The Leukemic Fly: Promises and Challenges
Abstract
1. Introduction
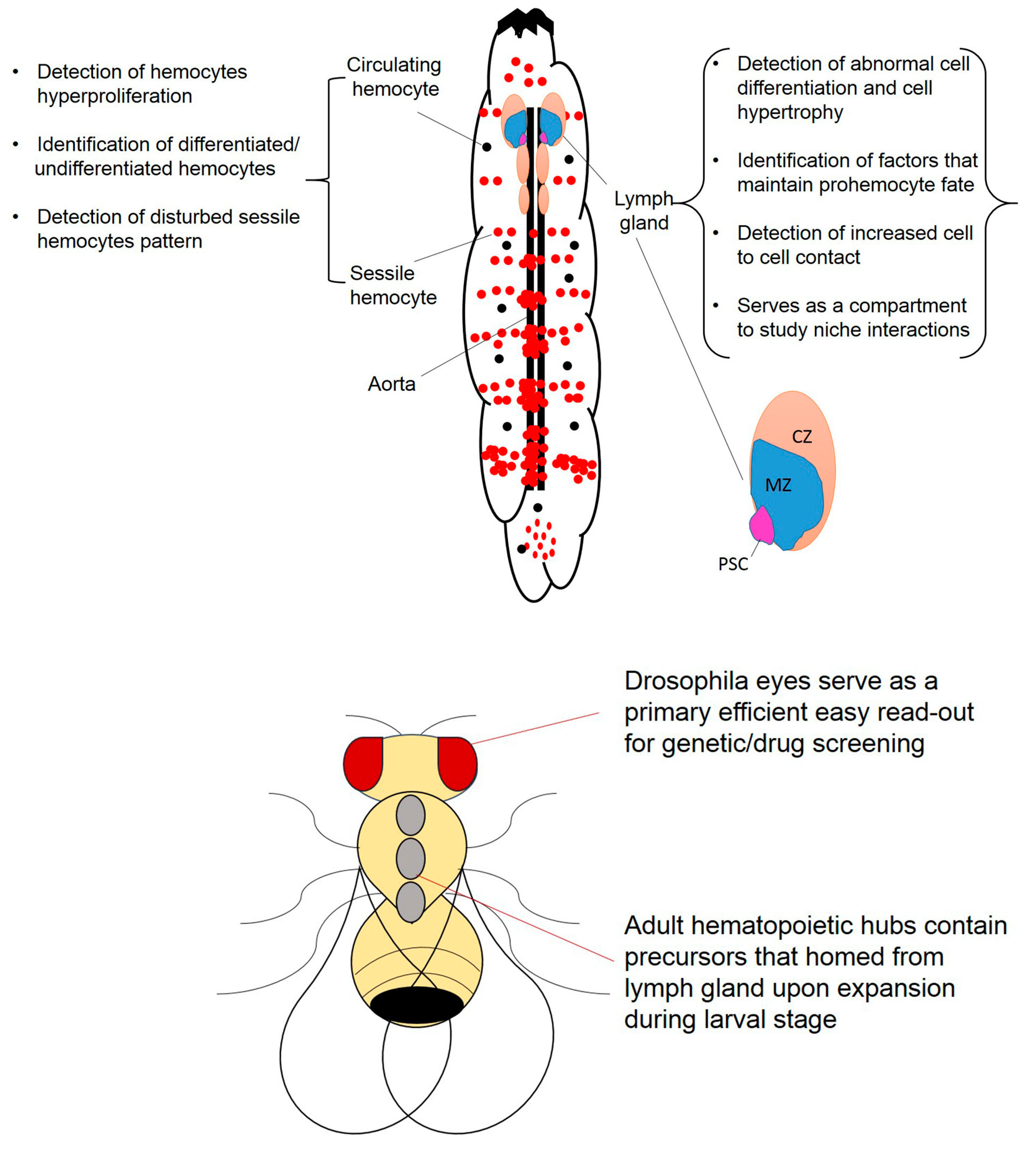
2. Drosophila melanogaster Hematopoiesis in a Glance
2.1. Circulating Hemocytes and Their Response to Oncogene Expression
2.2. Drosophila Sessile Hemocytes, Stem Cells and Their Response to Oncogene Expression
2.3. Drosophila Lymph Gland and Its Response to Oncogene Expression
3. Leukemia Models in Drosophila melanogaster
3.1. CML Models
3.2. Insights into Other Leukemia Models in Drosophila Melanogaster
3.3. Potential Avenues for Using the Leukemic Fly Model for Drug Discovery
4. Conclusion and Perspective
Author Contributions
Funding
Conflicts of Interest
References
- Galloway, J.L.; Zon, L.I. 3 Ontogeny of hematopoiesis: Examining the emergence of hematopoietic cells in the vertebrate embryo. In Current Topics in Developmental Biology; Academic Press: Cambridge, MA, USA, 2003; Volume 53, pp. 139–158. [Google Scholar]
- Palis, J.; Yoder, M.C. Yolk-sac hematopoiesis: The first blood cells of mouse and man. Exp. Hematol. 2001, 29, 927–936. [Google Scholar] [CrossRef]
- Godin, I.; Cumano, A. The hare and the tortoise: An embryonic haematopoietic race. Nat. Rev. Immunol. 2002, 2, 593–604. [Google Scholar] [CrossRef] [PubMed]
- Rotstein, B.; Paululat, A. On the Morphology of the Drosophila Heart. J. Cardiovasc. Dev. Dis. 2016, 3. [Google Scholar] [CrossRef] [PubMed]
- Evans, C.J.; Hartenstein, V.; Banerjee, U. Thicker than blood: Conserved mechanisms in Drosophila and vertebrate hematopoiesis. Dev. Cell 2003, 5, 673–690. [Google Scholar] [CrossRef]
- Salminen, T.S.; Vale, P.F. Drosophila as a Model System to Investigate the Effects of Mitochondrial Variation on Innate Immunity. Front. Immunol. 2020, 11. [Google Scholar] [CrossRef]
- Pham, L.N.; Dionne, M.S.; Shirasu-Hiza, M.; Schneider, D.S. A specific primed immune response in Drosophila is dependent on phagocytes. PLoS Pathog 2007, 3, e26. [Google Scholar] [CrossRef]
- Tassetto, M.; Kunitomi, M.; Andino, R. Circulating Immune Cells Mediate a Systemic RNAi-Based Adaptive Antiviral Response in Drosophila. Cell 2017, 169, 314–325.e13. [Google Scholar] [CrossRef] [PubMed]
- Markus, R.; Laurinyecz, B.; Kurucz, E.; Honti, V.; Bajusz, I.; Sipos, B.; Somogyi, K.; Kronhamn, J.; Hultmark, D.; Ando, I. Sessile hemocytes as a hematopoietic compartment in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2009, 106, 4805–4809. [Google Scholar] [CrossRef]
- Tepass, U.; Fessler, L.I.; Aziz, A.; Hartenstein, V. Embryonic origin of hemocytes and their relationship to cell death in Drosophila. Development 1994, 120, 1829–1837. [Google Scholar]
- Elrod-Erickson, M.; Mishra, S.; Schneider, D. Interactions between the cellular and humoral immune responses in Drosophila. Curr. Biol. CB 2000, 10, 781–784. [Google Scholar] [CrossRef]
- Rizki, T.M.; Rizki, R.M. The Cellular Defense System of Drosophila melanogaster. In Insect Ultrastructure: Volume 2; King, R.C., Akai, H., Eds.; Springer US: Boston, MA, USA, 1984; pp. 579–604. [Google Scholar] [CrossRef]
- Kocks, C.; Cho, J.H.; Nehme, N.; Ulvila, J.; Pearson, A.M.; Meister, M.; Strom, C.; Conto, S.L.; Hetru, C.; Stuart, L.M.; et al. Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila. Cell 2005, 123, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Shandala, T.; Woodcock, J.M.; Ng, Y.; Biggs, L.; Skoulakis, E.M.; Brooks, D.A.; Lopez, A.F. Drosophila 14-3-3epsilon has a crucial role in anti-microbial peptide secretion and innate immunity. J. Cell Sci. 2011, 124, 2165–2174. [Google Scholar] [CrossRef] [PubMed]
- Dimarcq, J.L.; Imler, J.L.; Lanot, R.; Ezekowitz, R.A.; Hoffmann, J.A.; Janeway, C.A.; Lagueux, M. Treatment of l(2)mbn Drosophila tumorous blood cells with the steroid hormone ecdysone amplifies the inducibility of antimicrobial peptide gene expression. Insect Biochem. Mol. Biol. 1997, 27, 877–886. [Google Scholar] [CrossRef]
