Epithelial Cell Adhesion Molecule: An Anchor to Isolate Clinically Relevant Circulating Tumor Cells
Abstract
1. Introduction
2. EpCAM in Cancer
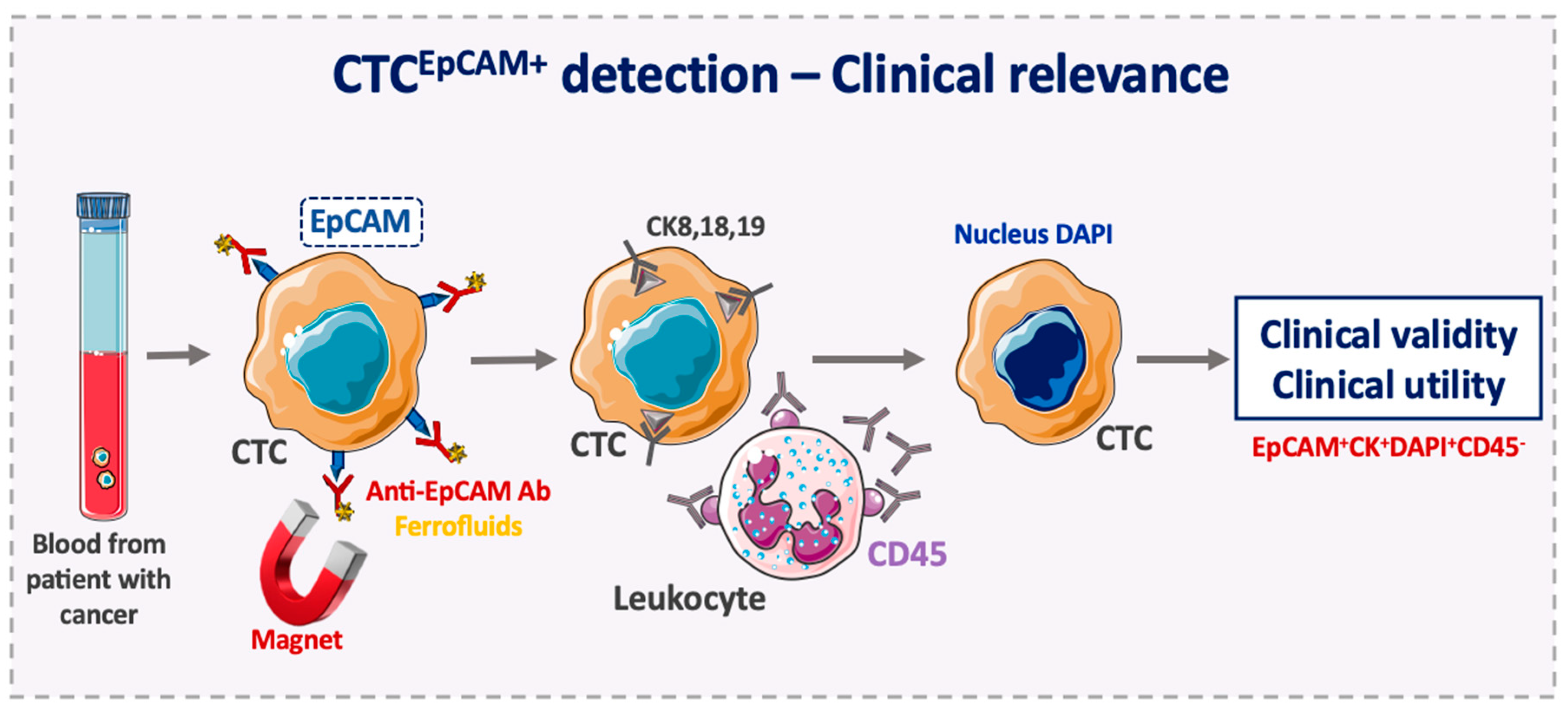
3. Advantages of EpCAM Use as a CTC Diagnostic Marker
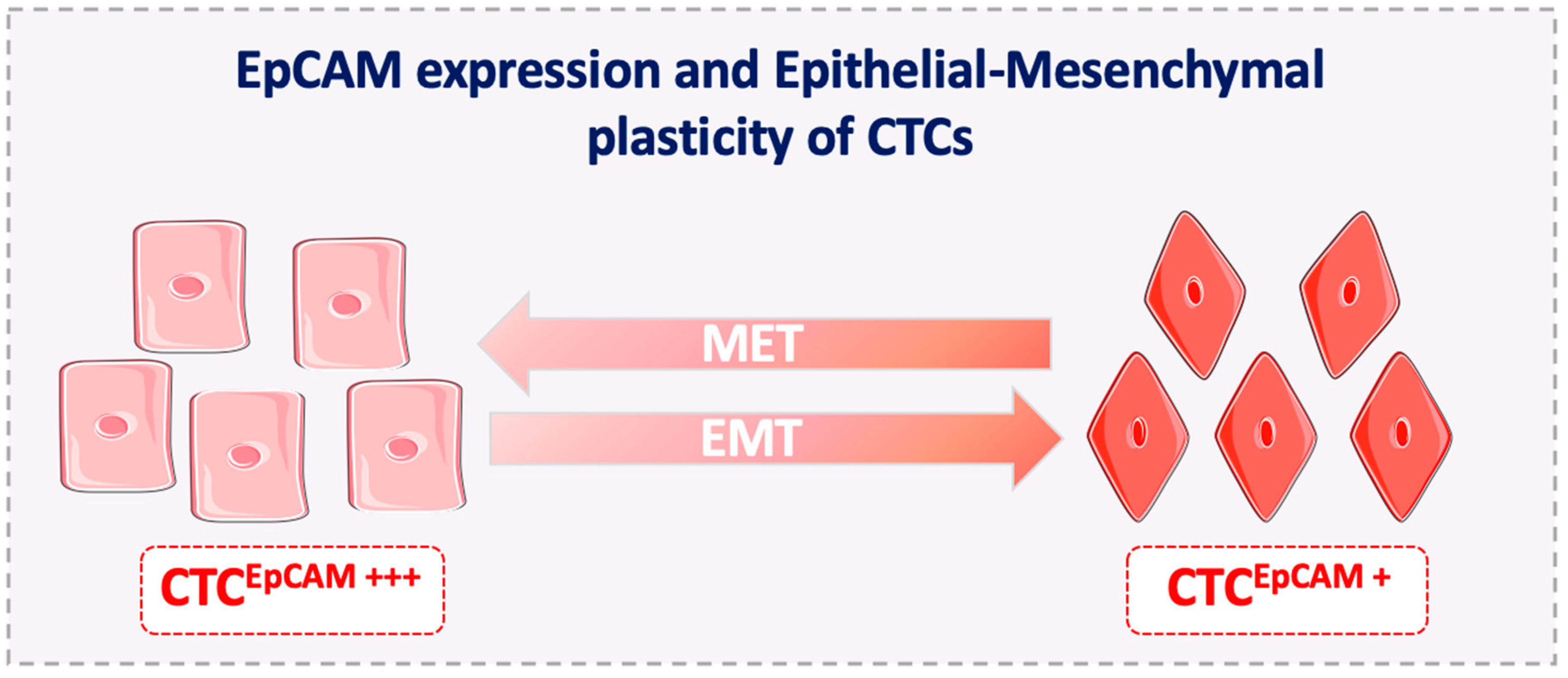
4. Disadvantages of EpCAM Use as a CTC Diagnostic Marker
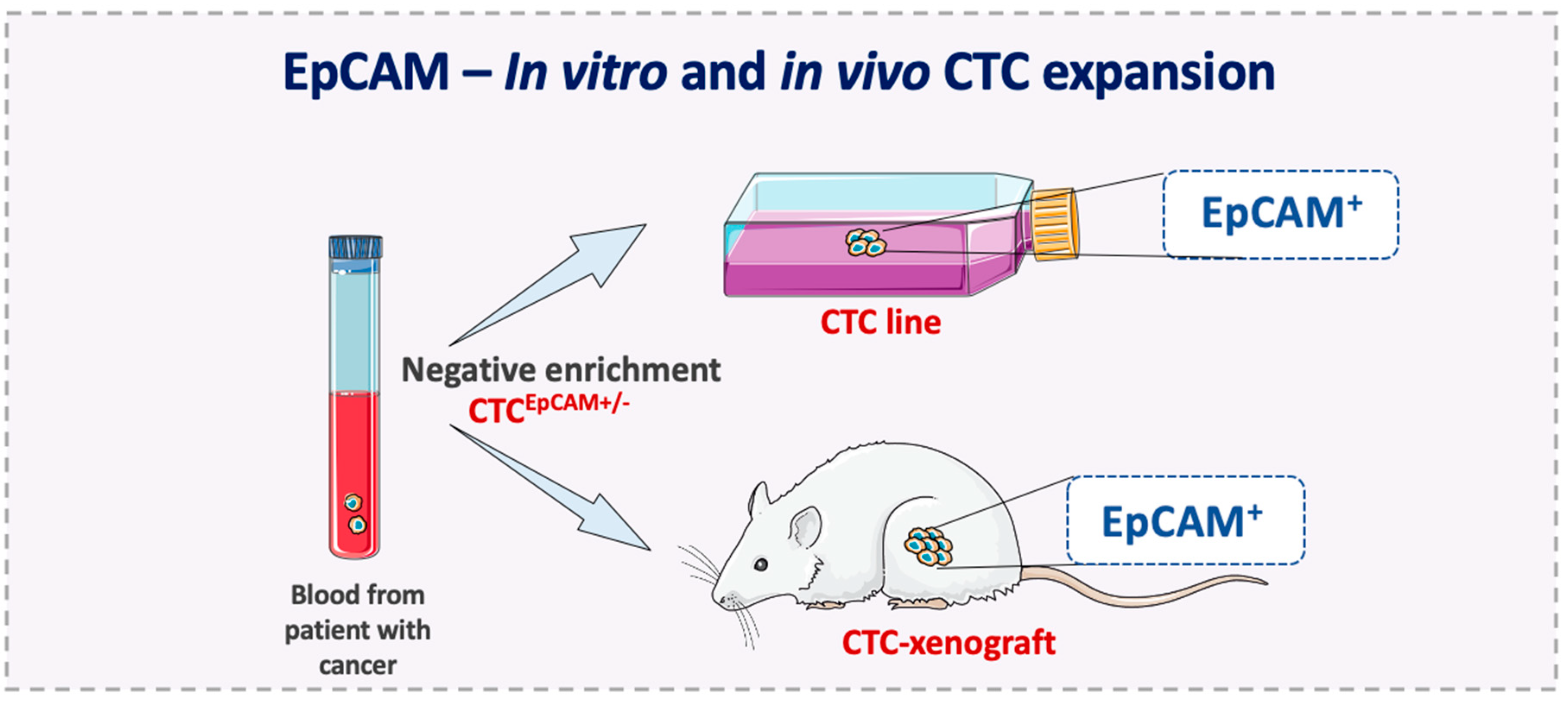
5. EpCAM Expression on Metastasis-Competent CTCs
6. EpCAM+ CTCs Might Predict the Outcome of EpCAM-Targeted Therapies
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lambert, A.W.; Pattabiraman, D.R.; Weinberg, R.A. Emerging Biological Principles of Metastasis. Cell 2017, 168, 670–691. [Google Scholar] [CrossRef] [PubMed]
- Ku, S.-Y.; Gleave, M.E.; Beltran, H. Towards precision oncology in advanced prostate cancer. Nat. Rev. Urol. 2019, 16, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.-T.; Shah, R.H.; Tegay, D.; Onel, K. Precision oncology: Lessons learned and challenges for the future. Cancer Manag. Res. 2019, 11, 7525–7536. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, P.R.; Verma, M.; Zhao, Y.; Srivastava, S. Proteomics for cancer biomarker discovery. Clin. Chem. 2002, 48, 1160–1169. [Google Scholar]
- Baeuerle, P.A.; Gires, O. EpCAM (CD326) finding its role in cancer. Br. J. Cancer 2007, 96, 417–423. [Google Scholar] [CrossRef]
- Herlyn, M.; Steplewski, Z.; Herlyn, D.; Koprowski, H. Colorectal carcinoma-specific antigen: Detection by means of monoclonal antibodies. Proc. Natl. Acad. Sci. USA 1979, 76, 1438–1442. [Google Scholar] [CrossRef]
- Imrich, S.; Hachmeister, M.; Gires, O. EpCAM and its potential role in tumor-initiating cells. Cell Adhes. Migr. 2012, 6, 30–38. [Google Scholar] [CrossRef]
- Yang, J.; Isaji, T.; Zhang, G.; Qi, F.; Duan, C.; Fukuda, T.; Gu, J. EpCAM associates with integrin and regulates cell adhesion in cancer cells. Biochem. Biophys. Res. Commun. 2020, 522, 903–909. [Google Scholar] [CrossRef]
- Juliano, R.L. Signal transduction by cell adhesion receptors and the cytoskeleton: Functions of integrins, cadherins, selectins, and immunoglobulin-superfamily members. Annu. Rev. Pharmacol. Toxicol. 2002, 42, 283–323. [Google Scholar] [CrossRef]
- Goossens-Beumer, I.J.; Zeestraten, E.C.M.; Benard, A.; Christen, T.; Reimers, M.S.; Keijzer, R.; Sier, C.F.M.; Liefers, G.J.; Morreau, H.; Putter, H.; et al. Clinical prognostic value of combined analysis of Aldh1, Survivin, and EpCAM expression in colorectal cancer. Br. J. Cancer 2014, 110, 2935–2944. [Google Scholar] [CrossRef]
- Spizzo, G.; Fong, D.; Wurm, M.; Ensinger, C.; Obrist, P.; Hofer, C.; Mazzoleni, G.; Gastl, G.; Went, P. EpCAM expression in primary tumour tissues and metastases: An immunohistochemical analysis. J. Clin. Pathol. 2011, 64, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.M.M.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabières, C.; Pantel, K. Challenges in circulating tumour cell research. Nat. Rev. Cancer 2014, 14, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Yap, T.A.; Lorente, D.; Omlin, A.; Olmos, D.; de Bono, J.S. Circulating tumor cells: A multifunctional biomarker. Clin. Cancer Res. 2014, 20, 2553–2568. [Google Scholar] [CrossRef] [PubMed]
- Gires, O.; Klein, C.A.; Baeuerle, P.A. On the abundance of EpCAM on cancer stem cells. Nat. Rev. Cancer 2009, 9, 143. [Google Scholar] [CrossRef] [PubMed]
- Visvader, J.E.; Lindeman, G.J. Cancer stem cells in solid tumours: Accumulating evidence and unresolved questions. Nat. Rev. Cancer 2008, 8, 755–768. [Google Scholar] [CrossRef]
- Spizzo, G.; Obrist, P.; Ensinger, C.; Theurl, I.; Dünser, M.; Ramoni, A.; Gunsilius, E.; Eibl, G.; Mikuz, G.; Gastl, G. Prognostic significance of Ep-CAM AND Her-2/neu overexpression in invasive breast cancer. Int. J. Cancer 2002, 98, 883–888. [Google Scholar] [CrossRef]
- Trzpis, M.; McLaughlin, P.M.J.; de Leij, L.M.F.H.; Harmsen, M.C. Epithelial cell adhesion molecule: More than a carcinoma marker and adhesion molecule. Am. J. Pathol. 2007, 171, 386–395. [Google Scholar] [CrossRef]
- Momburg, F.; Moldenhauer, G.; Hämmerling, G.J.; Möller, P. Immunohistochemical study of the expression of a Mr 34,000 human epithelium-specific surface glycoprotein in normal and malignant tissues. Cancer Res. 1987, 47, 2883–2891. [Google Scholar]
