biomarker discovery. Methods Mol. Biol. 2013, 1002, 151–165. [Google Scholar]
- Chevallet, M.; Diemer, H.; Van Dorssealer, A.; Villiers, C.; Rabilloud, T. Toward a better analysis of secreted proteins: The example of the myeloid cells secretome. Proteomics 2007, 7, 1757–1770. [Google Scholar] [CrossRef]
- Shevchenko, A.; Tomas, H.; Havlis, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Lanner, F.; Rossant, J. The role of FGF/Erk signaling in pluripotent cells. Development 2010, 137, 3351–3360. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ibrahimi, O.A.; Olsen, S.K.; Umemori, H.; Mohammadi, M.; Ornitz, D.M. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J. Biol. Chem. 2006, 281, 15694–15700. [Google Scholar] [CrossRef]
- Ornitz, D.M.; Xu, J.; Colvin, J.S.; McEwen, D.G.; MacArthur, C.A.; Coulier, F.; Gao, G.; Goldfarb, M. Receptor specificity of the fibroblast growth factor family. J. Biol. Chem. 1996, 271, 15292–15297. [Google Scholar] [CrossRef]
- Bansal, R.; Magge, S.; Winkler, S. Specific inhibitor of FGF receptor signaling: FGF-2-mediated effects on proliferation, differentiation, and MAPK activation are inhibited by PD173074 in oligodendrocyte-lineage cells. J. Neurosci. Res. 2003, 74, 486–493. [Google Scholar] [CrossRef]
- Ameri, J.; Stahlberg, A.; Pedersen, J.; Johansson, J.K.; Johannesson, M.M.; Artner, I.; Semb, H. FGF2 specifies hESC-derived definitive endoderm into foregut/midgut cell lineages in a concentration-dependent manner. Stem Cells 2010, 28, 45–56. [Google Scholar] [CrossRef]
- Watson, B.A.; Feenstra, J.M.; Van Arsdale, J.M.; Rai-Bhatti, K.S.; Kim, D.J.H.; Coggins, A.S.; Mattison, G.L.; Yoo, S.; Steinman, E.D.; Pira, C.U.; et al. LHX2 Mediates the FGF-to-SHH Regulatory Loop during Limb Development. J. Dev. Biol. 2018, 6, 13. [Google Scholar] [CrossRef]
- Vitelli, F.; Taddei, I.; Morishima, M.; Meyers, E.N.; Lindsay, E.A.; Baldini, A. A genetic link between Tbx1 and fibroblast growth factor signaling. Development 2002, 129, 4605–4611. [Google Scholar]
- Mitsiadis, T.A.; Tucker, A.S.; De Bari, C.; Cobourne, M.T.; Rice, D.P. A regulatory relationship between Tbx1 and FGF signaling during tooth morphogenesis and ameloblast lineage determination. Dev. Biol. 2008, 320, 39–48. [Google Scholar] [CrossRef]
- Kempf, H.; Olmer, R.; Haase, A.; Franke, A.; Bolesani, E.; Schwanke, K.; Robles-Diaz, D.; Coffee, M.; Gohring, G.; Drager, G.; et al. Bulk cell density and Wnt/TGFbeta signalling regulate mesendodermal patterning of human pluripotent stem cells. Nat. Commun. 2016, 7, 13602. [Google Scholar] [CrossRef] [PubMed]
- Rostovskaya, M.; Bredenkamp, N.; Smith, A. Towards consistent generation of pancreatic lineage progenitors from human pluripotent stem cells. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20140365. [Google Scholar] [CrossRef] [PubMed]
- Loo, L.S.W.; Vethe, H.; Soetedjo, A.A.P.; Paulo, J.A.; Jasmen, J.; Jackson, N.; Bjorlykke, Y.; Valdez, I.A.; Vaudel, M.; Barsnes, H.; et al. Dynamic proteome profiling of human pluripotent stem cell-derived pancreatic progenitors. Stem Cells 2020, 38, 542–555. [Google Scholar] [CrossRef] [PubMed]