- Ayyaz, A.; Li, H.; Jasper, H. Haemocytes control stem cell activity in the Drosophila intestine. Nat. Cell Biol. 2015, 17, 736–748. [Google Scholar] [CrossRef] [PubMed]
- Irving, P.; Ubeda, J.M.; Doucet, D.; Troxler, L.; Lagueux, M.; Zachary, D.; Hoffmann, J.A.; Hetru, C.; Meister, M. New insights into Drosophila larval haemocyte functions through genome-wide analysis. Cell. Microbiol. 2005, 7, 335–350. [Google Scholar] [CrossRef]
- Shia, A.K.; Glittenberg, M.; Thompson, G.; Weber, A.N.; Reichhart, J.M.; Ligoxygakis, P. Toll-dependent antimicrobial responses in Drosophila larval fat body require Spatzle secreted by haemocytes. J. Cell Sci. 2009, 122, 4505–4515. [Google Scholar] [CrossRef]
- Woodcock, K.J.; Kierdorf, K.; Pouchelon, C.A.; Vivancos, V.; Dionne, M.S.; Geissmann, F. Macrophage-derived upd3 cytokine causes impaired glucose homeostasis and reduced lifespan in Drosophila fed a lipid-rich diet. Immunity 2015, 42, 133–144. [Google Scholar] [CrossRef]
- Yang, H.; Hultmark, D. Tissue communication in a systemic immune response of Drosophila. Fly 2016, 10, 115–122. [Google Scholar] [CrossRef]
- Lanot, R.; Zachary, D.; Holder, F.; Meister, M. Postembryonic hematopoiesis in Drosophila. Dev. Biol. 2001, 230, 243–257. [Google Scholar] [CrossRef]
- Roshana, S.; Elisabeth, G. Ultrastructure and Cytochemistry of the Cell Types in the Larval Hematopoietic Organs and Hemolymph of Drosophila Melanogaster. Dev. Growth Differ. 1982, 24, 65–82. [Google Scholar] [CrossRef]
- Ramet, M.; Lanot, R.; Zachary, D.; Manfruelli, P. JNK signaling pathway is required for efficient wound healing in Drosophila. Dev. Biol. 2002, 241, 145–156. [Google Scholar] [CrossRef] [PubMed]
- De Gregorio, E.; Han, S.J.; Lee, W.J.; Baek, M.J.; Osaki, T.; Kawabata, S.; Lee, B.L.; Iwanaga, S.; Lemaitre, B.; Brey, P.T. An immune-responsive Serpin regulates the melanization cascade in Drosophila. Dev. Cell 2002, 3, 581–592. [Google Scholar] [CrossRef]
- Rizki, M.T.; Rizki, R.M. Functional significance of the crystal cells in the larva of Drosophila melanogaster. J. Biophys. Biochem. Cytol. 1959, 5, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Goto, A.; Kadowaki, T.; Kitagawa, Y. Drosophila hemolectin gene is expressed in embryonic and larval hemocytes and its knock down causes bleeding defects. Dev. Biol. 2003, 264, 582–591. [Google Scholar] [CrossRef]
- Goto, A.; Kumagai, T.; Kumagai, C.; Hirose, J.; Narita, H.; Mori, H.; Kadowaki, T.; Beck, K.; Kitagawa, Y. A Drosophila haemocyte-specific protein, hemolectin, similar to human von Willebrand factor. Biochem. J. 2001, 359, 99–108. [Google Scholar] [CrossRef]
- Scherfer, C.; Karlsson, C.; Loseva, O.; Bidla, G.; Goto, A.; Havemann, J.; Dushay, M.S.; Theopold, U. Isolation and Characterization of Hemolymph Clotting Factors in Drosophila melanogaster by a Pullout Method. Curr. Biol. 2004, 14, 625–629. [Google Scholar] [CrossRef]
- Rizki, T.M.; Rizki, R.M. Lamellocyte differentiation in Drosophila larvae parasitized by Leptopilina. Dev. Comp. Immunol. 1992, 16, 103–110. [Google Scholar] [CrossRef]
- Minakhina, S.; Steward, R. Melanotic mutants in Drosophila: Pathways and phenotypes. Genetics 2006, 174, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Claudio, B. Melanotic tumors in DROSOPHILA. J. Cell. Comp. Physiol. 1958, 52, 371–381. [Google Scholar] [CrossRef]
- Rizki, M.T. Melanotic tumor ormation in Drosophila. J. Morphol. 1960, 106, 147–157. [Google Scholar] [CrossRef]
- Oftedal, P. [The histogenesis of a new tumor in Drosophila melanogaster, and a comparison with tumors of five other stocks]. Z. Fur Indukt. Abstamm. Und Vererb. 1953, 85, 408–422. [Google Scholar] [CrossRef]
