Effect of the 3D Artificial Nichoid on the Morphology and Mechanobiological Response of Mesenchymal Stem Cells Cultured In Vitro
Abstract
1. Introduction
2. Materials and Methods
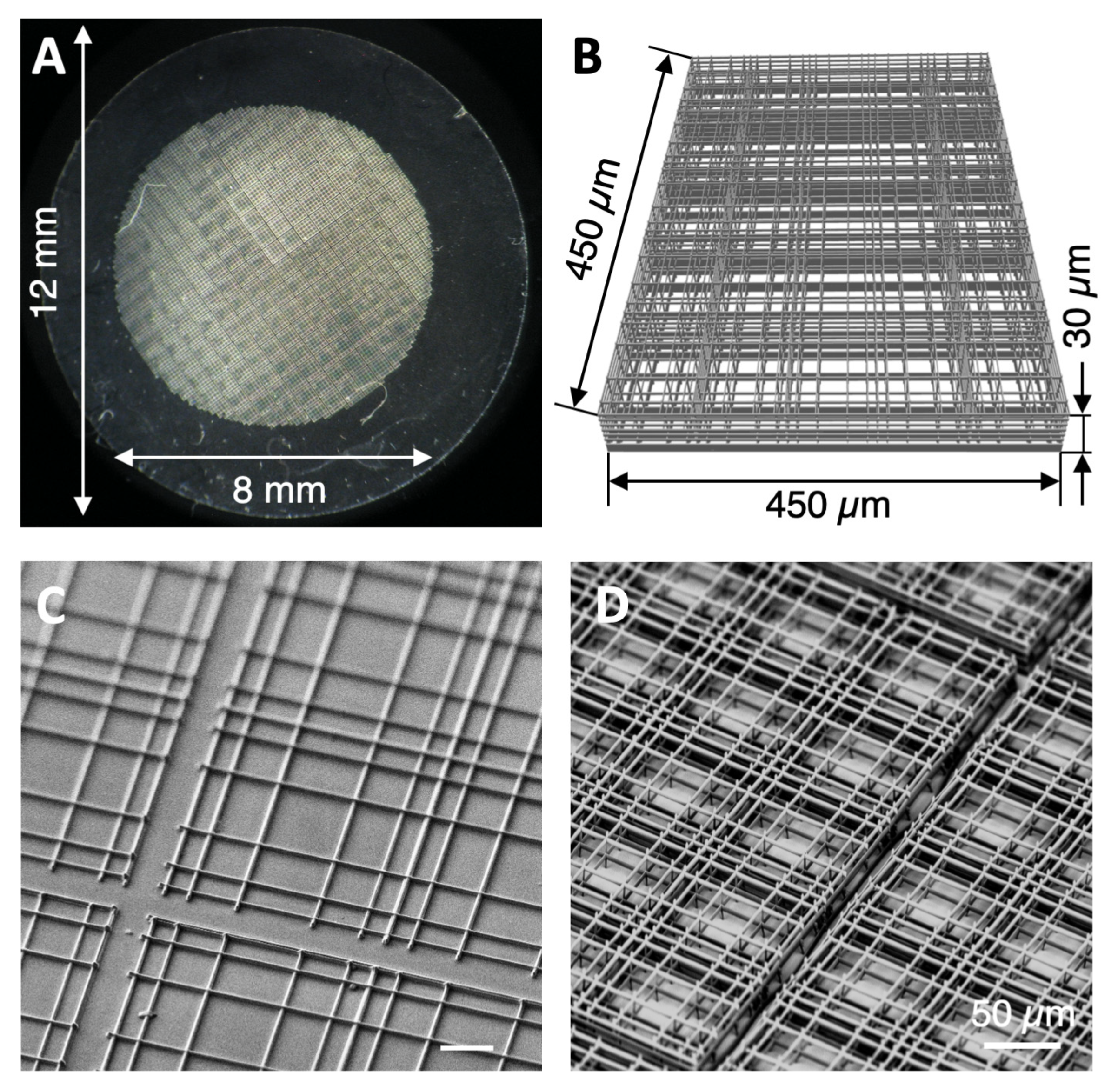
2.1. Fabrication of Nichoid Culture Substrates
2.2. Isolation of Rat MSCs
2.3. Substrate Preparation and MSCs Culture
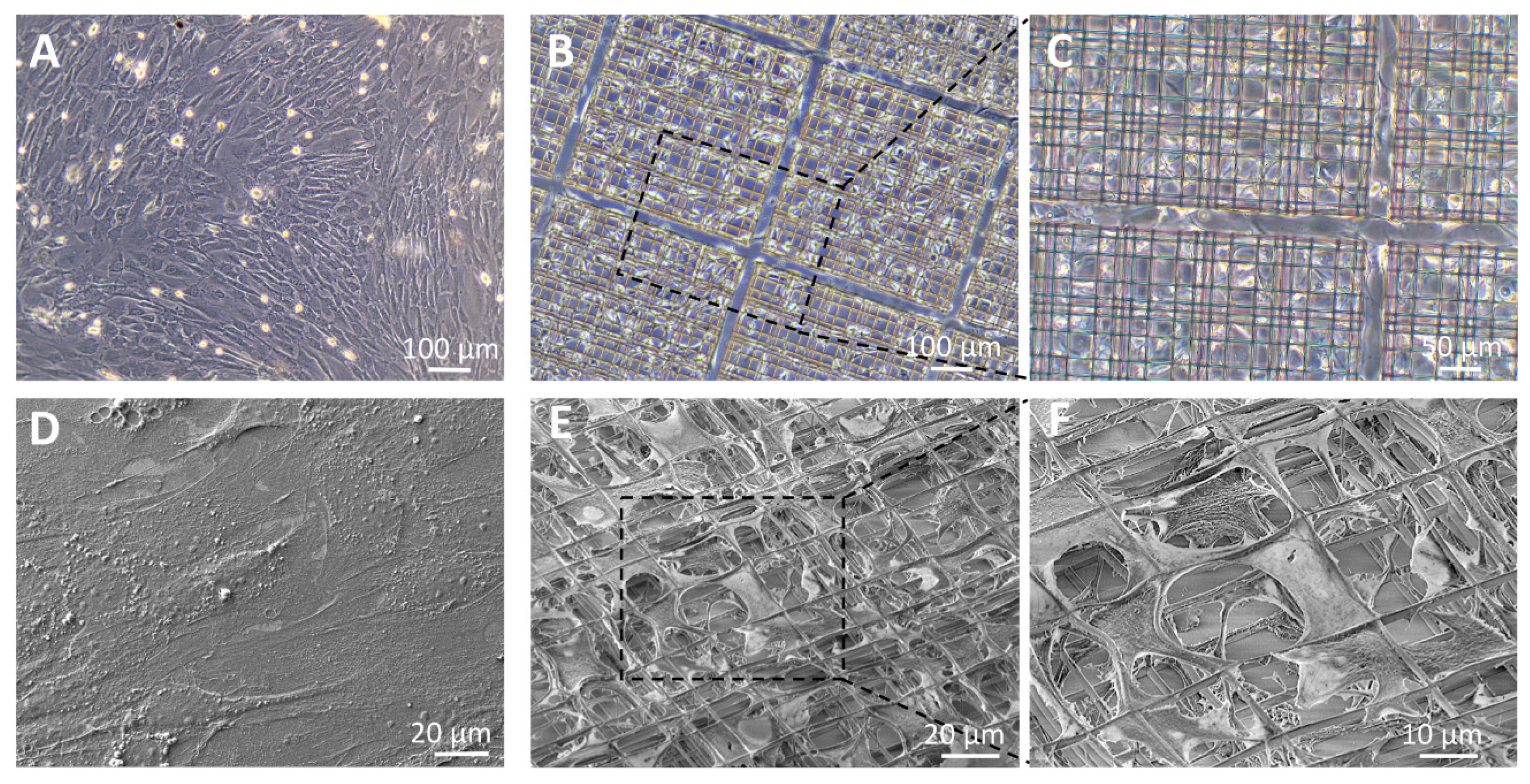
2.4. Scanning Electron Microscopy
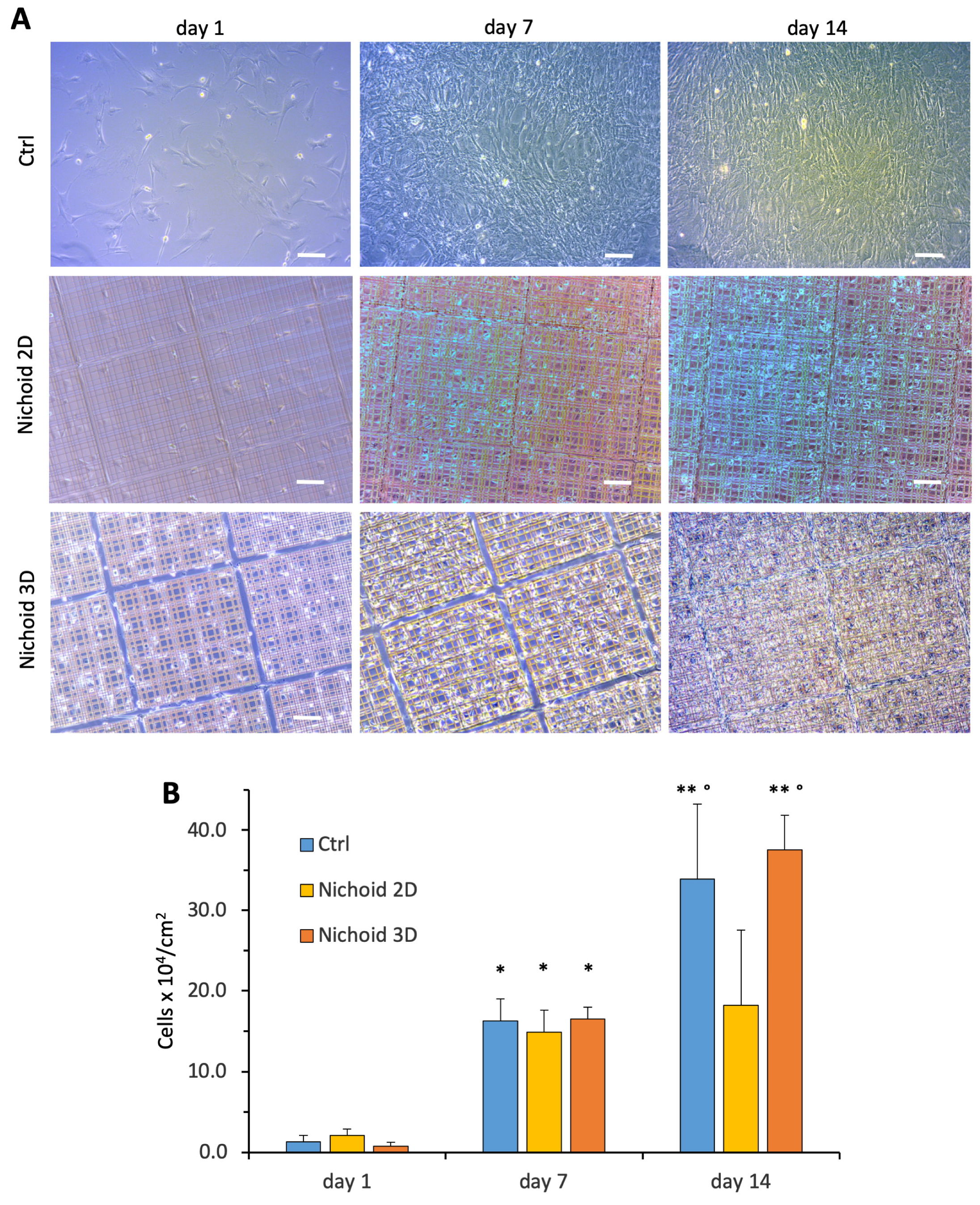
2.5. Cell Proliferation
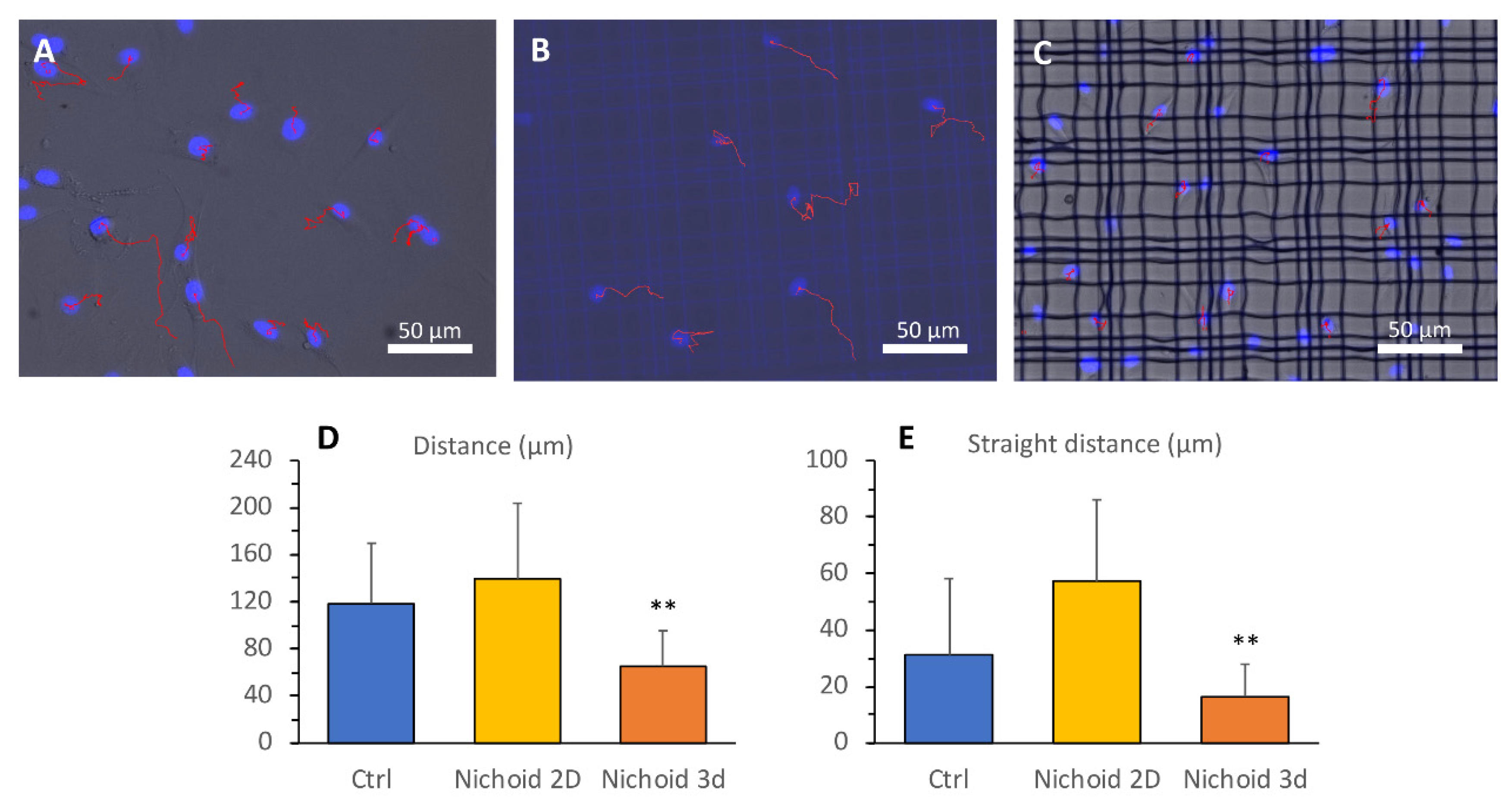
2.6. Analysis of Cellular Motility
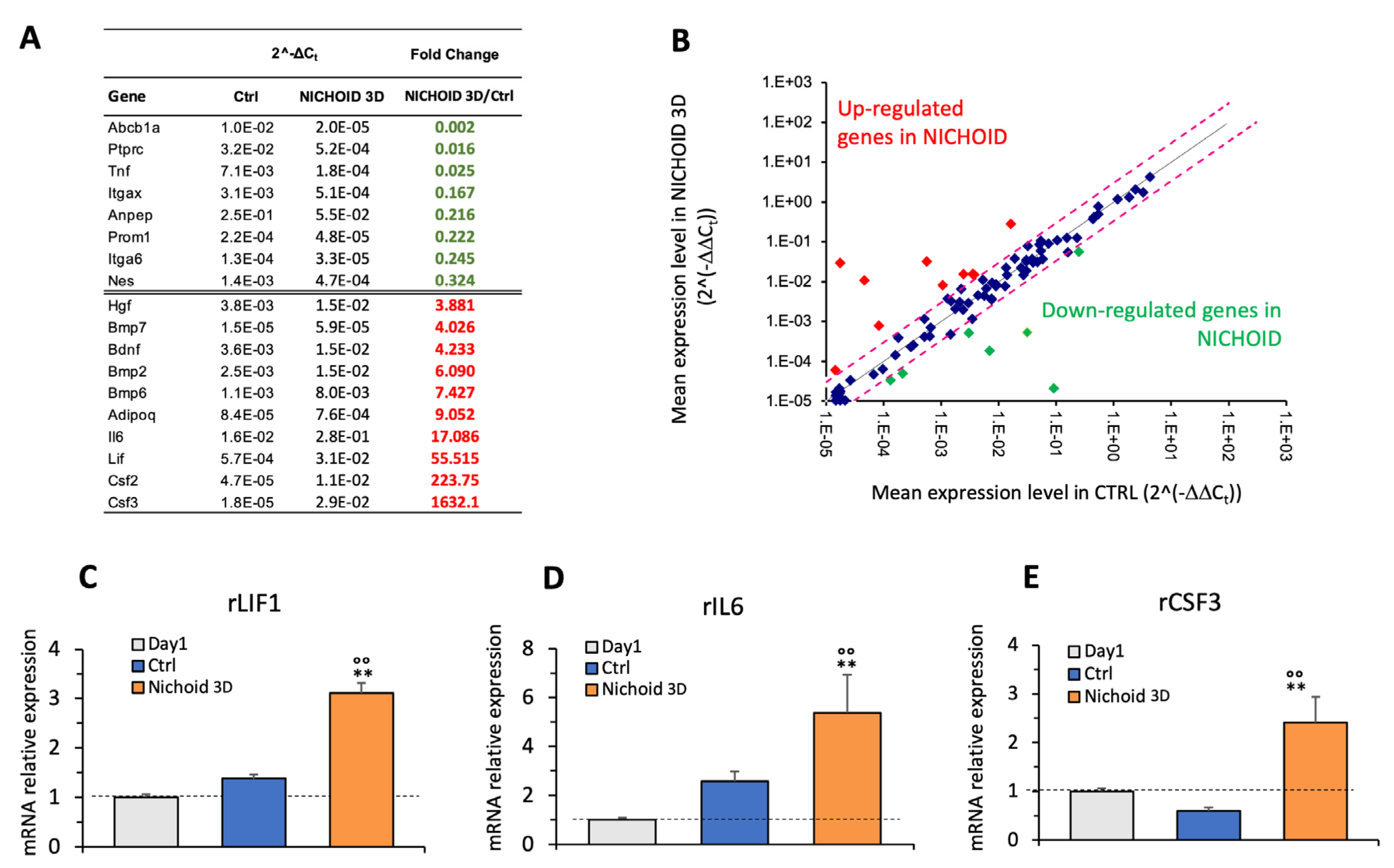
2.7. PCR Array
2.8. Real-Time PCR
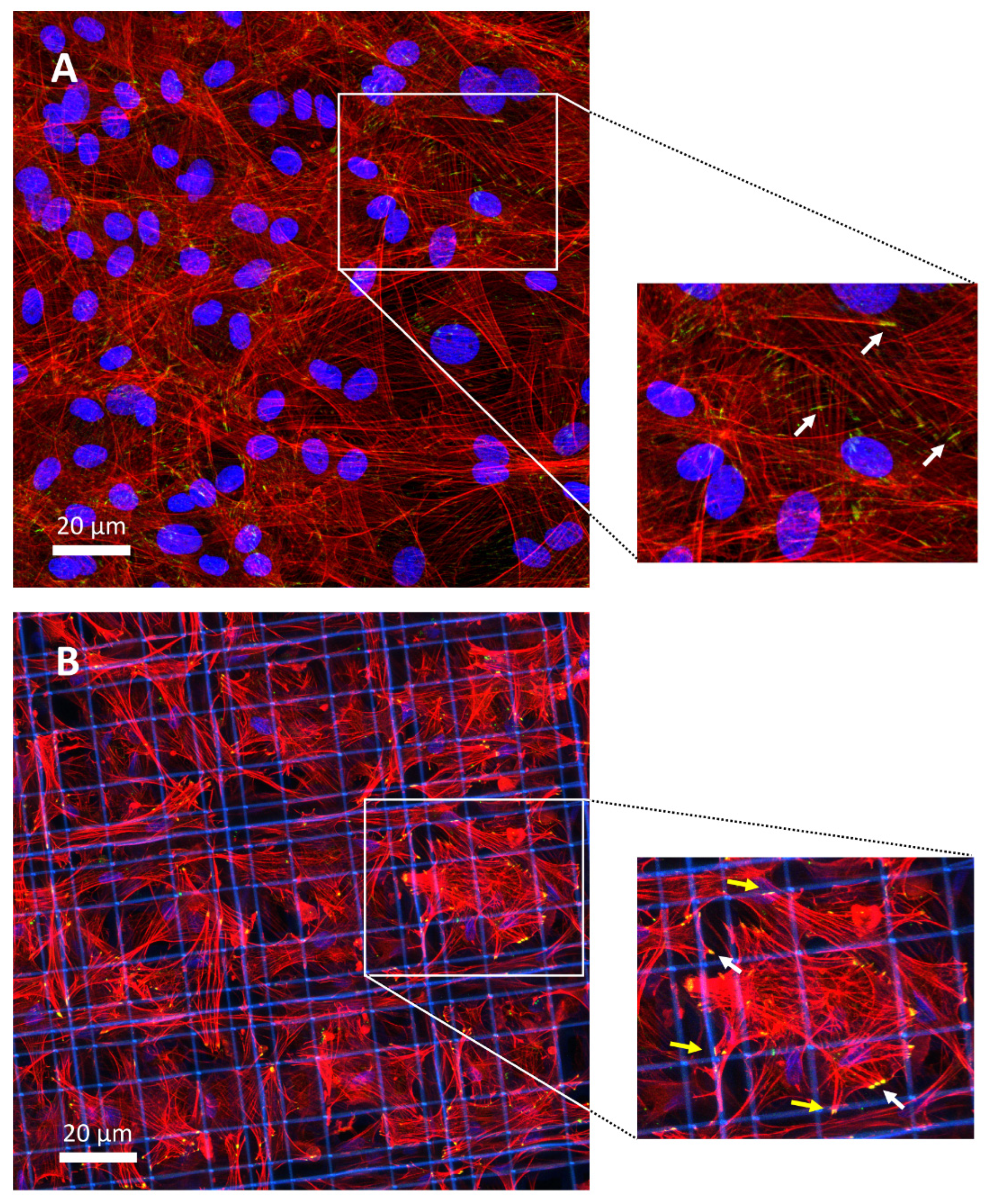
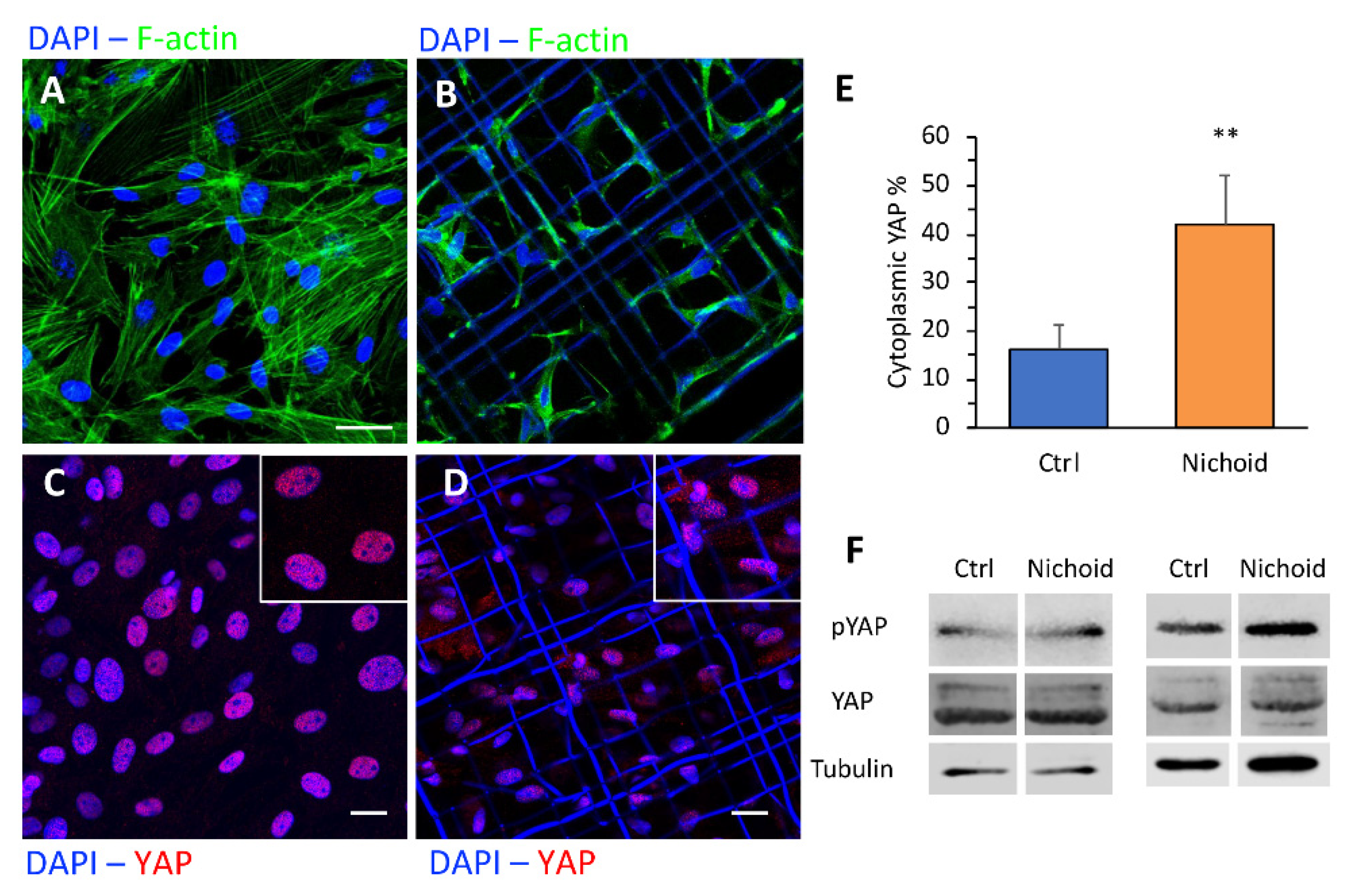
2.9. Immunofluorescence Analysis and Quantification of YAP and pFAK
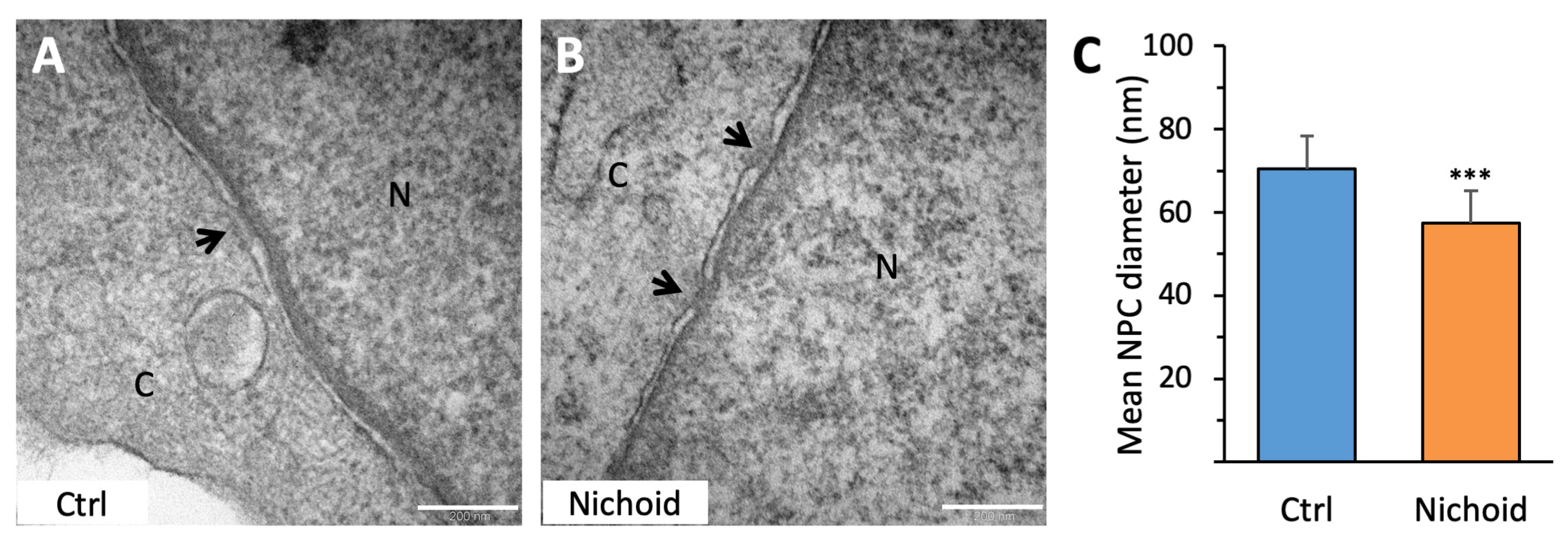
2.10. Transmission Electron Microscopy
2.11. Statistical Analysis
3. Results
3.1. Nichoid Structure Fabrication
3.2. Cell Morphology
3.3. MSC Growth and Proliferation
3.4. Focal Adhesion Contacts
3.5. Cell Motility
3.6. Gene Expression Profile
3.7. Immunofluorescence and Western Blot Analyses
3.8. MSC Ultrastructural Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ménard, C.; Tarte, K. Immunoregulatory properties of clinical grade mesenchymal stromal cells: Evidence, uncertainties, and clinical application. Stem Cell Res. Ther. 2013, 4, 64. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Baraniak, P.R.; McDevitt, T.C. Stem cell paracrine actions and tissue regeneration. Regen. Med. 2010, 5, 121–143. [Google Scholar] [CrossRef] [PubMed]
- Gnecchi, M.; Zhang, Z.; Ni, A.; Dzau, V.J. Paracrine mechanisms in adult stem cell signaling and therapy. Circ. Res. 2008, 103, 1204–1219. [Google Scholar] [CrossRef] [PubMed]
- Khubutiya, M.S.; Vagabov, A.V.; Temnov, A.; Sklifas, A.N. Paracrine mechanisms of proliferative, anti-apoptotic and anti-inflammatory effects of mesenchymal stromal cells in models of acute organ injury. Cytotherapy 2014, 16, 579–585. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-J.; Park, H.-J.; Lee, G.; Bang, O.Y.; Ahn, Y.H.; Joe, E.; Kim, H.O.; Lee, P.H. Neuroprotective effects of human mesenchymal stem cells on dopaminergic neurons through anti-inflammatory action. Glia 2009, 57, 13–23. [Google Scholar] [CrossRef]