- Besser, D. Expression of nodal, lefty-a, and lefty-B in undifferentiated human embryonic stem cells requires activation of Smad2/3. J. Biol. Chem. 2004, 279, 45076–45084. [Google Scholar] [CrossRef] [PubMed]
- Zorn, A.M.; Wells, J.M. Vertebrate endoderm development and organ formation. Annu. Rev. Cell Dev. Biol. 2009, 25, 221–251. [Google Scholar] [CrossRef] [PubMed]
- Meno, C.; Gritsman, K.; Ohishi, S.; Ohfuji, Y.; Heckscher, E.; Mochida, K.; Shimono, A.; Kondoh, H.; Talbot, W.S.; Robertson, E.J.; et al. Mouse Lefty2 and zebrafish antivin are feedback inhibitors of nodal signaling during vertebrate gastrulation. Mol. Cell 1999, 4, 287–298. [Google Scholar] [CrossRef]
- Manson, S.R.; Austin, P.F.; Guo, Q.; Moore, K.H. BMP-7 Signaling and its Critical Roles in Kidney Development, the Responses to Renal Injury, and Chronic Kidney Disease. Vitam. Horm. 2015, 99, 91–144. [Google Scholar]
- Chung, W.-S.; Shin, C.H.; Stainier, D.Y.R. Bmp2 Signaling Regulates the Hepatic versus Pancreatic Fate Decision. Dev. Cell. 2008, 15, 738–748. [Google Scholar] [CrossRef]
- Sgodda, M.; Dai, Z.; Zweigerdt, R.; Sharma, A.D.; Ott, M.; Cantz, T. A Scalable Approach for the Generation of Human Pluripotent Stem Cell-Derived Hepatic Organoids with Sensitive Hepatotoxicity Features. Stem Cells Dev. 2017, 26, 1490–1504. [Google Scholar] [CrossRef]
- Krejci, P.; Kunova, M.; Kubikova, I.; Trantirek, L.; Kozubik, A.; Dvorak, P. Expression of FGF19 in human embryonic stem cells. Stem Cells 2013, 31, 2582–2584. [Google Scholar] [CrossRef]
- Deutsch, G.; Jung, J.; Zheng, M.; Lora, J.; Zaret, K.S. A bipotential precursor population for pancreas and liver within the embryonic endoderm. Development 2001, 128, 871–881. [Google Scholar] [PubMed]
- Vegas, A.J.; Veiseh, O.; Gurtler, M.; Millman, J.R.; Pagliuca, F.W.; Bader, A.R.; Doloff, J.C.; Li, J.; Chen, M.; Olejnik, K.; et al. Long-term glycemic control using polymer-encapsulated human stem cell-derived beta cells in immune-competent mice. Nat. Med. 2016, 22, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Veres, A.; Faust, A.L.; Bushnell, H.L.; Engquist, E.N.; Kenty, J.H.; Harb, G.; Poh, Y.C.; Sintov, E.; Gurtler, M.; Pagliuca, F.W.; et al. Charting cellular identity during human in vitro beta-cell differentiation. Nature 2019, 569, 368–373. [Google Scholar] [CrossRef] [PubMed]


| Gene Names | Protein Names | Mean LFQ (d0) | Mean LFQ (d4) | Fold Change LFQ (d4/d0) | Fold Change Transcription (d4/d0) |
|---|---|---|---|---|---|
| FGF17 | Fibroblast growth factor 17 | 2.3 × 106 | 2.2 × 109 | 918.56 | 418.27 |
| SFRP1 | Secreted frizzled-related protein 1 | 4.0 × 106 | 1.6 × 109 | 392.12 | 2.43 |
| FGF8 | Fibroblast growth factor 8 | 1.8 × 106 | 6.2 × 108 | 338.51 | 64.36 |
| CER1 | Cerberus | 9.6 × 107 | 2.1 × 1010 | 223.01 | 816.30 |
| SFRP3 (FRZB) | Secreted frizzled-related protein 3 | 1.6 × 106 | 2.9 × 108 | 185.39 | 46.72 |
| DKK1 | Dickkopf-related protein 1 | 1.3 × 106 | 1.7 × 108 | 129.79 | 27.65 |
| FLRT3 | Leucine-rich repeat transmembrane protein FLRT3 | 2.6 × 106 | 3.0 × 108 | 117.66 | 38.37 |
| STC1 | Stanniocalcin-1 | 2.9 ×107 | 2.5 × 109 | 85.60 | 18.76 |
| NPPB | Natriuretic peptides | 2.5 × 106 | 1.5 × 108 | 60.52 | 890.54 |