- Zettervall, C.J.; Anderl, I.; Williams, M.J.; Palmer, R.; Kurucz, E.; Ando, I.; Hultmark, D. A directed screen for genes involved in Drosophila blood cell activation. Proc. Natl. Acad. Sci. USA 2004, 101, 14192–14197. [Google Scholar] [CrossRef] [PubMed]
- Qiu, P.; Pan, P.C.; Govind, S. A role for the Drosophila Toll/Cactus pathway in larval hematopoiesis. Development 1998, 125, 1909–1920. [Google Scholar] [PubMed]
- Lemaitre, B.; Meister, M.; Govind, S.; Georgel, P.; Steward, R.; Reichhart, J.M.; Hoffmann, J.A. Functional analysis and regulation of nuclear import of dorsal during the immune response in Drosophila. EMBO J. 1995, 14, 536–545. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Hanratty, W.P.; Dearolf, C.R. An amino acid substitution in the Drosophila hopTum-l Jak kinase causes leukemia-like hematopoietic defects. EMBO J. 1995, 14, 1412–1420. [Google Scholar] [CrossRef] [PubMed]
- Beaupre, D.M.; Kurzrock, R. RAS and Leukemia: From Basic Mechanisms to Gene-Directed Therapy. J. Clin. Oncol. 1999, 17, 1071–1079. [Google Scholar] [CrossRef] [PubMed]
- Asha, H.; Nagy, I.; Kovacs, G.; Stetson, D.; Ando, I.; Dearolf, C.R. Analysis of Ras-induced overproliferation in Drosophila hemocytes. Genetics 2003, 163, 203–215. [Google Scholar]
- Arefin, B.; Kunc, M.; Krautz, R.; Theopold, U. The Immune Phenotype of Three Drosophila Leukemia Models. G3 (BethesdaMd.) 2017, 7, 2139–2149. [Google Scholar] [CrossRef]
- Kurucz, É.; Váczi, B.; Márkus, R.; Laurinyecz, B.; Vilmos, P.; Zsámboki, J.; Csorba, K.; Gateff, E.; Hultmark, D.; Andó, I. Definition of Drosophila hemocyte subsets by cell-type specific antigens. Acta Biol. Hung. 2007, 58, 95–111. [Google Scholar] [CrossRef]
- Makhijani, K.; Alexander, B.; Tanaka, T.; Rulifson, E.; Brückner, K. The peripheral nervous system supports blood cell homing and survival in the Drosophila larva. Development (Camb. Engl.) 2011, 138, 5379–5391. [Google Scholar] [CrossRef]
- Stofanko, M.; Kwon, S.Y.; Badenhorst, P. A misexpression screen to identify regulators of Drosophila larval hemocyte development. Genetics 2008, 180, 253–267. [Google Scholar] [CrossRef]
- Williams, M.J.; Wiklund, M.-L.; Wikman, S.; Hultmark, D. Rac1 signalling in the Drosophila larval cellular immune response. J. Cell Sci. 2006, 119, 2015–2024. [Google Scholar] [CrossRef]
- Narita, R.; Yamashita, H.; Goto, A.; Imai, H.; Ichihara, S.; Mori, H.; Kitagawa, Y. Syndecan-dependent binding of Drosophila hemocytes to laminin α3/5 chain LG4-5 modules: Potential role in sessile hemocyte islets formation. FEBS Lett. 2004, 576, 127–132. [Google Scholar] [CrossRef][Green Version]
- Bretscher, A.J.; Honti, V.; Binggeli, O.; Burri, O.; Poidevin, M.; Kurucz, É.; Zsámboki, J.; Andó, I.; Lemaitre, B. The Nimrod transmembrane receptor Eater is required for hemocyte attachment to the sessile compartment in Drosophila melanogaster. Biol. Open 2015, 4, 355–363. [Google Scholar] [CrossRef]
- Sherri, N.; Salloum, N.; Mouawad, C.; Haidar-Ahmad, N.; Shirinian, M.; Rahal, E.A. Epstein-Barr Virus DNA Enhances Diptericin Expression and Increases Hemocyte Numbers in Drosophila melanogaster via the Immune Deficiency Pathway. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Ghosh, S.; Singh, A.; Mandal, S.; Mandal, L. Active hematopoietic hubs in Drosophila adults generate hemocytes and contribute to immune response. Dev. Cell 2015, 33, 478–488. [Google Scholar] [CrossRef]
- Micchelli, C.A.; Perrimon, N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 2006, 439, 475. [Google Scholar] [CrossRef] [PubMed]
- Margolis, J.; Spradling, A. Identification and behavior of epithelial stem cells in the Drosophila ovary. Development (Camb. Engl.) 1995, 121, 3797–3807. [Google Scholar]