- Breuhahn, K.; Baeuerle, P.A.; Peters, M.; Prang, N.; Töx, U.; Köhne-Volland, R.; Dries, V.; Schirmacher, P.; Leo, E. Expression of epithelial cellular adhesion molecule (Ep-CAM) in chronic (necro-)inflammatory liver diseases and hepatocellular carcinoma. Hepatol. Res. 2006, 34, 50–56. [Google Scholar] [CrossRef]
- Schnell, U.; Cirulli, V.; Giepmans, B.N.G. EpCAM: Structure and function in health and disease. Biochim. Biophys. Acta (BBA) Biomembr. 2013, 1828, 1989–2001. [Google Scholar] [CrossRef] [PubMed]
- Pavšič, M.; Gunčar, G.; Carugo, K.D.; Lenarčič, B. Crystal structure and its bearing towards an understanding of key biological functions of EpCAM. Nat. Commun. 2014, 5, 4764. [Google Scholar] [CrossRef]
- Paschkowsky, S.; Hsiao, J.M.; Young, J.C.; Munter, L.M. The discovery of proteases and intramembrane proteolysis. Biochem. Cell Biol. 2019, 97, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Maetzel, D.; Denzel, S.; Mack, B.; Canis, M.; Went, P.; Benk, M.; Kieu, C.; Papior, P.; Baeuerle, P.A.; Munz, M.; et al. Nuclear signalling by tumour-associated antigen EpCAM. Nat. Cell Biol. 2009, 11, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Munz, M.; Baeuerle, P.A.; Gires, O. The emerging role of EpCAM in cancer and stem cell signaling. Cancer Res. 2009, 69, 5627–5629. [Google Scholar] [CrossRef]
- Sadeghi, S.; Hojati, Z.; Tabatabaeian, H. Cooverexpression of EpCAM and c-myc genes in malignant breast tumours. J. Genet. 2017, 96, 109–118. [Google Scholar] [CrossRef]
- Chaves-Pérez, A.; Mack, B.; Maetzel, D.; Kremling, H.; Eggert, C.; Harréus, U.; Gires, O. EpCAM regulates cell cycle progression via control of cyclin D1 expression. Oncogene 2013, 32, 641–650. [Google Scholar] [CrossRef]
- Ralhan, R.; Cao, J.; Lim, T.; Macmillan, C.; Freeman, J.L.; Walfish, P.G. EpCAM nuclear localization identifies aggressive thyroid cancer and is a marker for poor prognosis. BMC Cancer 2010, 10, 331. [Google Scholar] [CrossRef]
- Srivastava, G.; Assi, J.; Kashat, L.; Matta, A.; Chang, M.; Walfish, P.G.; Ralhan, R. Nuclear Ep-ICD accumulation predicts aggressive clinical course in early stage breast cancer patients. BMC Cancer 2014, 14, 726. [Google Scholar] [CrossRef]
- Liang, K.-H.; Tso, H.-C.; Hung, S.-H.; Kuan, I.-I.; Lai, J.-K.; Ke, F.-Y.; Chuang, Y.-T.; Liu, I.-J.; Wang, Y.-P.; Chen, R.-H.; et al. Extracellular domain of EpCAM enhances tumor progression through EGFR signaling in colon cancer cells. Cancer Lett. 2018, 433, 165–175. [Google Scholar] [CrossRef]
- Went, P.T.; Lugli, A.; Meier, S.; Bundi, M.; Mirlacher, M.; Sauter, G.; Dirnhofer, S. Frequent EpCam protein expression in human carcinomas. Hum. Pathol. 2004, 35, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Kroepil, F.; Dulian, A.; Vallböhmer, D.; Geddert, H.; Krieg, A.; Vay, C.; Topp, S.A.; Esch, J.S.; Baldus, S.E.; Gires, O.; et al. High EpCAM expression is linked to proliferation and lauren classification in gastric cancer. BMC Res. Notes 2013, 6, 253. [Google Scholar] [CrossRef] [PubMed]
- Pan, M.; Schinke, H.; Luxenburger, E.; Kranz, G.; Shakhtour, J.; Libl, D.; Huang, Y.; Gaber, A.; Pavšič, M.; Lenarčič, B.; et al. EpCAM ectodomain EpEX is a ligand of EGFR that counteracts EGF-mediated epithelial-mesenchymal transition through modulation of phospho-ERK1/2 in head and neck cancers. PLoS Biol. 2018, 16, e2006624. [Google Scholar] [CrossRef] [PubMed]
- Soysal, S.D.; Muenst, S.; Barbie, T.; Fleming, T.; Gao, F.; Spizzo, G.; Oertli, D.; Viehl, C.T.; Obermann, E.C.; Gillanders, W.E. EpCAM expression varies significantly and is differentially associated with prognosis in the luminal B HER2(+), basal-like, and HER2 intrinsic subtypes of breast cancer. Br. J. Cancer 2013, 108, 1480–1487. [Google Scholar] [CrossRef] [PubMed]
- Keller, L.; Werner, S.; Pantel, K. Biology and clinical relevance of EpCAM. Cell Stress 2019, 3, 165–180. [Google Scholar] [CrossRef]
- Gires, O.; Pan, M.; Schinke, H.; Canis, M.; Baeuerle, P.A. Expression and function of epithelial cell adhesion molecule EpCAM: Where are we after 40 years? Cancer Metastasis Rev. 2020, 1–19. [Google Scholar]
- Litvinov, S.V.; van Driel, W.; van Rhijn, C.M.; Bakker, H.A.; van Krieken, H.; Fleuren, G.J.; Warnaar, S.O. Expression of Ep-CAM in cervical squamous epithelia correlates with an increased proliferation and the disappearance of markers for terminal differentiation. Am. J. Pathol. 1996, 148, 865–875. [Google Scholar]
