Derivation and Characterization of Immortalized Human Muscle Satellite Cell Clones from Muscular Dystrophy Patients and Healthy Individuals
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Immortalization of Primary Human MuSCs
2.3. Clonal Isolation and Selection of Clones
2.4. Assessment of the Myogenic Nature of iHMuSCs
2.5. Two-Dimensional Differentiation Assays
2.6. Three-Dimensional Differentiation Assays
2.7. Propidium Iodide Staining
2.8. Plasmid Construction
2.9. Test of Lentiviral Promoters
2.10. Quantitative RT-PCR
3. Results and Discussion
3.1. Immortalized Myogenic Cell Lines Generated from Duchenne Muscular Dystrophy, Congenital Muscular Dystrophies and Control Muscles
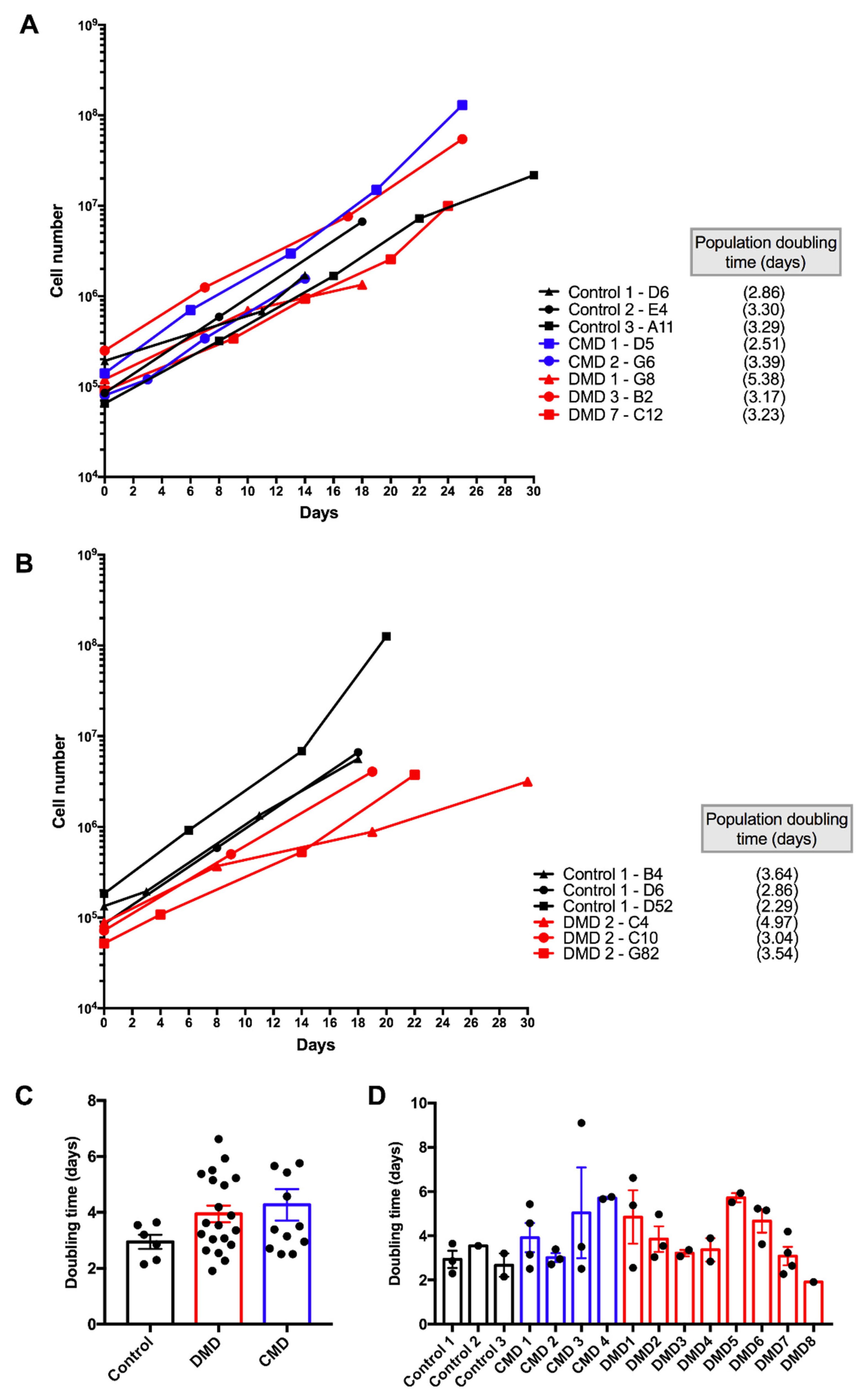
3.2. Selection of iHMuSCs Exhibiting Efficient Growth Capacity
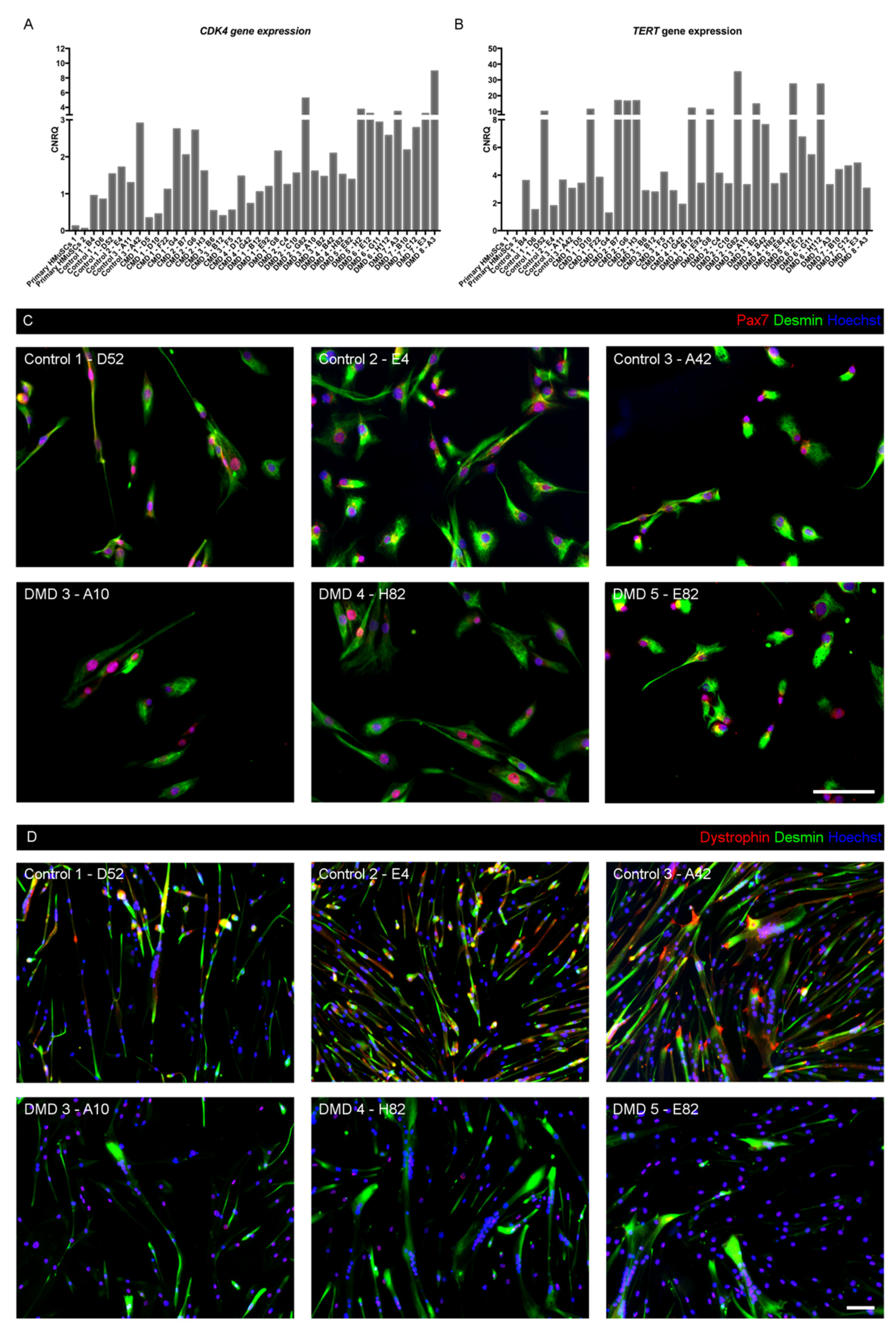
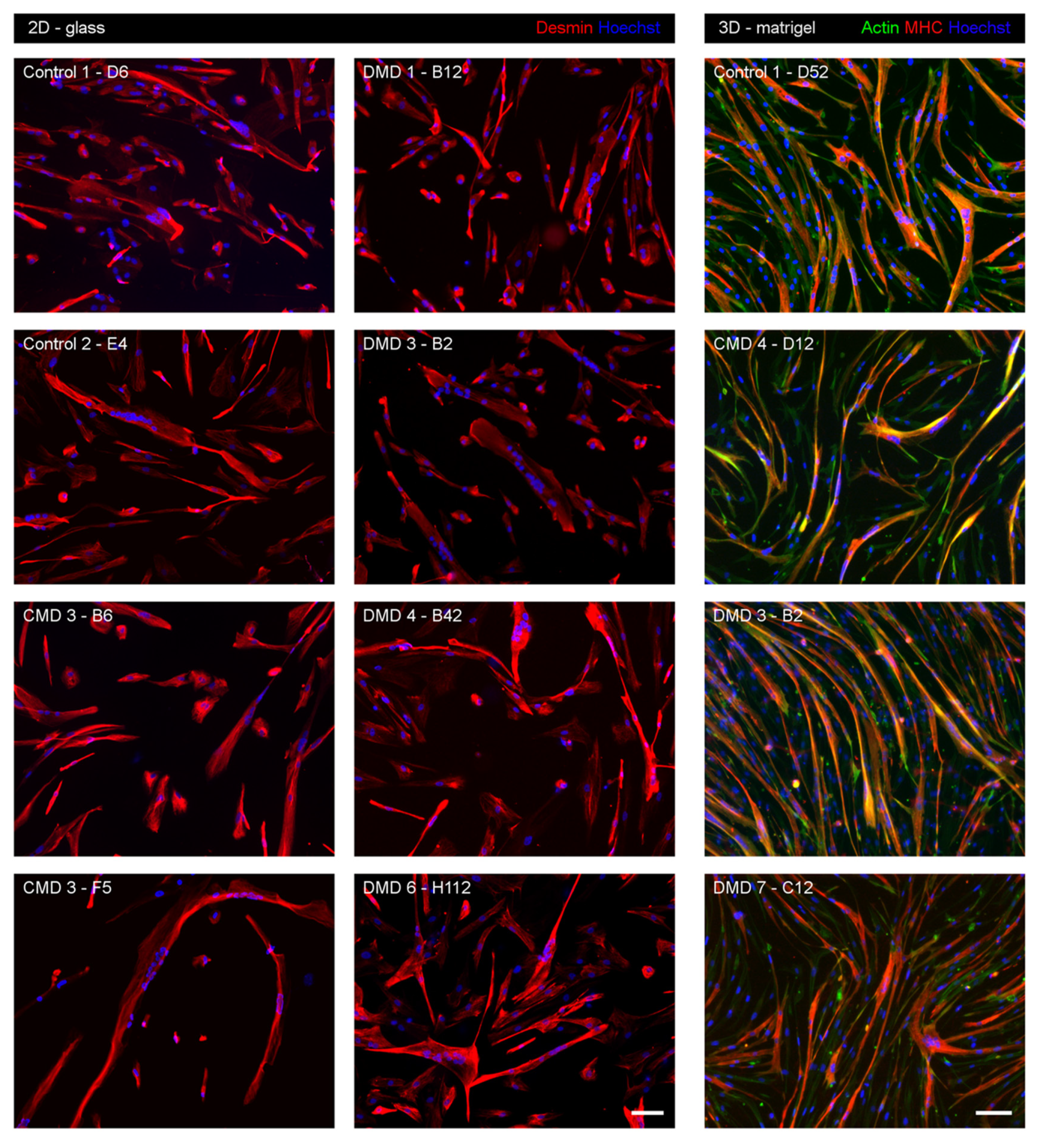
3.3. Myogenic Nature of iHMuSCs
3.4. Selection of iHMuSCs Exhibiting Efficient Myogenic Differentiation
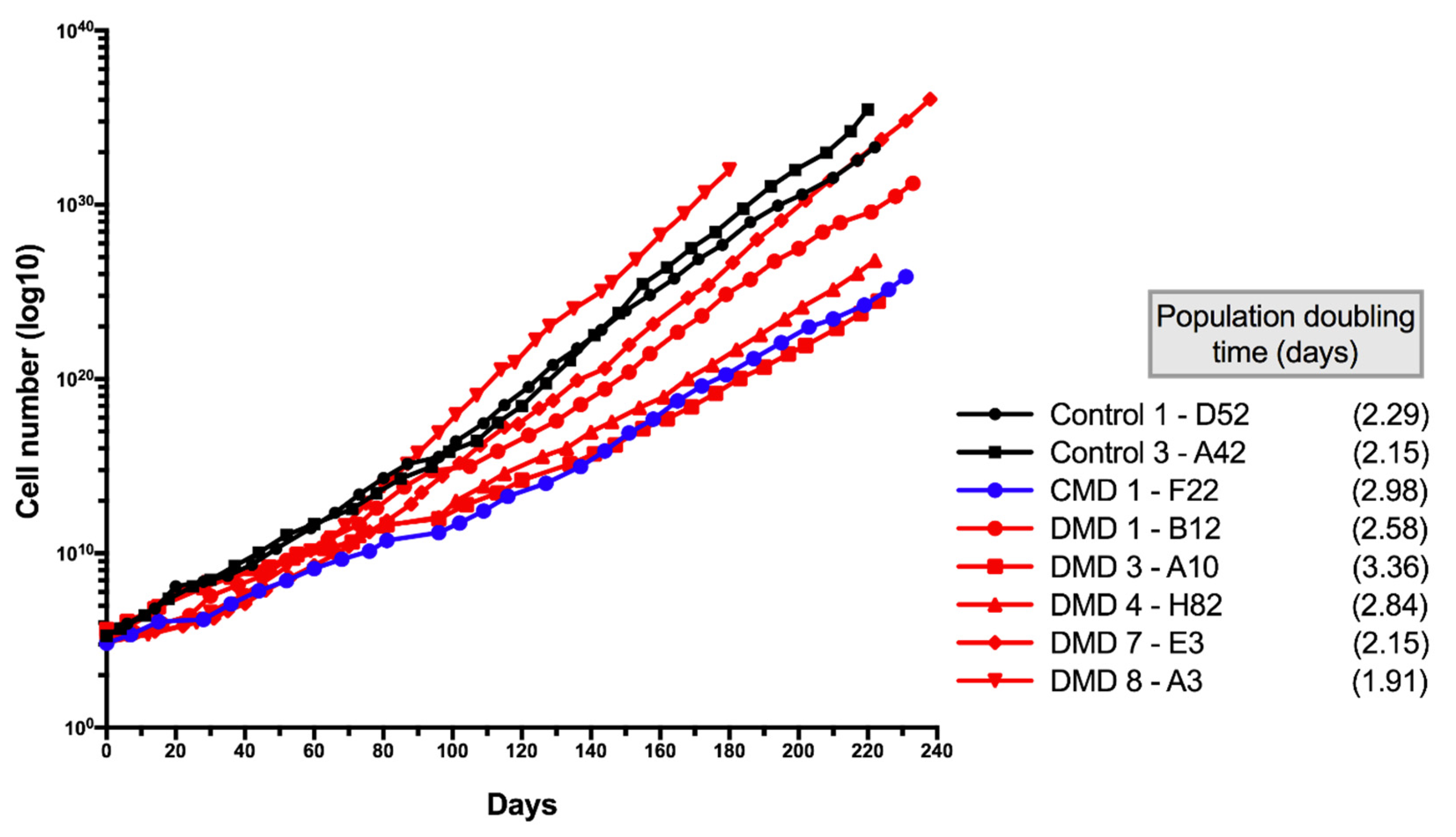
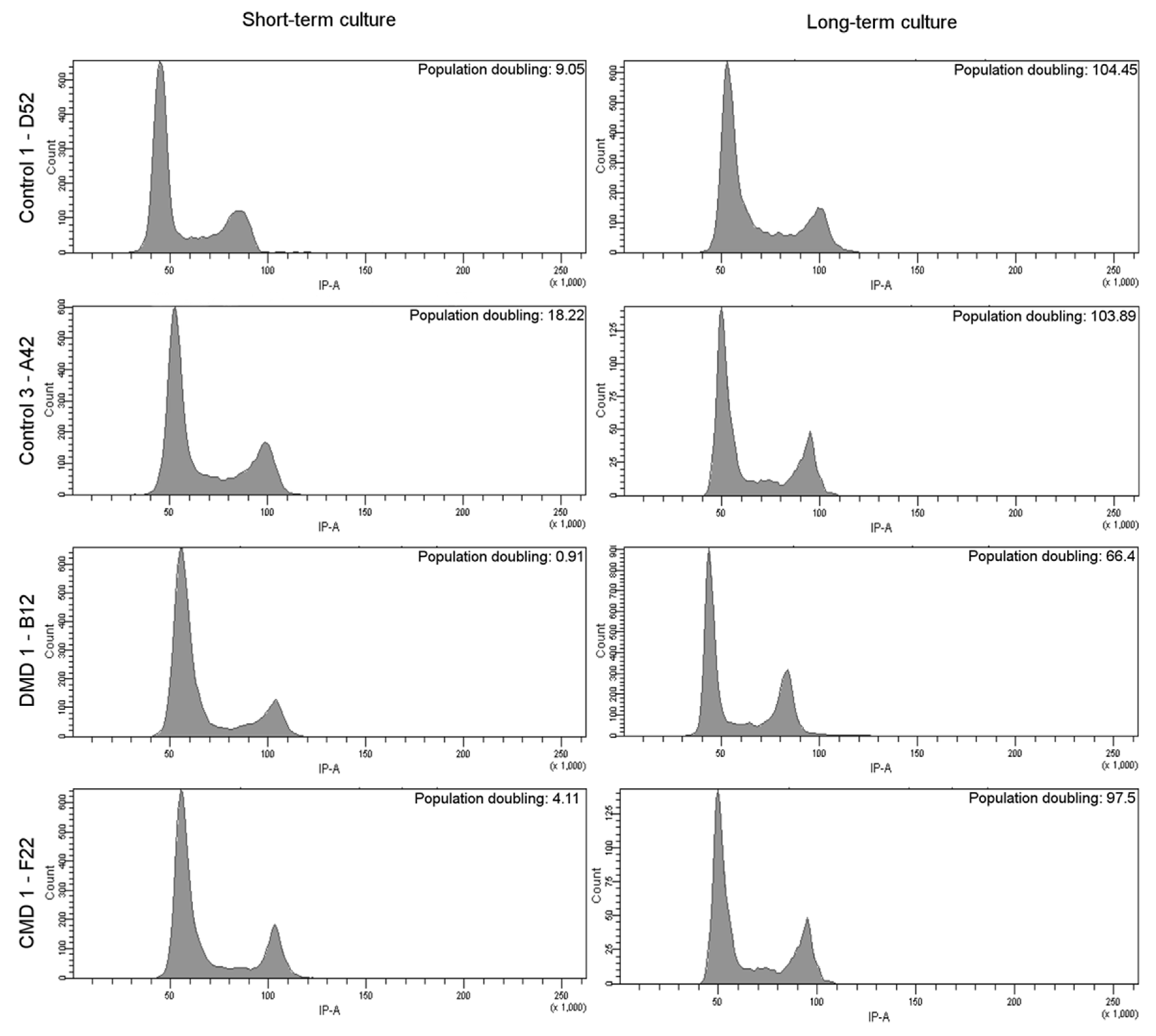
3.5. Characteristics of Long-Term Cultured iHMuSCs
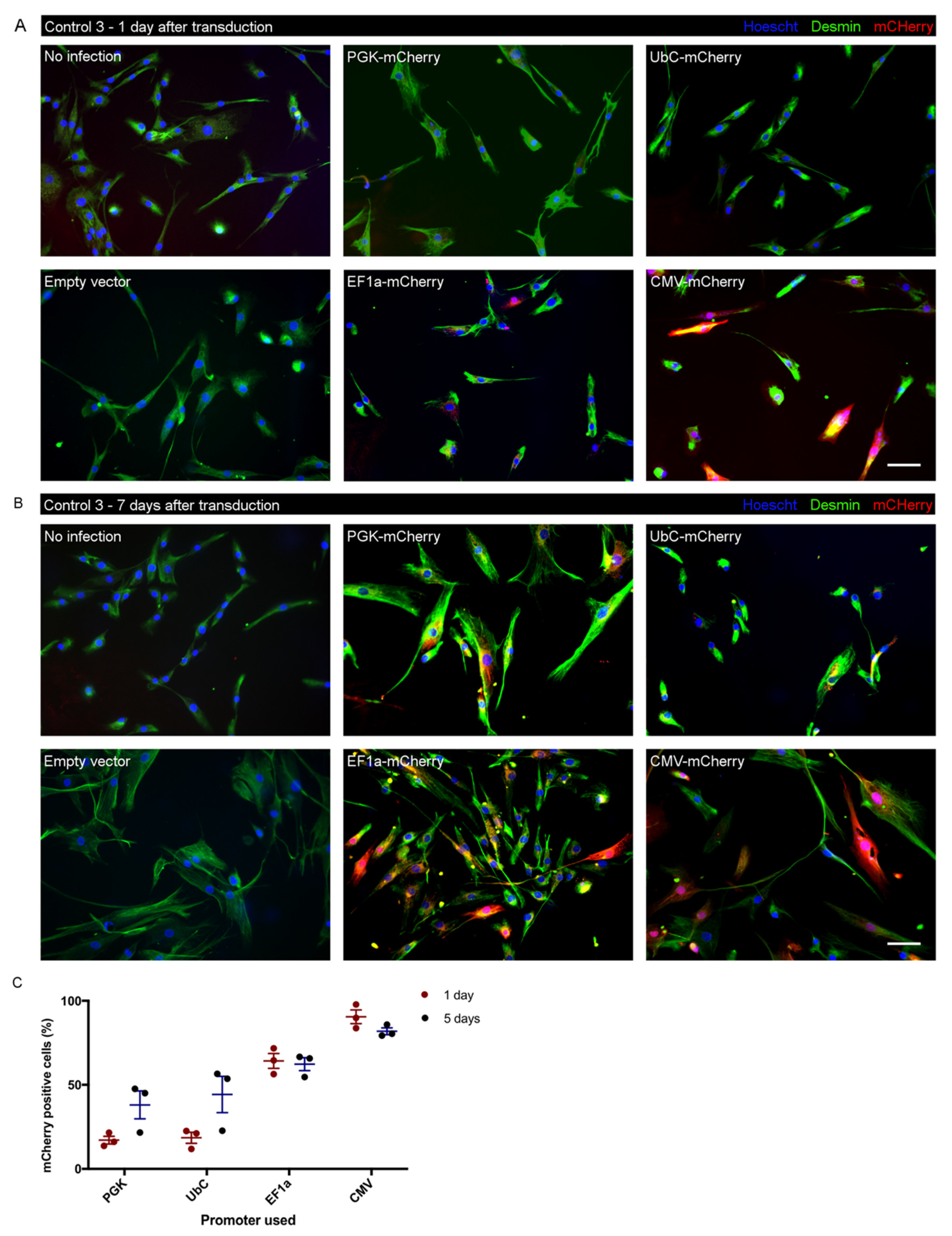
3.6. Test of Promoter Activity for Lentiviral Transduction in iHMuSCs
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mendell, J.R.; Lloyd-Puryear, M. Report of MDA muscle disease symposium on newborn screening for Duchenne muscular dystrophy: Issues & Opinions: Newborn Screening for DMD. Muscle Nerve 2013, 48, 21–26. [Google Scholar]
- Desguerre, I.; Mayer, M.; Leturcq, F.; Barbet, J.-P.; Gherardi, R.K.; Christov, C. Endomysial Fibrosis in Duchenne Muscular Dystrophy: A Marker of Poor Outcome Associated With Macrophage Alternative Activation. J. Neuropathol. Exp. Neurol. 2009, 68, 762–773. [Google Scholar] [CrossRef]
- Deconinck, A.E.; Rafael, J.A.; Skinner, J.A.; Brown, S.C.; Potter, A.C.; Metzinger, L.; Watt, D.J.; Dickson, J.G.; Tinsley, J.M.; Davies, K.E. Utrophin-Dystrophin-Deficient Mice as a Model for Duchenne Muscular Dystrophy. Cell 1997, 90, 717–727. [Google Scholar] [CrossRef]
- Sacco, A.; Mourkioti, F.; Tran, R.; Choi, J.; Llewellyn, M.; Kraft, P.; Shkreli, M.; Delp, S.; Pomerantz, J.H.; Artandi, S.E.; et al. Short Telomeres and Stem Cell Exhaustion Model Duchenne Muscular Dystrophy in mdx/mTR Mice. Cell 2010, 143, 1059–1071. [Google Scholar] [CrossRef] [PubMed]