- Markstein, M.; Dettorre, S.; Cho, J.; Neumüller, R.A.; Craig-Müller, S.; Perrimon, N. Systematic screen of chemotherapeutics in Drosophila stem cell tumors. Proc. Natl. Acad. Sci. USA 2014, 111, 4530–4535. [Google Scholar] [CrossRef]
- Minakhina, S.; Steward, R. Hematopoietic stem cells in Drosophila. Development (Camb. Engl.) 2010, 137, 27–31. [Google Scholar] [CrossRef]
- Dey, N.S.; Ramesh, P.; Chugh, M.; Mandal, S.; Mandal, L. Dpp dependent Hematopoietic stem cells give rise to Hh dependent blood progenitors in larval lymph gland of Drosophila. Elife 2016, 5, e18295. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.S.; Melnick, M.B.; Perrimon, N. Marelle acts downstream of the Drosophila HOP/JAK kinase and encodes a protein similar to the mammalian STATs. Cell 1996, 84, 411–419. [Google Scholar] [CrossRef]
- Mirzoyan, Z.; Sollazzo, M.; Allocca, M.; Valenza, A.M.; Grifoni, D.; Bellosta, P. Drosophila melanogaster: A Model Organism to Study Cancer. Front Genet 2019, 10, 51. [Google Scholar] [CrossRef] [PubMed]
- Boileau, M.; Shirinian, M.; Gayden, T.; Harutyunyan, A.S.; Chen, C.C.L.; Mikael, L.G.; Duncan, H.M.; Neumann, A.L.; Arreba-Tutusaus, P.; De Jay, N.; et al. Mutant H3 histones drive human pre-leukemic hematopoietic stem cell expansion and promote leukemic aggressiveness. Nat. Commun. 2019, 10, 2891. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.D.; Wagers, A.J. Dynamic niches in the origination and differentiation of haematopoietic stem cells. Nat. Rev. Mol. Cell Biol. 2011, 12, 643–655. [Google Scholar] [CrossRef] [PubMed]
- Mandal, L.; Banerjee, U.; Hartenstein, V. Evidence for a fruit fly hemangioblast and similarities between lymph-gland hematopoiesis in fruit fly and mammal aorta-gonadal-mesonephros mesoderm. Nat. Genet. 2004, 36, 1019–1023. [Google Scholar] [CrossRef]
- Crozatier, M.; Meister, M. Drosophila haematopoiesis. Cell. Microbiol. 2007, 9, 1117–1126. [Google Scholar] [CrossRef]
- Wang, L.; Kounatidis, I.; Ligoxygakis, P. Drosophila as a model to study the role of blood cells in inflammation, innate immunity and cancer. Front. Cell. Infect. Microbiol. 2014, 3, 113. [Google Scholar] [CrossRef]
- Daga, A.; Karlovich, C.A.; Dumstrei, K.; Banerjee, U. Patterning of cells in the Drosophila eye by Lozenge, which shares homologous domains with AML1. Genes Dev. 1996, 10, 1194–1205. [Google Scholar] [CrossRef]
- Rizki, T.; Rizki, R. Alleles of lz as suppressors of the Bc-phene in Drosophila melanogaster. Genetics 1981, 97, s90. [Google Scholar]
- Lebestky, T.; Chang, T.; Hartenstein, V.; Banerjee, U. Specification of Drosophila hematopoietic lineage by conserved transcription factors. Science (New York N.Y.) 2000, 288, 146–149. [Google Scholar] [CrossRef]
- Ferguson, G.B.; Martinez-Agosto, J.A. Kicking it up a Notch for the best in show: Scalloped leads Yorkie into the haematopoietic arena. Fly 2014, 8, 206–217. [Google Scholar] [CrossRef]
- Lutterbach, B.; Hiebert, S. Role of the transcription factor AML-1 in acute leukemia and hematopoietic differentiation. Gene 2000, 245, 223–235. [Google Scholar] [CrossRef]
- Speck, N.A.; Gilliland, D.G. Core-binding factors in haematopoiesis and leukaemia. Nat. Rev. Cancer 2002, 2, 502–513. [Google Scholar] [CrossRef] [PubMed]
- Reimels, T.A.; Pfleger, C.M. Methods to Examine the Lymph Gland and Hemocytes in Drosophila Larvae. J. Vis. Exp. 2016. [Google Scholar] [CrossRef] [PubMed]
- Rugendorff, A.; Younossi-Hartenstein, A.; Hartenstein, V. Embryonic origin and differentiation of the Drosophila heart. Roux’s Arch. Dev. Biol. Off. Organ EDBO 1994, 203, 266–280. [Google Scholar] [CrossRef]
- Jung, S.H.; Evans, C.J.; Uemura, C.; Banerjee, U. The Drosophila lymph gland as a developmental model of hematopoiesis. Development 2005, 132, 2521–2533. [Google Scholar] [CrossRef]