- Münz, M.; Kieu, C.; Mack, B.; Schmitt, B.; Zeidler, R.; Gires, O. The carcinoma-associated antigen EpCAM upregulates c-myc and induces cell proliferation. Oncogene 2004, 23, 5748–5758. [Google Scholar] [CrossRef]
- Osta, W.A.; Chen, Y.; Mikhitarian, K.; Mitas, M.; Salem, M.; Hannun, Y.A.; Cole, D.J.; Gillanders, W.E. EpCAM is overexpressed in breast cancer and is a potential target for breast cancer gene therapy. Cancer Res. 2004, 64, 5818–5824. [Google Scholar] [CrossRef]
- Driemel, C.; Kremling, H.; Schumacher, S.; Will, D.; Wolters, J.; Lindenlauf, N.; Mack, B.; Baldus, S.A.; Hoya, V.; Pietsch, J.M.; et al. Context-dependent adaption of EpCAM expression in early systemic esophageal cancer. Oncogene 2014, 33, 4904–4915. [Google Scholar] [CrossRef]
- Kuhn, S.; Koch, M.; Nübel, T.; Ladwein, M.; Antolovic, D.; Klingbeil, P.; Hildebrand, D.; Moldenhauer, G.; Langbein, L.; Franke, W.W.; et al. A complex of EpCAM, claudin-7, CD44 variant isoforms, and tetraspanins promotes colorectal cancer progression. Mol. Cancer Res. 2007, 5, 553–567. [Google Scholar] [CrossRef] [PubMed]
- Ladwein, M.; Pape, U.-F.; Schmidt, D.-S.; Schnölzer, M.; Fiedler, S.; Langbein, L.; Franke, W.W.; Moldenhauer, G.; Zöller, M. The cell-cell adhesion molecule EpCAM interacts directly with the tight junction protein claudin-7. Exp. Cell Res. 2005, 309, 345–357. [Google Scholar] [CrossRef] [PubMed]
- Eberlein, C.; Rooney, C.; Ross, S.J.; Farren, M.; Weir, H.M.; Barry, S.T. E-Cadherin and EpCAM expression by NSCLC tumour cells associate with normal fibroblast activation through a pathway initiated by integrin αvβ6 and maintained through TGFβ signalling. Oncogene 2015, 34, 704–716. [Google Scholar] [CrossRef] [PubMed]
- Winter, M.J.; Cirulli, V.; Briaire-de Bruijn, I.H.; Litvinov, S. V Cadherins are regulated by Ep-CAM via phosphaditylinositol-3 kinase. Mol. Cell. Biochem. 2007, 302, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Winter, M.J.; Nagelkerken, B.; Mertens, A.E.E.; Rees-Bakker, H.A.M.; Briaire-de Bruijn, I.H.; Litvinov, S. V Expression of Ep-CAM shifts the state of cadherin-mediated adhesions from strong to weak. Exp. Cell Res. 2003, 285, 50–58. [Google Scholar] [CrossRef]
- Jojović, M.; Adam, E.; Zangemeister-Wittke, U.; Schumacher, U. Epithelial glycoprotein-2 expression is subject to regulatory processes in epithelial-mesenchymal transitions during metastases: An investigation of human cancers transplanted into severe combined immunodeficient mice. Histochem. J. 1998, 30, 723–729. [Google Scholar] [CrossRef]
- Taube, J.H.; Herschkowitz, J.I.; Komurov, K.; Zhou, A.Y.; Gupta, S.; Yang, J.; Hartwell, K.; Onder, T.T.; Gupta, P.B.; Evans, K.W.; et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc. Natl. Acad. Sci. USA 2010, 107, 15449–15454. [Google Scholar] [CrossRef]
- Hyun, K.-A.; Koo, G.-B.; Han, H.; Sohn, J.; Choi, W.; Kim, S.-I.; Jung, H.-I.; Kim, Y.-S. Epithelial-to-mesenchymal transition leads to loss of EpCAM and different physical properties in circulating tumor cells from metastatic breast cancer. Oncotarget 2016, 7, 24677–24687. [Google Scholar] [CrossRef]
- Sankpal, N.V.; Fleming, T.P.; Sharma, P.K.; Wiedner, H.J.; Gillanders, W.E. A double-negative feedback loop between EpCAM and ERK contributes to the regulation of epithelial-mesenchymal transition in cancer. Oncogene 2017, 36, 3706–3717. [Google Scholar] [CrossRef]
- Hsu, Y.-T.; Osmulski, P.; Wang, Y.; Huang, Y.-W.; Liu, L.; Ruan, J.; Jin, V.X.; Kirma, N.B.; Gaczynska, M.E.; Huang, T.H.-M. EpCAM-Regulated Transcription Exerts Influences on Nanomechanical Properties of Endometrial Cancer Cells That Promote Epithelial-to-Mesenchymal Transition. Cancer Res. 2016, 76, 6171–6182. [Google Scholar] [CrossRef]
- Gires, O. EGFR-Dependent Regulated Intramembrane Proteolysis of EpCAM-Letter. Cancer Res. 2017, 77, 1775–1776. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lin, C.-W.; Liao, M.-Y.; Lin, W.-W.; Wang, Y.-P.; Lu, T.-Y.; Wu, H.-C. Epithelial cell adhesion molecule regulates tumor initiation and tumorigenesis via activating reprogramming factors and epithelial-mesenchymal transition gene expression in colon cancer. J. Biol. Chem. 2012, 287, 39449–39459. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Yan, Q.; Wang, J.; Liu, S.; Yang, X. Epithelial-to-mesenchymal transition induced by TGF-β1 is mediated by AP1-dependent EpCAM expression in MCF-7 cells. J. Cell. Physiol. 2015, 230, 775–782. [Google Scholar] [CrossRef] [PubMed]