- Le Blanc, K.; Frassoni, F.; Ball, L.; Locatelli, F.; Roelofs, H.; Lewis, I.; Lanino, E.; Sundberg, B.; Bernardo, M.E.; Remberger, M.; et al. Mesenchymal stem cells for treatment of steroid-resistant, severe, acute graft-versus-host disease: A phase II study. Lancet Lond. Engl. 2008, 371, 1579–1586. [Google Scholar] [CrossRef]
- Ranganath, S.H.; Levy, O.; Inamdar, M.S.; Karp, J.M. Harnessing the mesenchymal stem cell secretome for the treatment of cardiovascular disease. Cell Stem Cell 2012, 10, 244–258. [Google Scholar] [CrossRef]
- Phinney, D.G.; Pittenger, M.F. Concise Review: MSC-Derived Exosomes for Cell-Free Therapy. Stem Cells Dayt. Ohio. 2017, 35, 851–858. [Google Scholar] [CrossRef]
- Carelli, S.; Colli, M.; Vinci, V.; Caviggioli, F.; Klinger, M.; Gorio, A. Mechanical Activation of Adipose Tissue and Derived Mesenchymal Stem Cells: Novel Anti-Inflammatory Properties. Int. J. Mol. Sci. 2018, 19, 267. [Google Scholar] [CrossRef]
- Majumdar, M.K.; Thiede, M.A.; Haynesworth, S.E.; Bruder, S.P.; Gerson, S.L. Human marrow-derived mesenchymal stem cells (MSCs) express hematopoietic cytokines and support long-term hematopoiesis when differentiated toward stromal and osteogenic lineages. J. Hematotherapy Stem Cell Res. 2000, 9, 841–848. [Google Scholar] [CrossRef]
- A Noort, W.; Kruisselbrink, A.B.; Anker, P.S.I.; Kruger, M.; Van Bezooijen, R.L.; A De Paus, R.; Heemskerk, M.H.; Löwik, C.W.G.M.; Falkenburg, J.; Willemze, R.; et al. Mesenchymal stem cells promote engraftment of human umbilical cord blood-derived CD34(+) cells in NOD/SCID mice. Exp. Hematol. 2002, 30, 870–878. [Google Scholar] [CrossRef]