- Alsharidah, M.; Lazarus, N.R.; George, T.E.; Agley, C.C.; Velloso, C.P.; Harridge, S.D.R. Primary human muscle precursor cells obtained from young and old donors produce similar proliferative, differentiation and senescent profiles in culture. Aging Cell 2013, 12, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Illa, I.; Leon-Monzon, M.; Dalakas, M.C. Regenerating and denervated human muscle fibers and satellite cells express neural cell adhesion molecule recognized by monoclonal antibodies to natural killer cells. Ann. Neurol. 1992, 31, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Renault, V.; Thornell, L.-E.; Eriksson, P.-O.; Butler-Browne, G.; Mouly, V.; Thorne, L.-E. Regenerative potential of human skeletal muscle during aging. Aging Cell 2002, 1, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Simon, L.V.; Beauchamp, J.R.; O’Hare, M.; Olsen, I. Establishment of Long-Term Myogenic Cultures from Patients with Duchenne Muscular Dystrophy by Retroviral Transduction of a Temperature-Sensitive SV40 Large T Antigen. Exp. Cell Res. 1996, 224, 264–271. [Google Scholar] [CrossRef]
- Cudré-Mauroux, C.; Occhiodoro, T.; König, S.; Salmon, P.; Bernheim, L.; Trono, D. Lentivector-Mediated Transfer of Bmi-1 and Telomerase in Muscle Satellite Cells Yields a Duchenne Myoblast Cell Line with Long-Term Genotypic and Phenotypic Stability. Hum. Gene Ther. 2003, 14, 1525–1533. [Google Scholar] [CrossRef]
- Mouly, V.; Edom, F.; Decary, S.; Vicart, P.; Barbert, J.P.; Butler-Browne, G.S. SV40 large T antigen interferes with adult myosin heavy chain expression, but not with differentiation of human satellite cells. Exp. Cell Res. 1996, 225, 268–276. [Google Scholar] [CrossRef]
- Decary, S.; Mouly, V.; Butler-Browne, G.S. Telomere length as a tool to monitor satellite cell amplification for cell-mediated gene therapy. Hum. Gene Ther. 1996, 7, 1347–1350. [Google Scholar] [CrossRef] [PubMed]
- Halvorsen, T.L.; Leibowitz, G.; Levine, F. Telomerase activity is sufficient to allow transformed cells to escape from crisis. Mol. Cell. Biol. 1999, 19, 1864–1870. [Google Scholar] [CrossRef] [PubMed]
- Seigneurin-Venin, S.; Bernard, V.; Tremblay, J.P. Telomerase allows the immortalization of T antigen-positive DMD myoblasts: a new source of cells for gene transfer application. Gene Ther. 2000, 7, 619–623. [Google Scholar] [CrossRef]