- Bourbon, H.M.; Gonzy-Treboul, G.; Peronnet, F.; Alin, M.F.; Ardourel, C.; Benassayag, C.; Cribbs, D.; Deutsch, J.; Ferrer, P.; Haenlin, M.; et al. A P-insertion screen identifying novel X-linked essential genes in Drosophila. Mech. Dev. 2002, 110, 71–83. [Google Scholar] [CrossRef]
- Agaisse, H.; Petersen, U.M.; Boutros, M.; Mathey-Prevot, B.; Perrimon, N. Signaling role of hemocytes in Drosophila JAK/STAT-dependent response to septic injury. Dev. Cell 2003, 5, 441–450. [Google Scholar] [CrossRef]
- Mondal, B.C.; Mukherjee, T.; Mandal, L.; Evans, C.J.; Sinenko, S.A.; Martinez-Agosto, J.A.; Banerjee, U. Interaction between differentiating cell- and niche-derived signals in hematopoietic progenitor maintenance. Cell 2011, 147, 1589–1600. [Google Scholar] [CrossRef]
- Mandal, L.; Martinez-Agosto, J.A.; Evans, C.J.; Hartenstein, V.; Banerjee, U. A Hedgehog- and Antennapedia-dependent niche maintains Drosophila haematopoietic precursors. Nature 2007, 446, 320–324. [Google Scholar] [CrossRef]
- Krzemien, J.; Dubois, L.; Makki, R.; Meister, M.; Vincent, A.; Crozatier, M. Control of blood cell homeostasis in Drosophila larvae by the posterior signalling centre. Nature 2007, 446, 325–328. [Google Scholar] [CrossRef] [PubMed]
- Crozatier, M.; Ubeda, J.M.; Vincent, A.; Meister, M. Cellular immune response to parasitization in Drosophila requires the EBF orthologue collier. PLoS Biol. 2004, 2, E196. [Google Scholar] [CrossRef] [PubMed]
- Hagman, J.; Belanger, C.; Travis, A.; Turck, C.W.; Grosschedl, R. Cloning and functional characterization of early B-cell factor, a regulator of lymphocyte-specific gene expression. Genes Dev. 1993, 7, 760–773. [Google Scholar] [CrossRef] [PubMed]
- Nelson, R.E.; Fessler, L.I.; Takagi, Y.; Blumberg, B.; Keene, D.R.; Olson, P.F.; Parker, C.G.; Fessler, J.H. Peroxidasin: A novel enzyme-matrix protein of Drosophila development. EMBO J. 1994, 13, 3438–3447. [Google Scholar] [CrossRef]
- Pastor-Pareja, J.C.; Wu, M.; Xu, T. An innate immune response of blood cells to tumors and tissue damage in Drosophila. Dis. Models Mech. 2008, 1, 144–154; discussion 153. [Google Scholar] [CrossRef]
- Osman, D.; Gobert, V.; Ponthan, F.; Heidenreich, O.; Haenlin, M.; Waltzer, L. A Drosophila model identifies calpains as modulators of the human leukemogenic fusion protein AML1-ETO. Proc. Natl. Acad. Sci. USA 2009, 106, 12043–12048. [Google Scholar] [CrossRef] [PubMed]
- Sinenko, S.A.; Hung, T.; Moroz, T.; Tran, Q.M.; Sidhu, S.; Cheney, M.D.; Speck, N.A.; Banerjee, U. Genetic manipulation of AML1-ETO-induced expansion of hematopoietic precursors in a Drosophila model. Blood 2010, 116, 4612–4620. [Google Scholar] [CrossRef]
- Reitman, Z.J.; Sinenko, S.A.; Spana, E.P.; Yan, H. Genetic dissection of leukemia-associated IDH1 and IDH2 mutants and D-2-hydroxyglutarate in Drosophila. Blood 2015, 125, 336–345. [Google Scholar] [CrossRef]
- Terriente-Félix, A.; Pérez, L.; Bray, S.J.; Nebreda, A.R.; Milán, M. A Drosophila model of myeloproliferative neoplasm reveals a feed-forward loop in the JAK pathway mediated by p38 MAPK signalling. Dis. Models Mech. 2017, 10, 399–407. [Google Scholar] [CrossRef]
- Lacout, C.; Pisani, D.F.; Tulliez, M.; Gachelin, F.M.; Vainchenker, W.; Villeval, J.L. JAK2V617F expression in murine hematopoietic cells leads to MPD mimicking human PV with secondary myelofibrosis. Blood 2006, 108, 1652–1660. [Google Scholar] [CrossRef]
- Tan, K.L.; Goh, S.C.; Minakhina, S. Genetic screen for regulators of lymph gland homeostasis and hemocyte maturation in Drosophila. G3 (BethesdaMd.) 2012, 2, 393–405. [Google Scholar] [CrossRef] [PubMed]
- Harrison, D.A.; Binari, R.; Nahreini, T.S.; Gilman, M.; Perrimon, N. Activation of a Drosophila Janus kinase (JAK) causes hematopoietic neoplasia and developmental defects. EMBO J. 1995, 14, 2857–2865. [Google Scholar] [CrossRef]