- Nava, M.M.; Raimondi, M.T.; Pietrabissa, R. Controlling self-renewal and differentiation of stem cells via mechanical cues. J. Biomed. Biotechnol. 2012, 2012, 797410. [Google Scholar] [CrossRef] [PubMed]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D. Matrix elasticity directs stem cell lineage specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zhou, J.; Chowdhury, F.; Cheng, J.; Wang, N.; Wang, F. Role of mechanical factors in fate decisions of stem cells. Regen. Med. 2011, 6, 229–240. [Google Scholar] [CrossRef]
- Chen, C.S.; Mrksich, M.; Huang, S.; Whitesides, G.M.; Ingber, D.E. Geometric control of cell life and death. Science 1997, 276, 1425–1428. [Google Scholar] [CrossRef]
- McBeath, R.; Pirone, D.M.; Nelson, C.M.; Bhadriraju, K.; Chen, C.S. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev. Cell 2004, 6, 483–495. [Google Scholar] [CrossRef]
- Wang, N.; Tytell, J.D.; Ingber, D.E. Mechanotransduction at a distance: Mechanically coupling the extracellular matrix with the nucleus. Nat. Rev. Mol. Cell Biol. 2009, 10, 75–82. [Google Scholar] [CrossRef]
- Yeung, T.; Georges, P.; Flanagan, L.A.; Marg, B.; Ortiz, M.; Funaki, M.; Zahir, N.; Ming, W.; Weaver, V.M.; Janmey, P.A. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil. Cytoskelet. 2004, 60, 24–34. [Google Scholar] [CrossRef]
- Gilbert, P.M.; Havenstrite, K.L.; Magnusson, K.E.G.; Sacco, A.; Leonardi, N.A.; Kraft, P.; Nguyen, N.K.; Thrun, S.; Lutolf, M.P.; Blau, H.M. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science 2010, 329, 1078–1081. [Google Scholar] [CrossRef]
- Flores-Figueroa, E.; Gratzinger, D. Beyond the Niche: Myelodysplastic Syndrome Topobiology in the Laboratory and in the Clinic. Int. J. Mol. Sci. 2016, 17, 553. [Google Scholar] [CrossRef]
- Reilly, G.C.; Engler, A.J. Intrinsic extracellular matrix properties regulate stem cell differentiation. J. Biomech. 2010, 43, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Vining, K.H.; Mooney, D.J. Mechanical forces direct stem cell behaviour in development and regeneration. Nat. Rev. Mol. Cell Biol. 2017, 18, 728–742. [Google Scholar] [CrossRef] [PubMed]