- Runz, S.; Keller, S.; Rupp, C.; Stoeck, A.; Issa, Y.; Koensgen, D.; Mustea, A.; Sehouli, J.; Kristiansen, G.; Altevogt, P. Malignant ascites-derived exosomes of ovarian carcinoma patients contain CD24 and EpCAM. Gynecol. Oncol. 2007, 107, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Yang, Y.; Zeng, Y.; He, M. A microfluidic ExoSearch chip for multiplexed exosome detection towards blood-based ovarian cancer diagnosis. Lab Chip 2016, 16, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Im, H.; Shao, H.; Park, Y., II; Peterson, V.M.; Castro, C.M.; Weissleder, R.; Lee, H. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat. Biotechnol. 2014, 32, 490–495. [Google Scholar] [CrossRef]
- Taylor, D.D.; Gercel-Taylor, C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008, 110, 13–21. [Google Scholar] [CrossRef]
- Tauro, B.J.; Greening, D.W.; Mathias, R.A.; Mathivanan, S.; Ji, H.; Simpson, R.J. Two distinct populations of exosomes are released from LIM1863 colon carcinoma cell-derived organoids. Mol. Cell. Proteom. 2013, 12, 587–598. [Google Scholar] [CrossRef]
- Nanou, A.; Miller, M.C.; Zeune, L.L.; de Wit, S.; Punt, C.J.A.; Groen, H.J.M.; Hayes, D.F.; de Bono, J.S.; Terstappen, L.W.M.M. Tumour-derived extracellular vesicles in blood of metastatic cancer patients associate with overall survival. Br. J. Cancer 2020, 122, 801–811. [Google Scholar] [CrossRef]
- Pantel, K.; Alix-Panabières, C. Liquid biopsy and minimal residual disease—Latest advances and implications for cure. Nat. Rev. Clin. Oncol. 2019, 16, 409–424. [Google Scholar] [CrossRef]
- Allard, W.J.; Matera, J.; Miller, M.C.; Repollet, M.; Connelly, M.C.; Rao, C.; Tibbe, A.G.J.; Uhr, J.W.; Terstappen, L.W.M.M. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 2004, 10, 6897–6904. [Google Scholar] [CrossRef] [PubMed]
- De Bono, J.S.; Scher, H.I.; Montgomery, R.B.; Parker, C.; Miller, M.C.; Tissing, H.; Doyle, G.V.; Terstappen, L.W.W.M.; Pienta, K.J.; Raghavan, D. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin. Cancer Res. 2008, 14, 6302–6309. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.J.; Punt, C.J.A.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.; Mitchell, E.; Miller, M.C.; et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J. Clin. Oncol. 2008, 26, 3213–3221. [Google Scholar] [CrossRef] [PubMed]
- Cristofanilli, M.; Hayes, D.F.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Reuben, J.M.; Doyle, G.V.; Matera, J.; Allard, W.J.; Miller, M.C.; et al. Circulating tumor cells: A novel prognostic factor for newly diagnosed metastatic breast cancer. J. Clin. Oncol. 2005, 23, 1420–1430. [Google Scholar] [CrossRef]
- Danila, D.C.; Heller, G.; Gignac, G.A.; Gonzalez-Espinoza, R.; Anand, A.; Tanaka, E.; Lilja, H.; Schwartz, L.; Larson, S.; Fleisher, M.; et al. Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer. Clin. Cancer Res. 2007, 13, 7053–7058. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Pantel, K. Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as Liquid Biopsy. Cancer Discov. 2016, 6, 479–491. [Google Scholar] [CrossRef]
- Talasaz, A.H.; Powell, A.A.; Huber, D.E.; Berbee, J.G.; Roh, K.-H.; Yu, W.; Xiao, W.; Davis, M.M.; Pease, R.F.; Mindrinos, M.N.; et al. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proc. Natl. Acad. Sci. USA 2009, 106, 3970–3975. [Google Scholar] [CrossRef]
- Saucedo-Zeni, N.; Mewes, S.; Niestroj, R.; Gasiorowski, L.; Murawa, D.; Nowaczyk, P.; Tomasi, T.; Weber, E.; Dworacki, G.; Morgenthaler, N.G.; et al. A novel method for the in vivo isolation of circulating tumor cells from peripheral blood of cancer patients using a functionalized and structured medical wire. Int. J. Oncol. 2012, 41, 1241–1250. [Google Scholar]
- Harb, W.; Fan, A.; Tran, T.; Danila, D.C.; Keys, D.; Schwartz, M.; Ionescu-Zanetti, C. Mutational Analysis of Circulating Tumor Cells Using a Novel Microfluidic Collection Device and qPCR Assay. Transl. Oncol. 2013, 6, 528–538. [Google Scholar] [CrossRef]
- Stott, S.L.; Hsu, C.-H.; Tsukrov, D.I.; Yu, M.; Miyamoto, D.T.; Waltman, B.A.; Rothenberg, S.M.; Shah, A.M.; Smas, M.E.; Korir, G.K.; et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl. Acad. Sci. USA 2010, 107, 18392–18397. [Google Scholar] [CrossRef]