| MDK | Midkine | 1.7 × 107 | 7.6 × 108 | 43.79 | 2.77 |
| INHBA | Inhibin beta A chain | 6.0 × 106 | 2.6 × 108 | 43.67 | 0.02 |
| BMP2 | Bone morphogenetic protein 2 | 1.8 × 106 | 6.9 × 107 | 38.04 | 116.29 |
| CPE | Carboxypeptidase E | 9.5 × 107 | 2.8 × 109 | 29.74 | 15.76 |
| LEFTY2 | Left-right determination factor 2 | 8.9 × 108 | 2.0× 1010 | 23.03 | 1757.26 |
| LEFTY1 | Left-right determination factor 1 | 9.2 × 108 | 2.0 × 1010 | 21.16 | 679.87 |
| GRN | Granulins | 1.0 × 107 | 1.7 × 108 | 17.12 | 1.54 |
| BMP7 | Bone morphogenetic protein 7 | 3.0 × 106 | 4.4 × 107 | 14.70 | 3.16 |
| NRP2 | Neuropilin-2 | 1.1 × 107 | 1.1 × 108 | 9.73 | 0.40 |
| ROR2 | Tyrosine-protein kinase transmembrane receptor ROR2 | 1.3 ×107 | 9.8 × 107 | 7.31 | 19.48 |
| ITGB5 | Integrin beta-5 | 1.1 × 107 | 7.4 ×107 | 6.71 | 2.81 |
| HSPB1 | Heat shock protein beta-1 | 5.9 × 107 | 2.7 × 108 | 4.54 | 2.00 |
| PTK7 | Inactive tyrosine-protein kinase 7 | 2.9 × 108 | 1.3 × 109 | 4.54 | 1.10 |
| NXN | Nucleoredoxin | 5.7 ×107 | 2.1 × 108 | 3.64 | 1.70 |
| GPC4 | Glypican-4 | 6.7 × 108 | 2.4 × 109 | 3.63 | 1.61 |
| NCBP1 | Nuclear cap-binding protein subunit 1 | 2.5 × 107 | 9.1× 107 | 3.59 | 0.81 |
| SNW1 | SNW domain-containing protein 1 | 1.5 × 106 | 5.0 × 106 | 3.25 | 0.83 |
| FKBP8 | Peptidyl-prolyl cis-trans isomerase FKBP8 | 4.9 × 106 | 1.4 × 107 | 2.93 | 0.76 |
| GTF2F2 | General transcription factor IIF subunit 2 | 8.6 × 106 | 1.9 × 107 | 2.22 | 0.87 |
| ADAM10 | Disintegrin and metalloproteinase domain-containing protein 10 | 7.2 × 107 | 1.5 × 108 | 2.11 | 1.36 |
| TIAL1 | Nucleolysin TIAR | 1.9 × 108 | 1.1 × 108 | 0.57 | 0.82 |
| DDB1 | DNA damage-binding protein 1 | 1.6 × 109 | 7.4 × 108 | 0.45 | 0.95 |
| CTGF | Connective tissue growth factor | 1.2 × 109 | 4.7 × 108 | 0.392 | 0.39 |
| WDR12 | Ribosome biogenesis protein WDR12 | 1.5 × 108 | 4.3 × 107 | 0.290 | 0.53 |
| PARP1 | Poly [ADP-ribose] polymerase 1 | 3.0 × 109 | 5.8 × 108 | 0.191 | 0.51 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dettmer, R.; Cirksena, K.; Münchhoff, J.; Kresse, J.; Diekmann, U.; Niwolik, I.; Buettner, F.F.R.; Naujok, O. FGF2 Inhibits Early Pancreatic Lineage Specification during Differentiation of Human Embryonic Stem Cells. Cells 2020, 9, 1927. https://doi.org/10.3390/cells9091927
Dettmer R, Cirksena K, Münchhoff J, Kresse J, Diekmann U, Niwolik I, Buettner FFR, Naujok O. FGF2 Inhibits Early Pancreatic Lineage Specification during Differentiation of Human Embryonic Stem Cells. Cells. 2020; 9(9):1927. https://doi.org/10.3390/cells9091927
Chicago/Turabian StyleDettmer, Rabea, Karsten Cirksena, Julia Münchhoff, Jasmin Kresse, Ulf Diekmann, Isabell Niwolik, Falk F. R. Buettner, and Ortwin Naujok. 2020. "FGF2 Inhibits Early Pancreatic Lineage Specification during Differentiation of Human Embryonic Stem Cells" Cells 9, no. 9: 1927. https://doi.org/10.3390/cells9091927
APA StyleDettmer, R., Cirksena, K., Münchhoff, J., Kresse, J., Diekmann, U., Niwolik, I., Buettner, F. F. R., & Naujok, O. (2020). FGF2 Inhibits Early Pancreatic Lineage Specification during Differentiation of Human Embryonic Stem Cells. Cells, 9(9), 1927. https://doi.org/10.3390/cells9091927