- Castillo, A.B.; Jacobs, C.R. Mesenchymal stem cell mechanobiology. Curr. Osteoporos. Rep. 2010, 8, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Tičkūnas, T.; Perrenoud, M.; Butkus, S.; Gadonas, R.; Tyte, S.R.; Malinauskas, M.; Paipulas, D.; Bellouard, Y.; Sirutkaitis, V. Combination of Additive and Subtractive Laser 3D Microprocessing in Hybrid Glass/Polymer Microsystems for Chemical Sensing Applications. Opt. Express 2017, 25, 26280–26288. [Google Scholar] [CrossRef]
- Ricci, D.; Nava, M.M.; Zandrini, T.; Cerullo, G.; Raimondi, M.T.; Osellame, R. Scaling-Up Techniques for the Nanofabrication of Cell Culture Substrates via Two-Photon Polymerization for Industrial-Scale Expansion of Stem Cells. Materials 2017, 10, 66. [Google Scholar] [CrossRef] [PubMed]
- Raimondi, M.T.; Eaton, S.M.; Laganà, M.; Aprile, V.; Nava, M.M.; Cerullo, G.; Osellame, R. Three-dimensional structural niches engineered via two-photon laser polymerization promote stem cell homing. Acta Biomater. 2013, 9, 4579–4584. [Google Scholar] [CrossRef]
- Raimondi, M.T.; Nava, M.M.; Eaton, S.M.; Bernasconi, A.; Vishnubhatla, K.C.; Cerullo, G.; Osellame, R. Optimization of Femtosecond Laser Polymerized Structural Niches to Control Mesenchymal Stromal Cell Fate in Culture. Micromachines 2014, 5, 341–358. [Google Scholar] [CrossRef]
- Nava, M.M.; Di Maggio, N.; Zandrini, T.; Cerullo, G.; Osellame, R.; Martin, I.; Raimondi, M.T. Synthetic niche substrates engineered via two-photon laser polymerization for the expansion of human mesenchymal stromal cells. J. Tissue Eng. Regen. Med. 2017, 11, 2836–2845. [Google Scholar] [CrossRef]
- Tajik, A.; Zhang, Y.; Wei, F.; Sun, J.; Jia, Q.; Zhou, W.; Singh, R.; Khanna, N.; Belmont, A.S.; Wang, N. Transcription upregulation via force-induced direct stretching of chromatin. Nat. Mater. 2016, 15, 1287–1296. [Google Scholar] [CrossRef]
- Piccolo, S.; Dupont, S.; Cordenonsi, M. The biology of YAP/TAZ: Hippo signaling and beyond. Physiol. Rev. 2014, 94, 1287–12312. [Google Scholar] [CrossRef]