- Lin, P.P.; Li, J.; Wang, D.; Gires, O.; Wang, H. Abstract 1059: EpCAM differentially expressed on circulating and disseminated tumor cells promotes tumor metastasis in breast cancer patients. Cancer Res. 2018, 78, 1059. [Google Scholar]
- Fehm, T.; Müller, V.; Aktas, B.; Janni, W.; Schneeweiss, A.; Stickeler, E.; Lattrich, C.; Löhberg, C.R.; Solomayer, E.; Rack, B.; et al. HER2 status of circulating tumor cells in patients with metastatic breast cancer: A prospective, multicenter trial. Breast Cancer Res. Treat. 2010, 124, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Gervasoni, A.; Sandri, M.T.; Nascimbeni, R.; Zorzino, L.; Cassatella, M.C.; Baglioni, L.; Panigara, S.; Gervasi, M.; Di Lorenzo, D.; Parolini, O. Comparison of three distinct methods for the detection of circulating tumor cells in colorectal cancer patients. Oncol. Rep. 2011, 25, 1669–1703. [Google Scholar] [PubMed]
- Krebs, M.G.; Hou, J.-M.; Sloane, R.; Lancashire, L.; Priest, L.; Nonaka, D.; Ward, T.H.; Backen, A.; Clack, G.; Hughes, A.; et al. Analysis of circulating tumor cells in patients with non-small cell lung cancer using epithelial marker-dependent and -independent approaches. J. Thorac. Oncol. 2012, 7, 306–315. [Google Scholar] [CrossRef] [PubMed]
- Antolovic, D.; Galindo, L.; Carstens, A.; Rahbari, N.; Büchler, M.W.; Weitz, J.; Koch, M. Heterogeneous detection of circulating tumor cells in patients with colorectal cancer by immunomagnetic enrichment using different EpCAM-specific antibodies. BMC Biotechnol. 2010, 10, 35. [Google Scholar] [CrossRef] [PubMed]
- Cortes-Hernandez, L.E.; Eslami-S, Z.; Alix-Panabieres, C. Circulating tumor cell as the functional aspect of liquid biopsy to understand the metastatic cascade in solid cancer. Mol. Asp. Med. 2019, 72, 100816. [Google Scholar] [CrossRef] [PubMed]
- Gorges, T.M.; Tinhofer, I.; Drosch, M.; Röse, L.; Zollner, T.M.; Krahn, T.; von Ahsen, O. Circulating tumour cells escape from EpCAM-based detection due to epithelial-to-mesenchymal transition. BMC Cancer 2012, 12, 178. [Google Scholar] [CrossRef]
- Eslami-S, Z.; Cortés-Hernández, L.E.; Alix-Panabières, C. Circulating tumor cells: Moving forward into clinical applications. Precis. Cancer Med. 2019, 3, 4. [Google Scholar] [CrossRef]
- Denève, E.; Riethdorf, S.; Ramos, J.; Nocca, D.; Coffy, A.; Daurès, J.-P.; Maudelonde, T.; Fabre, J.-M.; Pantel, K.; Alix-Panabières, C. Capture of Viable Circulating Tumor Cells in the Liver of Colorectal Cancer Patients. Clin. Chem. 2013, 59, 1384–1392. [Google Scholar] [CrossRef]
- Rao, C.G.; Chianese, D.; Doyle, G.V.; Miller, M.C.; Russell, T.; Sanders, R.A.J.; Terstappen, L.W.M.M. Expression of epithelial cell adhesion molecule in carcinoma cells present in blood and primary and metastatic tumors. Int. J. Oncol. 2005, 27, 49–57. [Google Scholar] [CrossRef]
- Yu, M.; Bardia, A.; Wittner, B.S.; Stott, S.L.; Smas, M.E.; Ting, D.T.; Isakoff, S.J.; Ciciliano, J.C.; Wells, M.N.; Shah, A.M.; et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013, 339, 580–584. [Google Scholar] [CrossRef] [PubMed]
- Fischer, K.R.; Durrans, A.; Lee, S.; Sheng, J.; Li, F.; Wong, S.T.C.; Choi, H.; El Rayes, T.; Ryu, S.; Troeger, J.; et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 2015, 527, 472–476. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Carstens, J.L.; Kim, J.; Scheible, M.; Kaye, J.; Sugimoto, H.; Wu, C.-C.; LeBleu, V.S.; Kalluri, R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 2015, 527, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Antin, P.; Berx, G.; Blanpain, C.; Brabletz, T.; Bronner, M.; Campbell, K.; Cano, A.; Casanova, J.; Christofori, G.; et al. Guidelines and definitions for research on epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2020, 21, 341–352. [Google Scholar] [CrossRef]
- Gabriel, M.T.; Calleja, L.R.; Chalopin, A.; Ory, B.; Heymann, D. Circulating Tumor Cells: A Review of Non-EpCAM—Based Approaches for Cell Enrichment and Isolation. Clin. Chem. 2016, 62, 571–581. [Google Scholar] [CrossRef]
- Lampignano, R.; Schneck, H.; Neumann, M.; Fehm, T.; Neubauer, H. Enrichment, Isolation and Molecular Characterization of EpCAM-Negative Circulating Tumor Cells. Adv. Exp. Med. Biol. 2017, 994, 181–203. [Google Scholar]
- Agerbæk, M.Ø.; Bang-Christensen, S.R.; Yang, M.-H.; Clausen, T.M.; Pereira, M.A.; Sharma, S.; Ditlev, S.B.; Nielsen, M.A.; Choudhary, S.; Gustavsson, T.; et al. The VAR2CSA malaria protein efficiently retrieves circulating tumor cells in an EpCAM-independent manner. Nat. Commun. 2018, 9, 3279. [Google Scholar] [CrossRef]