- Lemaitre, B.; Hoffmann, J. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007, 25, 697–743. [Google Scholar] [CrossRef]
- Hanratty, W.P.; Ryerse, J.S. A genetic melanotic neoplasm of Drosophila melanogaster. Dev. Biol. 1981, 83, 238–249. [Google Scholar] [CrossRef]
- Baril, C.; Gavory, G.; Bidla, G.; Knaevelsrud, H.; Sauvageau, G.; Therrien, M. Human NUP98-HOXA9 promotes hyperplastic growth of hematopoietic tissues in Drosophila. Dev. Biol. 2017, 421, 16–26. [Google Scholar] [CrossRef]
- Giordani, G.; Barraco, M.; Giangrande, A.; Martinelli, G.; Guadagnuolo, V.; Simonetti, G.; Perini, G.; Bernardoni, R. The human Smoothened inhibitor PF-04449913 induces exit from quiescence and loss of multipotent Drosophila hematopoietic progenitor cells. Oncotarget 2016, 7, 55313–55327. [Google Scholar] [CrossRef] [PubMed]
- Fogerty, F.J.; Juang, J.L.; Petersen, J.; Clark, M.J.; Hoffmann, F.M.; Mosher, D.F. Dominant effects of the bcr-abl oncogene on Drosophila morphogenesis. Oncogene 1999, 18, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Clarke, C.J.; Holyoake, T.L. Preclinical approaches in chronic myeloid leukemia: From cells to systems. Exp. Hematol. 2017, 47, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Pophali, P.A.; Patnaik, M.M. The Role of New Tyrosine Kinase Inhibitors in Chronic Myeloid Leukemia. Cancer J. 2016, 22, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Suryanarayan, K.; Hunger, S.P.; Kohler, S.; Carroll, A.J.; Crist, W.; Link, M.P.; Cleary, M.L. Consistent involvement of the bcr gene by 9;22 breakpoints in pediatric acute leukemias. Blood 1991, 77, 324–330. [Google Scholar] [CrossRef] [PubMed]
- Chan, L.; Karhi, K.; Rayter, S.; Heisterkamp, N.; Eridani, S.; Powle, R.; Lawler, S.; Groffen, J.; Foulkes, J.; Greaves, M. A novel abl protein expressed in Philadelphia chromosome positive acute lymphoblastic leukaemia positive acute lymphoblastic leukaemia. Nature 1987, 325, 635. [Google Scholar] [CrossRef] [PubMed]
- Stevens, T.L.; Rogers, E.M.; Koontz, L.M.; Fox, D.T.; Homem, C.C.; Nowotarski, S.H.; Artabazon, N.B.; Peifer, M. Using Bcr-Abl to examine mechanisms by which abl kinase regulates morphogenesis in Drosophila. Mol. Biol. Cell 2008, 19, 378–393. [Google Scholar] [CrossRef] [PubMed]
- Salgia, R.; Li, J.L.; Ewaniuk, D.S.; Pear, W.; Pisick, E.; Burky, S.A.; Ernst, T.; Sattler, M.; Chen, L.B.; Griffin, J.D. BCR/ABL induces multiple abnormalities of cytoskeletal function. J. Clin. Investig. 1997, 100, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Bernardoni, R.; Giordani, G.; Signorino, E.; Monticelli, S.; Messa, F.; Pradotto, M.; Rosso, V.; Bracco, E.; Giangrande, A.; Perini, G.; et al. A new BCR-ABL1 Drosophila model as a powerful tool to elucidate the pathogenesis and progression of chronic myeloid leukemia. Haematologica 2019, 104, 717–728. [Google Scholar] [CrossRef]
- Nieborowska-Skorska, M.; Wasik, M.A.; Slupianek, A.; Salomoni, P.; Kitamura, T.; Calabretta, B.; Skorski, T. Signal transducer and activator of transcription (STAT)5 activation by BCR/ABL is dependent on intact Src homology (SH)3 and SH2 domains of BCR/ABL and is required for leukemogenesis. J. Exp. Med. 1999, 189, 1229–1242. [Google Scholar] [CrossRef]
- Outa, A.A.; Abubaker, D.; Bazarbachi, A.; Sabban, M.E.; Shirinian, M.; Nasr, R. Validation of a Drosophila model of wild-type and T315I mutated BCR-ABL1 in chronic myeloid leukemia: An effective platform for treatment screening. Haematologica 2020, 105, 387–397. [Google Scholar] [CrossRef] [PubMed]
- SEER. Cancer Stat Facts: Leukemia—Acute Myeloid Leukemia (AML). Available online: https://seer.cancer.gov/statfacts/html/amyl.html (accessed on 2 December 2019).