- Pricola, K.L.; Kuhn, N.Z.; Haleem-Smith, H.; Song, Y.; Tuan, R.S. Interleukin-6 maintains bone marrow-derived mesenchymal stem cell stemness by an ERK1/2-dependent mechanism. J. Cell Biochem. 2009, 108, 577–588. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.-L.; Chen, J.-F.; Qiu, W.-H.; Wang, K.-W.; Xie, D.-Y.; Chen, X.-Y.; Liu, Q.L.; Peng, L.; Li, J.G.; Mei, Y.Y.; et al. Allogeneic bone marrow-derived mesenchymal stromal cells for hepatitis B virus-related acute-on-chronic liver failure: A randomized controlled trial. Hepatol. Baltim. Md. 2017, 66, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Morigi, M.; Rota, C.; Remuzzi, G. Mesenchymal Stem Cells in Kidney Repair. Methods Mol. Biol. Clifton. NJ. 2016, 1416, 89–107. [Google Scholar]
- Plock, J.A.; Schnider, J.T.; Schweizer, R.; Zhang, W.; Tsuji, W.; Waldner, M.; Waldner, M.; Solari, M.G.; Marra, K.G.; Rubin, J.P.; et al. The Influence of Timing and Frequency of Adipose-Derived Mesenchymal Stem Cell Therapy on Immunomodulation Outcomes After Vascularized Composite Allotransplantation. Transplantation 2017, 101, e1–e11. [Google Scholar] [CrossRef]
- Yu, P.; Wang, Z.; Liu, Y.; Xiao, Z.; Guo, Y.; Li, M.; Zhao, M. Marrow Mesenchymal Stem Cells Effectively Reduce Histologic Changes in a Rat Model of Chronic Renal Allograft Rejection. Transpl. Proc. 2017, 49, 2194–2203. [Google Scholar] [CrossRef]
- Wang, H.; Qi, F.; Dai, X.; Tian, W.; Liu, T.; Han, H.; Zhang, B.; Li, H.; Zhang, Z.; Du, C. Requirement of B7-H1 in mesenchymal stem cells for immune tolerance to cardiac allografts in combination therapy with rapamycin. Transpl. Immunol. 2014, 31, 65–74. [Google Scholar] [CrossRef]
- Park, J.S.; Burckhardt, C.J.; Lazcano, R.; Solis, L.M.; Isogai, T.; Li, L.; Chen, C.S.; Gao, B.; Minna, J.D.; Bachoo, R.; et al. Mechanical regulation of glycolysis via cytoskeleton architecture. Nature 2020, 578, 621–632. [Google Scholar] [CrossRef]
- Elosegui-Artola, A.; Andreu, I.; Beedle, A.E.M.; Lezamiz, A.; Uroz, M.; Kosmalska, A.J.; Oria, R.; Kechagia, J.Z.; Rico-Lastres, P.; Le Roux, A.L.; et al. Force Triggers YAP Nuclear Entry by Regulating Transport across Nuclear Pores. Cell 2017, 171, 1397–1410. [Google Scholar] [CrossRef]