- Yoneda, K.; Chikaishi, Y.; Kuwata, T.; Ohnaga, T.; Tanaka, F. Capture of mesothelioma cells with “universal” CTC-chip. Oncol. Lett. 2018, 15, 2635–2640. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.-T.; Kim, Y.J.; Bu, J.; Chen, S.; Cho, Y.-H.; Lee, H.M.; Ryu, C.J.; Lim, Y.; Han, S.-W. Epithelial and mesenchymal circulating tumor cell isolation and discrimination using dual-immunopatterned device with newly-developed anti-63B6 and anti-EpCAM. Sens. Actuators B Chem. 2018, 260, 320–330. [Google Scholar] [CrossRef]
- Thege, F.I.; Lannin, T.B.; Saha, T.N.; Tsai, S.; Kochman, M.L.; Hollingsworth, M.A.; Rhim, A.D.; Kirby, B.J. Microfluidic immunocapture of circulating pancreatic cells using parallel EpCAM and MUC1 capture: Characterization, optimization and downstream analysis. Lab Chip 2014, 14, 1775–1784. [Google Scholar] [CrossRef] [PubMed]
- Ghazani, A.A.; Castro, C.M.; Gorbatov, R.; Lee, H.; Weissleder, R. Sensitive and direct detection of circulating tumor cells by multimarker µ-nuclear magnetic resonance. Neoplasia 2012, 14, 388–395. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Scharpenseel, H.; Hanssen, A.; Loges, S.; Mohme, M.; Bernreuther, C.; Peine, S.; Lamszus, K.; Goy, Y.; Petersen, C.; Westphal, M.; et al. EGFR and HER3 expression in circulating tumor cells and tumor tissue from non-small cell lung cancer patients. Sci. Rep. 2019, 9, 7406. [Google Scholar] [CrossRef] [PubMed]
- Satelli, A.; Brownlee, Z.; Mitra, A.; Meng, Q.H.; Li, S. Circulating tumor cell enumeration with a combination of epithelial cell adhesion molecule- and cell-surface vimentin-based methods for monitoring breast cancer therapeutic response. Clin. Chem. 2015, 61, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Po, J.W.; Roohullah, A.; Lynch, D.; DeFazio, A.; Harrison, M.; Harnett, P.R.; Kennedy, C.; de Souza, P.; Becker, T.M. Improved ovarian cancer EMT-CTC isolation by immunomagnetic targeting of epithelial EpCAM and mesenchymal N-cadherin. J. Circ. Biomark. 2018, 7, 1–10. [Google Scholar] [CrossRef]
- Zhao, X.-H.; Wang, Z.-R.; Chen, C.-L.; Di, L.; Bi, Z.-F.; Li, Z.-H.; Liu, Y.-M. Molecular detection of epithelial-mesenchymal transition markers in circulating tumor cells from pancreatic cancer patients: Potential role in clinical practice. World J. Gastroenterol. 2019, 25, 138–150. [Google Scholar] [CrossRef]
- Gorges, T.M.; Stein, A.; Quidde, J.; Hauch, S.; Röck, K.; Riethdorf, S.; Joosse, S.A.; Pantel, K. Improved Detection of Circulating Tumor Cells in Metastatic Colorectal Cancer by the Combination of the CellSearch® System and the AdnaTest®. PLoS ONE 2016, 11, e0155126. [Google Scholar] [CrossRef]
- Gorges, T.M.; Kuske, A.; Röck, K.; Mauermann, O.; Müller, V.; Peine, S.; Verpoort, K.; Novosadova, V.; Kubista, M.; Riethdorf, S.; et al. Accession of Tumor Heterogeneity by Multiplex Transcriptome Profiling of Single Circulating Tumor Cells. Clin. Chem. 2016, 62, 1504–1515. [Google Scholar] [CrossRef] [PubMed]
- Hofman, V.; Ilie, M.I.; Long, E.; Selva, E.; Bonnetaud, C.; Molina, T.; Vénissac, N.; Mouroux, J.; Vielh, P.; Hofman, P. Detection of circulating tumor cells as a prognostic factor in patients undergoing radical surgery for non-small-cell lung carcinoma: Comparison of the efficacy of the CellSearch AssayTM and the isolation by size of epithelial tumor cell method. Int. J. Cancer 2011, 129, 1651–1660. [Google Scholar] [CrossRef] [PubMed]
- Farace, F.; Massard, C.; Vimond, N.; Drusch, F.; Jacques, N.; Billiot, F.; Laplanche, A.; Chauchereau, A.; Lacroix, L.; Planchard, D.; et al. A direct comparison of CellSearch and ISET for circulating tumour-cell detection in patients with metastatic carcinomas. Br. J. Cancer 2011, 105, 847–853. [Google Scholar] [CrossRef] [PubMed]
- Schiechl, H.; Dohr, G. Immunohistochemical studies of the distribution of a basolateral-membrane protein in intestinal epithelial cells (GZ1-Ag) in rats using monoclonal antibodies. Histochemistry 1987, 87, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Ng, V.Y.; Ang, S.N.; Chan, J.X.; Choo, A.B.H. Characterization of epithelial cell adhesion molecule as a surface marker on undifferentiated human embryonic stem cells. Stem Cells 2010, 28, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Yang, Y.; Yang, F.; Liu, S.; Zhu, Z.; Lei, Z.; Guo, J. Functions of EpCAM in physiological processes and diseases (Review). Int. J. Mol. Med. 2018, 42, 1771–1785. [Google Scholar] [CrossRef] [PubMed]