- Shiomi, K.; Kiyono, T.; Okamura, K.; Uezumi, M.; Goto, Y.; Yasumoto, S.; Shimizu, S.; Hashimoto, N. CDK4 and cyclin D1 allow human myogenic cells to recapture growth property without compromising differentiation potential. Gene Ther. 2011, 18, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Stadler, G.; Chen, J.C.; Wagner, K.; Robin, J.D.; Shay, J.W.; Emerson Jr, C.P.; Wright, W.E. Establishment of clonal myogenic cell lines from severely affected dystrophic muscles - CDK4 maintains the myogenic population. Skeletal Muscle 2011, 1, 12. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.-H.; Mouly, V.; Cooper, R.N.; Mamchaoui, K.; Bigot, A.; Shay, J.W.; Di Santo, J.P.; Butler-Browne, G.S.; Wright, W.E. Cellular senescence in human myoblasts is overcome by human telomerase reverse transcriptase and cyclin-dependent kinase 4: consequences in aging muscle and therapeutic strategies for muscular dystrophies. Aging Cell 2007, 6, 515–523. [Google Scholar] [CrossRef]
- Donai, K.; Kiyono, T.; Eitsuka, T.; Guo, Y.; Kuroda, K.; Sone, H.; Isogai, E.; Fukuda, T. Bovine and porcine fibroblasts can be immortalized with intact karyotype by the expression of mutant cyclin dependent kinase 4, cyclin D, and telomerase. J. Biotechnol. 2014, 176, 50–57. [Google Scholar] [CrossRef]
- Pantic, B.; Borgia, D.; Giunco, S.; Malena, A.; Kiyono, T.; Salvatori, S.; De Rossi, A.; Giardina, E.; Sangiuolo, F.; Pegoraro, E.; et al. Reliable and versatile immortal muscle cell models from healthy and myotonic dystrophy type 1 primary human myoblasts. Exp. Cell Res. 2016, 342, 39–51. [Google Scholar] [CrossRef]
- Thorley, M.; Duguez, S.; Mazza, E.M.C.; Valsoni, S.; Bigot, A.; Mamchaoui, K.; Harmon, B.; Voit, T.; Mouly, V.; Duddy, W. Skeletal muscle characteristics are preserved in hTERT/cdk4 human myogenic cell lines. Skeletal Muscle 2016, 6, 43. [Google Scholar] [CrossRef]
- Mamchaoui, K.; Trollet, C.; Bigot, A.; Negroni, E.; Chaouch, S.; Wolff, A.; Kandalla, P.K.; Marie, S.; Di Santo, J.; St Guily, J.; et al. Immortalized pathological human myoblasts: towards a universal tool for the study of neuromuscular disorders. Skeletal Muscle 2011, 1, 34. [Google Scholar] [CrossRef]