- Nava, M.M.; Piuma, A.; Figliuzzi, M.; Cattaneo, I.; Bonandrini, B.; Zandrini, T.; Cerullo, G.; Osellame, R.; Remuzzi, A.; Raimondi, M.T. Two-photon polymerized “nichoid” substrates maintain function of pluripotent stem cells when expanded under feeder-free conditions. Stem Cell Res. Ther. 2016, 7, 132. [Google Scholar] [CrossRef]
- Nava, M.M.; Raimondi, M.T.; Credi, C.; De Marco, C.; Turri, S.; Cerullo, G.; Osellame, R. Interactions between structural and chemical biomimetism in synthetic stem cell niches. Biomed. Mater. Bristol. Engl. 2015, 10, 15012. [Google Scholar] [CrossRef]
- Zandrini, T.; Shan, O.; Parodi, V.; Cerullo, G.; Raimondi, M.T.; Osellame, R. Multi-foci laser microfabrication of 3D polymeric scaffolds for stem cell expansion in regenerative medicine. Sci. Rep. 2019, 9, 11761. [Google Scholar] [CrossRef] [PubMed]
- Ghibaudo, M.; Trichet, L.; Le Digabel, J.; Richert, A.; Hersen, P.; Ladoux, B. Substrate topography induces a crossover from 2D to 3D behavior in fibroblast migration. Biophys. J. 2009, 97, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Dupont, S.; Morsut, L.; Aragona, M.; Enzo, E.; Giulitti, S.; Cordenonsi, M.; Zanconato, F.; Le Digabel, J.; Forcato, M.; Bicciato, S.; et al. Role of YAP/TAZ in mechanotransduction. Nature 2011, 474, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Ege, N.; Dowbaj, A.; Jiang, M.; Howell, M.; Hooper, S.; Foster, C.; Jenkins, R.P.; Sahai, E. Quantitative Analysis Reveals that Actin and Src-Family Kinases Regulate Nuclear YAP1 and Its Export. Cell Syst. 2018, 6, 692–708. [Google Scholar] [CrossRef]
- Kolf, C.M.; Cho, E.; Tuan, R.S. Mesenchymal stromal cells. Biology of adult mesenchymal stem cells: Regulation of niche, self-renewal and differentiation. Arthritis Res. Ther. 2007, 9, 204. [Google Scholar] [CrossRef][Green Version]
- Popa-Wagner, A.; Stöcker, K.; Balseanu, A.T.; Rogalewski, A.; Diederich, K.; Minnerup, J.; Margaritescu, C.; Schabitz, W.-R. Effects of granulocyte-colony stimulating factor after stroke in aged rats. Stroke 2010, 41, 1027–1031. [Google Scholar] [CrossRef]