- Spizzo, G.; Went, P.; Dirnhofer, S.; Obrist, P.; Moch, H.; Baeuerle, P.A.; Mueller-Holzner, E.; Marth, C.; Gastl, G.; Zeimet, A.G. Overexpression of epithelial cell adhesion molecule (Ep-CAM) is an independent prognostic marker for reduced survival of patients with epithelial ovarian cancer. Gynecol. Oncol. 2006, 103, 483–488. [Google Scholar] [CrossRef] [PubMed]
- Went, P.; Vasei, M.; Bubendorf, L.; Terracciano, L.; Tornillo, L.; Riede, U.; Kononen, J.; Simon, R.; Sauter, G.; Baeuerle, P.A. Frequent high-level expression of the immunotherapeutic target Ep-CAM in colon, stomach, prostate and lung cancers. Br. J. Cancer 2006, 94, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Bardia, A.; Aceto, N.; Bersani, F.; Madden, M.W.; Donaldson, M.C.; Desai, R.; Zhu, H.; Comaills, V.; Zheng, Z.; et al. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science 2014, 345, 216–220. [Google Scholar] [CrossRef] [PubMed]
- Cayrefourcq, L.; Mazard, T.; Joosse, S.; Solassol, J.; Ramos, J.; Assenat, E.; Schumacher, U.; Costes, V.; Maudelonde, T.; Pantel, K.; et al. Establishment and characterization of a cell line from human circulating colon cancer cells. Cancer Res. 2015, 75, 892–901. [Google Scholar] [CrossRef]
- Hamilton, G.; Hochmair, M.; Rath, B.; Klameth, L.; Zeillinger, R. Small cell lung cancer: Circulating tumor cells of extended stage patients express a mesenchymal-epithelial transition phenotype. Cell Adhes. Migr. 2016, 10, 360–367. [Google Scholar] [CrossRef]
- Que, Z.; Luo, B.; Zhou, Z.; Dong, C.; Jiang, Y.; Wang, L.; Shi, Q.; Tian, J. Establishment and characterization of a patient-derived circulating lung tumor cell line in vitro and in vivo. Cancer Cell Int. 2019, 19, 21. [Google Scholar] [CrossRef]
- Baccelli, I.; Schneeweiss, A.; Riethdorf, S.; Stenzinger, A.; Schillert, A.; Vogel, V.; Klein, C.; Saini, M.; Bäuerle, T.; Wallwiener, M.; et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat. Biotechnol. 2013, 31, 539–544. [Google Scholar] [CrossRef]
- Edd, J.F.; Mishra, A.; Dubash, T.D.; Herrera, S.; Mohammad, R.; Williams, E.K.; Hong, X.; Mutlu, B.R.; Walsh, J.R.; de Carvalho, F.M.; et al. Microfluidic concentration and separation of circulating tumor cell clusters from large blood volumes. Lab Chip 2020, 20, 558–567. [Google Scholar] [CrossRef]
- Hong, X.; Sullivan, R.J.; Kalinich, M.; Kwan, T.T.; Giobbie-Hurder, A.; Pan, S.; LiCausi, J.A.; Milner, J.D.; Nieman, L.T.; Wittner, B.S.; et al. Molecular signatures of circulating melanoma cells for monitoring early response to immune checkpoint therapy. Proc. Natl. Acad. Sci. USA 2018, 115, 2467–2472. [Google Scholar] [CrossRef] [PubMed]
- Koch, C.; Kuske, A.; Joosse, S.A.; Yigit, G.; Sflomos, G.; Thaler, S.; Smit, D.J.; Werner, S.; Borgmann, K.; Gärtner, S.; et al. Characterization of circulating breast cancer cells with tumorigenic and metastatic capacity. EMBO Mol. Med. 2020, e11908. [Google Scholar] [CrossRef] [PubMed]
- Faugeroux, V.; Pailler, E.; Oulhen, M.; Deas, O.; Brulle-Soumare, L.; Hervieu, C.; Marty, V.; Alexandrova, K.; Andree, K.C.; Stoecklein, N.H.; et al. Genetic characterization of a unique neuroendocrine transdifferentiation prostate circulating tumor cell-derived eXplant model. Nat. Commun. 2020, 11, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ridgway, L.D.; Wetzel, M.D.; Ngo, J.; Yin, W.; Kumar, D.; Goodman, J.C.; Groves, M.D.; Marchetti, D. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci. Transl. Med. 2013, 5, 180ra48. [Google Scholar] [CrossRef] [PubMed]
- Dongre, A.; Weinberg, R.A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 69–84. [Google Scholar] [CrossRef]
- Riethmüller, G.; Schneider-Gädicke, E.; Schlimok, G.; Schmiegel, W.; Raab, R.; Höffken, K.; Gruber, R.; Pichlmaier, H.; Hirche, H.; Pichlmayr, R. Randomised trial of monoclonal antibody for adjuvant therapy of resected Dukes’ C colorectal carcinoma. German Cancer Aid 17-1A Study Group. Lancet 1994, 343, 1177–1183. [Google Scholar] [CrossRef]
- Riethmüller, G.; Holz, E.; Schlimok, G.; Schmiegel, W.; Raab, R.; Höffken, K.; Gruber, R.; Funke, I.; Pichlmaier, H.; Hirche, H.; et al. Monoclonal antibody therapy for resected Dukes’ C colorectal cancer: Seven-year outcome of a multicenter randomized trial. J. Clin. Oncol. 1998, 16, 1788–1794. [Google Scholar] [CrossRef]