- Yoon, S.; Stadler, G.; Beermann, M.; Schmidt, E.V.; Windelborn, J.A.; Schneiderat, P.; Wright, W.E.; Miller, J. Immortalized myogenic cells from congenital muscular dystrophy type1A patients recapitulate aberrant caspase activation in pathogenesis: a new tool for MDC1A research. Skeletal Muscle 2013, 3, 28. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Krom, Y.D.; Dumonceaux, J.; Mamchaoui, K.; den Hamer, B.; Mariot, V.; Negroni, E.; Geng, L.N.; Martin, N.; Tawil, R.; Tapscott, S.J.; et al. Generation of Isogenic D4Z4 Contracted and Noncontracted Immortal Muscle Cell Clones from a Mosaic Patient. Am. J. Pathol. 2012, 181, 1387–1401. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Hua, R.; Wei, M.; Li, C.; Qiu, Z.; Yang, X.; Zhang, C. An optimized method for high-titer lentivirus preparations without ultracentrifugation. Sci. Rep. 2015, 5, 13875. [Google Scholar] [CrossRef] [PubMed]
- Hildyard, J.C.W.; Wells, D.J. Identification and Validation of Quantitative PCR Reference Genes Suitable for Normalizing Expression in Normal and Dystrophic Cell Culture Models of Myogenesis. PLoS Curr. 2014. [Google Scholar] [CrossRef]
- Blau, H.M.; Webster, C.; Pavlath, G.K. Defective myoblasts identified in Duchenne muscular dystrophy. Proc. Nat. Acad. Sci. 1983, 80, 4856–4860. [Google Scholar] [CrossRef]
- Jasmin, G.; Tautu, C.; Vanasse, M.; Brochu, P.; Simoneau, R. Impaired muscle differentiation in explant cultures of Duchenne muscular dystrophy. Lab. Invest. 1984, 50, 197–207. [Google Scholar]
- Delaporte, C.; Dehaupas, M.; Fardeau, M. Comparison between the growth pattern of cell cultures from normal and Duchenne dystrophy muscle. J. Neurol. Sci. 1984, 64, 149–160. [Google Scholar] [CrossRef]
- Iannaccone, S.T.; Nagy, B.; Samaha, F.J. Decreased Creatine Kinase Activity in Cultured Duchenne Dystrophic Muscle Cells. J. Child Neurol. 1987, 2, 17–21. [Google Scholar] [CrossRef]
- Blau, H. Differentiation properties of pure populations of human dystrophic muscle cells. Exp. Cell Res. 1983, 144, 495–503. [Google Scholar] [CrossRef]
- Zanotti, S.; Saredi, S.; Ruggieri, A.; Fabbri, M.; Blasevich, F.; Romaggi, S.; Morandi, L.; Mora, M. Altered extracellular matrix transcript expression and protein modulation in primary Duchenne muscular dystrophy myotubes. Matrix Biol. 2007, 26, 615–624. [Google Scholar] [CrossRef]