- Menzie, J.; Gharibani, P.; Modi, J.; Ma, Z.; Tao, R.; Prentice, H.; Wu, J.-Y. Granulocyte-colony stimulating factor protects against endoplasmic reticulum stress in an experimental model of stroke. Brain Res. 2018, 1682, 1–13. [Google Scholar] [CrossRef]
- Castro, E.R.; Cunningham, C.; Miller, J.; Brown, H.; Allan, S.M.; Pinteaux, E. Changes in the secretome of tri-dimensional spheroid-cultured human mesenchymal stem cells in vitro by interleukin-1 priming. Stem Cell Res. Ther. 2018, 9, 11. [Google Scholar] [CrossRef]
- Bartosh, T.J.; Ylöstalo, J.; Mohammadipoor, A.; Bazhanov, N.; Coble, K.; Claypool, K.; Lee, R.H.; Choi, H.; Prockop, D.J. Aggregation of human mesenchymal stromal cells (MSCs) into 3D spheroids enhances their antiinflammatory properties. Proc. Natl. Acad. Sci. USA 2010, 107, 13724–13729. [Google Scholar] [CrossRef]
- Santos, J.M.; Camões, S.P.; Filipe, E.C.; Cipriano, M.; Bárcia, R.N.; Filipe, M.; Teixeira, M.; Simões, S.; Gaspar, M.M.; Mosqueira, D.; et al. Three-dimensional spheroid cell culture of umbilical cord tissue-derived mesenchymal stromal cells leads to enhanced paracrine induction of wound healing. Stem Cell Res. Ther. 2015, 6, 90. [Google Scholar] [CrossRef]
- Choi, H.J.; Kim, N.E.; Kim, B.M.; Seo, M.; Heo, J.H. TNF-a-Induced YAP/TAZ Activity Mediates Leukocyte-Endothelial Adhesion by Regulating VCAM1 Expression in Endothelial Cells. Int. J. Mol. Sci. 2018, 19, 3428. [Google Scholar] [CrossRef] [PubMed]
- Ai, L.Q.Y.; Yuan, R.D.; Chen, X.; Liu, Y.J.; Liu, W.Y.; Zhu, J.Y.; Ye, J. Retinal Blood Vessel-Origin Yes-Associated Protein (YAP) Governs Astrocytic Maturation via Leukaemia Inhibitory Factor (LIF). Cell Prolif. 2020, 53, e12757. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Song, T.; Zhou, S.; Kong, X. YAP promotes the malignancy of endometrial cancer cells via regulation of IL-6 and IL-1. Mol. Med. 2019, 25, 32. [Google Scholar] [CrossRef]