- Watts, J.; Nimer, S. Recent advances in the understanding and treatment of acute myeloid leukemia. F1000Res 2018, 7, F1000 Faculty Rev-1196. [Google Scholar] [CrossRef]
- McCormack, E.; Bruserud, O.; Gjertsen, B.T. Review: Genetic models of acute myeloid leukaemia. Oncogene 2008, 27, 3765–3779. [Google Scholar] [CrossRef]
- Erickson, P.; Gao, J.; Chang, K.S.; Look, T.; Whisenant, E.; Raimondi, S.; Lasher, R.; Trujillo, J.; Rowley, J.; Drabkin, H. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood 1992, 80, 1825–1831. [Google Scholar] [CrossRef]
- Nisson, P.E.; Watkins, P.C.; Sacchi, N. Transcriptionally active chimeric gene derived from the fusion of the AML1 gene and a novel gene on chromosome 8 in t(8;21) leukemic cells. Cancer Genet. Cytogenet. 1992, 63, 81–88. [Google Scholar] [CrossRef]
- Shimizu, K.; Miyoshi, H.; Kozu, T.; Nagata, J.; Enomoto, K.; Maseki, N.; Kaneko, Y.; Ohki, M. Consistent disruption of the AML1 gene occurs within a single intron in the t(8;21) chromosomal translocation. Cancer Res. 1992, 52, 6945–6948. [Google Scholar]
- Miyoshi, H.; Kozu, T.; Shimizu, K.; Enomoto, K.; Maseki, N.; Kaneko, Y.; Kamada, N.; Ohki, M. The t(8;21) translocation in acute myeloid leukemia results in production of an AML1-MTG8 fusion transcript. EMBO J. 1993, 12, 2715–2721. [Google Scholar] [CrossRef] [PubMed]
- Look, A.T. Oncogenic transcription factors in the human acute leukemias. Science (New York N.Y.) 1997, 278, 1059–1064. [Google Scholar] [CrossRef] [PubMed]
- Canon, J.; Banerjee, U. In vivo analysis of a developmental circuit for direct transcriptional activation and repression in the same cell by a Runx protein. Genes Dev. 2003, 17, 838–843. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wildonger, J.; Mann, R.S. The t(8;21) translocation converts AML1 into a constitutive transcriptional repressor. Development 2005, 132, 2263–2272. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Schwieger, M.; Löhler, J.; Friel, J.; Scheller, M.; Horak, I.; Stocking, C. AML1-ETO inhibits maturation of multiple lymphohematopoietic lineages and induces myeloblast transformation in synergy with ICSBP deficiency. J. Exp. Med. 2002, 196, 1227–1240. [Google Scholar] [CrossRef] [PubMed]
- Breig, O.; Bras, S.; Martinez Soria, N.; Osman, D.; Heidenreich, O.; Haenlin, M.; Waltzer, L. Pontin is a critical regulator for AML1-ETO-induced leukemia. Leukemia 2014, 28, 1271–1279. [Google Scholar] [CrossRef]
- Peterson, L.F.; Zhang, D.E. The 8;21 translocation in leukemogenesis. Oncogene 2004, 23, 4255–4262. [Google Scholar] [CrossRef]
- Gobert, V.; Haenlin, M.; Waltzer, L. Myeloid leukemia factor: A return ticket from human leukemia to fly hematopoiesis. Transcription 2012, 3, 250–254. [Google Scholar] [CrossRef]
- Bras, S.; Martin-Lanneree, S.; Gobert, V.; Auge, B.; Breig, O.; Sanial, M.; Yamaguchi, M.; Haenlin, M.; Plessis, A.; Waltzer, L. Myeloid leukemia factor is a conserved regulator of RUNX transcription factor activity involved in hematopoiesis. Proc. Natl. Acad. Sci. USA 2012, 109, 4986–4991. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, A. Transcriptional activation by MLL fusion proteins in leukemogenesis. Exp. Hematol. 2017, 46, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Collins, E.C.; Appert, A.; Ariza-McNaughton, L.; Pannell, R.; Yamada, Y.; Rabbitts, T.H. Mouse Af9 is a controller of embryo patterning, like Mll, whose human homologue fuses with Af9 after chromosomal translocation in leukemia. Mol. Cell. Biol. 2002, 22, 7313–7324. [Google Scholar] [CrossRef]
- Muyrers-Chen, I.; Rozovskaia, T.; Lee, N.; Kersey, J.H.; Nakamura, T.; Canaani, E.; Paro, R. Expression of leukemic MLL fusion proteins in Drosophila affects cell cycle control and chromosome morphology. Oncogene 2004, 23, 8639–8648. [Google Scholar] [CrossRef]
- Alharbi, R.A.; Pettengell, R.; Pandha, H.S.; Morgan, R. The role of HOX genes in normal hematopoiesis and acute leukemia. Leukemia 2013, 27, 1000–1008. [Google Scholar] [CrossRef] [PubMed]
- De Braekeleer, E.; Douet-Guilbert, N.; Basinko, A.; Le Bris, M.J.; Morel, F.; De Braekeleer, M. Hox gene dysregulation in acute myeloid leukemia. Future Oncol. (Lond. Engl.) 2014, 10, 475–495. [Google Scholar] [CrossRef] [PubMed]