- Xu, X.; Wilschut, K.J.; Kouklis, G.; Tian, H.; Hesse, R.; Garland, C.; Sbitany, H.; Hansen, S.; Seth, R.; Knott, P.D.; et al. Human Satellite Cell Transplantation and Regeneration from Diverse Skeletal Muscles. Stem Cell Rep. 2015, 5, 419–434. [Google Scholar] [CrossRef] [PubMed]
- Barruet, E.; Garcia, S.M.; Striedinger, K.; Wu, J.; Lee, S.; Byrnes, L.; Wong, A.; Xuefeng, S.; Tamaki, S.; Brack, A.S.; et al. Functionally heterogeneous human satellite cells identified by single cell RNA sequencing. Elife. 2020, 9, e51576. [Google Scholar] [CrossRef] [PubMed]
- Dumont, N.A.; Wang, Y.X.; von Maltzahn, J.; Pasut, A.; Bentzinger, C.F.; Brun, C.E.; Rudnicki, M.A. Dystrophin expression in muscle stem cells regulates their polarity and asymmetric division. Nat. Med. 2015, 21, 1455–1463. [Google Scholar] [CrossRef] [PubMed]
- Muntoni, F.; Torelli, S.; Ferlini, A. Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol. 2003, 2, 731–740. [Google Scholar] [CrossRef]
- Capkovic, K.L.; Stevenson, S.; Johnson, M.C.; Thelen, J.J.; Cornelison, D.D.W. Neural cell adhesion molecule (NCAM) marks adult myogenic cells committed to differentiation. Exp. Cell Res. 2008, 314, 1553–1565. [Google Scholar] [CrossRef]
- Mackey, A.L.; Kjaer, M.; Charifi, N.; Henriksson, J.; Bojsen-Moller, J.; Holm, L.; Kadi, F. Assessment of satellite cell number and activity status in human skeletal muscle biopsies. Muscle Nerve 2009, 40, 455–465. [Google Scholar] [CrossRef]
- Rokach, O.; Ullrich, N.D.; Rausch, M.; Mouly, V.; Zhou, H.; Muntoni, F.; Zorzato, F.; Treves, S. Establishment of a human skeletal muscle-derived cell line: biochemical, cellular and electrophysiological characterization. Biochem. J. 2013, 455, 169–177. [Google Scholar] [CrossRef]
- Gilbert, P.M.; Havenstrite, K.L.; Magnusson, K.E.G.; Sacco, A.; Leonardi, N.A.; Kraft, P.; Nguyen, N.K.; Thrun, S.; Lutolf, M.P.; Blau, H.M. Substrate Elasticity Regulates Skeletal Muscle Stem Cell Self-Renewal in Culture. Science 2010, 329, 1078–1081. [Google Scholar] [CrossRef]
- Duchêne, B.L.; Cherif, K.; Iyombe-Engembe, J.-P.; Guyon, A.; Rousseau, J.; Ouellet, D.L.; Barbeau, X.; Lague, P.; Tremblay, J.P. CRISPR-Induced Deletion with SaCas9 Restores Dystrophin Expression in Dystrophic Models In Vitro and In Vivo. Mol. Ther. 2018, 26, 2604–2616. [Google Scholar] [CrossRef]