- Punt, C.J.A.; Nagy, A.; Douillard, J.-Y.; Figer, A.; Skovsgaard, T.; Monson, J.; Barone, C.; Fountzilas, G.; Riess, H.; Moylan, E.; et al. Edrecolomab alone or in combination with fluorouracil and folinic acid in the adjuvant treatment of stage III colon cancer: A randomised study. Lancet 2002, 360, 671–677. [Google Scholar] [CrossRef]
- Schmidt, M.; Scheulen, M.E.; Dittrich, C.; Obrist, P.; Marschner, N.; Dirix, L.; Schmidt, M.; Rüttinger, D.; Schuler, M.; Reinhardt, C.; et al. An open-label, randomized phase II study of adecatumumab, a fully human anti-EpCAM antibody, as monotherapy in patients with metastatic breast cancer. Ann. Oncol. 2010, 21, 275–282. [Google Scholar] [CrossRef]
- Münz, M.; Murr, A.; Kvesic, M.; Rau, D.; Mangold, S.; Pflanz, S.; Lumsden, J.; Volkland, J.; Fagerberg, J.; Riethmüller, G.; et al. Side-by-side analysis of five clinically tested anti-EpCAM monoclonal antibodies. Cancer Cell Int. 2010, 10, 44. [Google Scholar] [CrossRef]
- Bidard, F.-C.; Fehm, T.; Ignatiadis, M.; Smerage, J.B.; Alix-Panabières, C.; Janni, W.; Messina, C.; Paoletti, C.; Müller, V.; Hayes, D.F.; et al. Clinical application of circulating tumor cells in breast cancer: Overview of the current interventional trials. Cancer Metastasis Rev. 2013, 32, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Bidard, F.-C.; Jacot, W.; Dureau, S.; Brain, E.; Bachelot, T.; Bourgeois, H.; Goncalves, A.; Ladoire, S.; Naman, H.; Dalenc, F.; et al. Abstract GS3-07: Clinical utility of circulating tumor cell count as a tool to chose between first line hormone therapy and chemotherapy for ER+ HER2- metastatic breast cancer: Results of the phase III STIC CTC trial. Cancer Res. 2019, 79. [Google Scholar] [CrossRef]
- Bidard, F.-C.; Peeters, D.J.; Fehm, T.; Nolé, F.; Gisbert-Criado, R.; Mavroudis, D.; Grisanti, S.; Generali, D.; Garcia-Saenz, J.A.; Stebbing, J.; et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: A pooled analysis of individual patient data. Lancet Oncol. 2014, 15, 406–414. [Google Scholar] [CrossRef]
- Li, H.; Meng, Q.H.; Noh, H.; Batth, I.S.; Somaiah, N.; Torres, K.E.; Xia, X.; Wang, R.; Li, S. Detection of circulating tumor cells from cryopreserved human sarcoma peripheral blood mononuclear cells. Cancer Lett. 2017, 403, 216–223. [Google Scholar] [CrossRef]
- Königsberg, R.; Obermayr, E.; Bises, G.; Pfeiler, G.; Gneist, M.; Wrba, F.; de Santis, M.; Zeillinger, R.; Hudec, M.; Dittrich, C. Detection of EpCAM positive and negative circulating tumor cells in metastatic breast cancer patients. Acta Oncol. 2011, 50, 700–710. [Google Scholar] [CrossRef]
- Liao, M.-Y.; Lai, J.-K.; Kuo, M.Y.-P.; Lu, R.-M.; Lin, C.-W.; Cheng, P.-C.; Liang, K.-H.; Wu, H.-C. An anti-EpCAM antibody EpAb2-6 for the treatment of colon cancer. Oncotarget 2015, 6, 24947–24968. [Google Scholar] [CrossRef]
- Eyvazi, S.; Farajnia, S.; Dastmalchi, S.; Kanipour, F.; Zarredar, H.; Bandehpour, M. Antibody Based EpCAM Targeted Therapy of Cancer, Review and Update. Curr. Cancer Drug Targets 2018, 18, 857–868. [Google Scholar] [CrossRef]
- Hiraga, T.; Ito, S.; Nakamura, H. EpCAM expression in breast cancer cells is associated with enhanced bone metastasis formation. Int. J. Cancer 2016, 138, 1698–1708. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eslami-S, Z.; Cortés-Hernández, L.E.; Alix-Panabières, C. Epithelial Cell Adhesion Molecule: An Anchor to Isolate Clinically Relevant Circulating Tumor Cells. Cells 2020, 9, 1836. https://doi.org/10.3390/cells9081836
Eslami-S Z, Cortés-Hernández LE, Alix-Panabières C. Epithelial Cell Adhesion Molecule: An Anchor to Isolate Clinically Relevant Circulating Tumor Cells. Cells. 2020; 9(8):1836. https://doi.org/10.3390/cells9081836
Chicago/Turabian StyleEslami-S, Zahra, Luis Enrique Cortés-Hernández, and Catherine Alix-Panabières. 2020. "Epithelial Cell Adhesion Molecule: An Anchor to Isolate Clinically Relevant Circulating Tumor Cells" Cells 9, no. 8: 1836. https://doi.org/10.3390/cells9081836
APA StyleEslami-S, Z., Cortés-Hernández, L. E., & Alix-Panabières, C. (2020). Epithelial Cell Adhesion Molecule: An Anchor to Isolate Clinically Relevant Circulating Tumor Cells. Cells, 9(8), 1836. https://doi.org/10.3390/cells9081836