- Neto, F.; Klaus-Bergmann, A.; Ong, A.T.; Alt, S.; Vion, A.C.; Szymborska, A.; Potente, M. YAP and TAZ regulate adherens junction dynamics and endothelial cell distribution during vascular development. eLife 2018, 7, e31037. [Google Scholar] [CrossRef] [PubMed]
- Pountos, I.; Georgouli, T.; Henshaw, K.; Bird, H.; Jones, E.; Giannoudis, P.V. The effect of bone morphogenetic protein-2, bone morphogenetic protein-7, parathyroid hormone, and platelet-derived growth factor on the proliferation and osteogenic differentiation of mesenchymal stem cells derived from osteoporotic bone. J. Orthop. Trauma 2010, 24, 552–556. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Remuzzi, A.; Bonandrini, B.; Tironi, M.; Longaretti, L.; Figliuzzi, M.; Conti, S.; Zandrini, T.; Osellame, R.; Cerullo, G.; Raimondi, M.T. Effect of the 3D Artificial Nichoid on the Morphology and Mechanobiological Response of Mesenchymal Stem Cells Cultured In Vitro. Cells 2020, 9, 1873. https://doi.org/10.3390/cells9081873
Remuzzi A, Bonandrini B, Tironi M, Longaretti L, Figliuzzi M, Conti S, Zandrini T, Osellame R, Cerullo G, Raimondi MT. Effect of the 3D Artificial Nichoid on the Morphology and Mechanobiological Response of Mesenchymal Stem Cells Cultured In Vitro. Cells. 2020; 9(8):1873. https://doi.org/10.3390/cells9081873
Chicago/Turabian StyleRemuzzi, Andrea, Barbara Bonandrini, Matteo Tironi, Lorena Longaretti, Marina Figliuzzi, Sara Conti, Tommaso Zandrini, Roberto Osellame, Giulio Cerullo, and Manuela Teresa Raimondi. 2020. "Effect of the 3D Artificial Nichoid on the Morphology and Mechanobiological Response of Mesenchymal Stem Cells Cultured In Vitro" Cells 9, no. 8: 1873. https://doi.org/10.3390/cells9081873
APA StyleRemuzzi, A., Bonandrini, B., Tironi, M., Longaretti, L., Figliuzzi, M., Conti, S., Zandrini, T., Osellame, R., Cerullo, G., & Raimondi, M. T. (2020). Effect of the 3D Artificial Nichoid on the Morphology and Mechanobiological Response of Mesenchymal Stem Cells Cultured In Vitro. Cells, 9(8), 1873. https://doi.org/10.3390/cells9081873