- Zarrin, A.A.; Malkin, L.; Fong, I.; Luk, K.D.; Ghose, A.; Berinstein, N.L. Comparison of CMV, RSV, SV40 viral and Vλ1 cellular promoters in B and T lymphoid and non-lymphoid cell lines. Biochim. Biophys. Acta 1999, 1446, 135–139. [Google Scholar] [CrossRef]
- Ebadat, S.; Ahmadi, S.; Ahmadi, M.; Nematpour, F.; Barkhordari, F.; Mahdian, R.; Davami, F.; Mahboudi, F. Evaluating the efficiency of CHEF and CMV promoter with IRES and Furin/2A linker sequences for monoclonal antibody expression in CHO cells. PLoS ONE 2017, 12, e0185967. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chusainow, J.M.; Yap, M.G.S. DNA methylation contributes to loss in productivity of monoclonal antibody-producing CHO cell lines. J. Biotechnol. 2010, 147, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Hotta, A. Genome Editing Gene Therapy for Duchenne Muscular Dystrophy. J Neuromuscul. Dis. 2015, 2, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Gee, P.; Xu, H.; Hotta, A. Cellular Reprogramming, Genome Editing, and Alternative CRISPR Cas9 Technologies for Precise Gene Therapy of Duchenne Muscular Dystrophy. Stem Cells Int. 2017, 2017, 8765154. [Google Scholar] [CrossRef]
| Primer Name | Sequence | Anneling Temperature |
|---|---|---|
| mCherry NheI Forward | GAGATCGCTAGCGGGCCCGCCACCATGGTGAGCAAGGGCGAGGAG | 60.9 °C |
| mCherry XhoI CMV Reverse | GAGATCCTCGAGCTACTTGTACAGCTCGTCCATG | 60.9 °C |
| Promoter EF1ɑ Forward | TCGGGTTTTTCGAACGGCTCCGGTGCCCGTCAG | 60 °C |
| Promoter EF1ɑ Reverse | ATGGTGGCGGGCCCGTCACGACACCTGAAATGGAAG | 60 °C |
| Promoter PGK/UBC Forward | GATCTCGACGGTATCGAAAGC | 60 °C |
| Promoter PGK Reverse | CCATGGTGGCGGGCCCGAATTCGATCTCGGATCCGAAAGCGAAGGAGCAAAGCTG | 60 °C |
| Promoter UbC Reverse | CCATGGTGGCGGGCCCGAATTCGATCTCGGATCCGTCTAACAAAAAAGCCAAAAAC | 60 °C |
| AP3D1 Forward | GGCATCCGTAACCACAAGGA | 60 °C |
| AP3D1 Reverse | TTGTCCTGCTTCAGCTCCTG | 60 °C |
| CDK4 Forward | GGCTGAAATTGGTGTCGGTG | 60 °C |
| CDK4 Reverse | CACGAACTGTGCTGATGGGA | 60 °C |
| TERT Forward | TGTACTTTGTCAAGGTGGATGTGA | 60 °C |
| TERT Reverse | GCTGGAGGTCTGTCAAGGTAGAG | 60 °C |
| Patient | Sex (M/F) | Age at the Time of Biopsy (m) | Diagnosis | Genetic Results | CK Blood Level at Diagnosis (IU/L) | Age at First Walking (m) | Cardiomyopathy Age at Onset (m) | ScoliosisAge at Onset (m) | Scoliosis Surgery/Age at Surgery |
|---|---|---|---|---|---|---|---|---|---|
| Control 1 | M | 41 | control | / | nl | 24 | N | N | N |
| Control 2 | M | 14 | control | / | nl | 24 | N | N | N |
| Control 3 | M | 19 | control | / | nl | 18 | N | N | N |
| CMD 1 | M | 144 | Collagen VI related myopathy | C6210 + 5G > A | nd | 72 | N | Y | Y/145 |
| CMD 2 | F | 144 | CMD | genetic unknown causes | nl | 18 | N | Y/195 | Y/231 |
| CMD 3 | F | 39 | Laminopathy LMNA | c.94–96 deletion; p.lys32 deletion | 860 | never walking | N | N | N |
| CMD 4 | F | 12 | LAMA2 related myopathy | c.1553 deletion GTT; pCys518 deletion and c.2866 deletion T | 14400 | never walking | N | N | N |
| DMD 1 | M | 54 | DMD | c.3–26 duplication | 18000 | 24 | N | N | N |
| DMD 2 | M | 87 | DMD | c.8–43 deletion | nd | 13 | Y/109 | Y/141 | Y/148 |
| DMD 3 | M | 97 | DMD | c.433 C > T substitution (p.R145X) | 12881 | 15 | N | Y/161 | Y/161 |
| DMD 4 | M | 25 | DMD | c.8562 deletion A; p.Glu2854Asp fs X2 | 19000 | 24 | N | N | N |
| DMD 5 | M | 79 | DMD | c.5758 C > T substitution; p.Gln1920X | 8041 | 15 | N | N | N |
| DMD 6 | M | nd | DMD | Exon skipping 19 / IVS 19 +1 G > C/c.2380 + 1 G >C | nd | nd | Y/82 | N | N |
| DMD 7 | M | 89 | DMD | c.50–59 dup | 47270 | 15 | N | N | N |
| DMD 8 | M | nd | DMD | nd | nd | nd | nd | nd | nd |
| Patients | Clones | Population Doublings a | CD56 pos Cells (%) | Differentiation | Doubling Time (days) b |
|---|---|---|---|---|---|
| Control 1 | B4 | 3.33 | 99.50 | ++ | 3.64 |
| D6 | 2.80 | 99.80 | +++ | 2.86 | |
| D52 long term | 5.22 | 99.70 | +++ | 2.3 | |
| 73.23 | 99.90 | ++ | |||
| Control 2 | E4 | 4.69 | 99.40 | ++ | 3.55 |
| Control 3 | A11 | 1.82 | 98.07 | ++ | 3.2 |
| A42 long term | 3.87 | 98.20 | ++ | 2.15 | |
| 73.83 | 99.90 | +++ | |||
| CMD 1 | D5 | 6.50 | 99.00 | +++ | 2.51 |
| D10 | 8.73 | 98.90 | ++ | 5.43 | |
| F22 long term | 4.53 | 98.50 | +++ | 3.15 | |
| 69.86 | 99.80 | +++ | |||
| G4 | 5.15 | 98.91 | +++ | 4.57 | |
| CMD 2 | B7 | 4.74 | 99.80 | +++ | 2.95 |
| G6 | 4.29 | 99.80 | +++ | 3.39 | |
| H3 | 5.80 | 99.70 | +++ | 2.71 | |
| CMD 3 | B6 | 4.42 | 96.00 | ++ | 3.5 |
| B12 | 2.00 | 97.30 | +++ | 2.51 | |
| F5 | 1.96 | 90.00 | +++ | 9.1 | |
| CMD 4 | D12 | 5.40 | 93.30 | +++ | 5.76 |
| G42 | 2.51 | 91.00 | +++ | 5.66 | |
| DMD 1 | B12 long term | 5.17 | 99.00 | ++ | 2.55 |
| 66.39 | 93.40 | ++ | |||
| E92 | 3.39 | 99.00 | ++ | 6.62 | |
| G8 | 3.48 | 97.20 | +++ | 5.38 | |
| DMD 2 | C4 | 2.07 | 86.00 | +++ | 4.97 |
| C10 | 5.82 | 93.10 | ++ | 3.04 | |
| G82 | 6.18 | 98.50 | ++ | 3.54 | |
| aDMD 3 | A10 long term | 3.90 | 99.30 | ++ | 3.36 |
| 42.39 | 95.10 | + | |||
| B2 | 7.77 | 89.00 | +++ | 3.07 | |
| DMD 4 | B42 | 3.61 | 98.10 | ++ | 3.89 |
| H82 long term | 5.39 | 99.80 | ++ | 2.84 | |
| 52.30 | nd | +++ | |||
| DMD 5 | E82 | 4.32 | 98.00 | ++ | 5.93 |
| H2 | 2.56 | 100.00 | ++ | 5.51 | |
| DMD 6 | E12 | 2.65 | 96.70 | +++ | 3.62 |
| G11 | 6.45 | 93.20 | +++ | 5.23 | |
| H112 | 3.39 | 91.70 | ++ | 5.15 | |
| DMD 7 | A3 | 5.80 | 93.70 | +++ | 2.64 |
| B10 | 8.40 | 98.00 | +++ | 4.19 | |
| C12 | 6.80 | 99.30 | +++ | 3.23 | |
| E3 long term | 15.96 | 99.00 | ++ | 2.27 | |
| 119.30 | 82.90 | nd | |||
| DMD 8 | A3 long term | 5.72 | 99.20 | ++ | 1.91 |
| 63.55 | 98.00 | nd |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Massenet, J.; Gitiaux, C.; Magnan, M.; Cuvellier, S.; Hubas, A.; Nusbaum, P.; Dilworth, F.J.; Desguerre, I.; Chazaud, B. Derivation and Characterization of Immortalized Human Muscle Satellite Cell Clones from Muscular Dystrophy Patients and Healthy Individuals. Cells 2020, 9, 1780. https://doi.org/10.3390/cells9081780
Massenet J, Gitiaux C, Magnan M, Cuvellier S, Hubas A, Nusbaum P, Dilworth FJ, Desguerre I, Chazaud B. Derivation and Characterization of Immortalized Human Muscle Satellite Cell Clones from Muscular Dystrophy Patients and Healthy Individuals. Cells. 2020; 9(8):1780. https://doi.org/10.3390/cells9081780
Chicago/Turabian StyleMassenet, Jimmy, Cyril Gitiaux, Mélanie Magnan, Sylvain Cuvellier, Arnaud Hubas, Patrick Nusbaum, F Jeffrey Dilworth, Isabelle Desguerre, and Bénédicte Chazaud. 2020. "Derivation and Characterization of Immortalized Human Muscle Satellite Cell Clones from Muscular Dystrophy Patients and Healthy Individuals" Cells 9, no. 8: 1780. https://doi.org/10.3390/cells9081780
APA StyleMassenet, J., Gitiaux, C., Magnan, M., Cuvellier, S., Hubas, A., Nusbaum, P., Dilworth, F. J., Desguerre, I., & Chazaud, B. (2020). Derivation and Characterization of Immortalized Human Muscle Satellite Cell Clones from Muscular Dystrophy Patients and Healthy Individuals. Cells, 9(8), 1780. https://doi.org/10.3390/cells9081780