- Kroon, E.; Krosl, J.; Thorsteinsdottir, U.; Baban, S.; Buchberg, A.M.; Sauvageau, G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998, 17, 3714–3725. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, M.; Kuwata, T.; Yamazaki, Y.; Jenkins, N.A.; Copeland, N.G.; Osato, M.; Ito, Y.; Kroon, E.; Sauvageau, G.; Nakamura, T. Identification of cooperative genes for NUP98-HOXA9 in myeloid leukemogenesis using a mouse model. Blood 2005, 105, 784–793. [Google Scholar] [CrossRef][Green Version]
- Berlandi, J.; Chaouch, A.; De Jay, N.; Tegeder, I.; Thiel, K.; Shirinian, M.; Kleinman, C.L.; Jeibmann, A.; Lasko, P.; Jabado, N.; et al. Identification of genes functionally involved in the detrimental effects of mutant histone H3.3-K27M in Drosophila melanogaster. Neuro Oncol. 2019, 21, 628–639. [Google Scholar] [CrossRef]
- Shirinian, M.; Kambris, Z.; Hamadeh, L.; Grabbe, C.; Journo, C.; Mahieux, R.; Bazarbachi, A. A Transgenic Drosophila melanogaster Model To Study Human T-Lymphotropic Virus Oncoprotein Tax-1-Driven Transformation In Vivo. J. Virol. 2015, 89, 8092–8095. [Google Scholar] [CrossRef]
- Azran, I.; Schavinsky-Khrapunsky, Y.; Aboud, M. Role of Tax protein in human T-cell leukemia virus type-I leukemogenicity. Retrovirology 2004, 1, 20. [Google Scholar] [CrossRef] [PubMed]
- Shirinian, M.; Kfoury, Y.; Dassouki, Z.; El-Hajj, H.; Bazarbachi, A. Tax-1 and Tax-2 similarities and differences: Focus on post-translational modifications and NF-κB activation. Front. Microbiol. 2013, 4, 231. [Google Scholar] [CrossRef] [PubMed]
- Lavorgna, A.; Harhaj, E.W. Regulation of HTLV-1 tax stability, cellular trafficking and NF-κB activation by the ubiquitin-proteasome pathway. Viruses 2014, 6, 3925–3943. [Google Scholar] [CrossRef] [PubMed]
- Kohnken, R.; Porcu, P.; Mishra, A. Overview of the Use of Murine Models in Leukemia and Lymphoma Research. Front. Oncol. 2017, 7, 22. [Google Scholar] [CrossRef]
- Strange, K. Drug Discovery in Fish, Flies, and Worms. ILAR J. 2016, 57, 133–143. [Google Scholar] [CrossRef]
- Bangi, E.; Ang, C.; Smibert, P.; Uzilov, A.V.; Teague, A.G.; Antipin, Y.; Chen, R.; Hecht, C.; Gruszczynski, N.; Yon, W.J.; et al. A personalized platform identifies trametinib plus zoledronate for a patient with KRAS-mutant metastatic colorectal cancer. Sci. Adv. 2019, 5, eaav6528. [Google Scholar] [CrossRef]
- Manon Boulet, M.M.; Laurence, V.; Lucas, W. From Drosophila Blood Cells to Human Leukemia. In Drosophila Models for Human Diseases; Yamaguchi, M., Ed.; Springer: Singapore, 2018; pp. 195–214. [Google Scholar]
| Type of Leukemia | Transgene | Phenotype | Site of Oncogene Expression | Reference |
|---|---|---|---|---|
| CML /Ph+ ALL | Human/fly BCR-ABL1P210 /BCR-ABL1P185 | - CNS and eye defects and increase in the phosphorylation of the dAbl substrate “Ena” | CNS and eye imaginal discs | [90] |
| - embryonic lethality and disruption of morphogenesis (disruption of head involution, segment grooves and dorsal closure) | Various embryonic sites | [95] | ||
| Human BCR-ABL1P210/ BCR-ABL1T315I | - Altered differentiation in Drosophila eyes and interference with dAbl signaling - Increase in circulating hemocytes | Eye imaginal discs and hemocytes | [97] | |
| - T315I resulted in a more severe rough eye phenotype - The model was validated for drug screening by feeding flies TKIs | Eye imaginal discs | [99] | ||
| ALL/AML | MLL, MLL-AF9, and MLL-AF4 | - MLL-AF9, and MLL-AF4 cause larval/pupal lethality upon expression in blood lineage and during early and late development - The fusions showed differing effects on proliferation and chromosome condensation in larval brain | Ubiquitously, all imaginal discs and in hematopoietic system | [117] |
| AML | Human AML1-ETO | - AML1-ETO acts as a transcriptional repressor of Lozenge target genes in Drosophila eyes | Eye imaginal discs | [109] |
| - Expression in Lz+ blood cells inhibited the differentiation of crystal cells, and induced an increase in circulating Lz+ progenitors - Identification of calpain B as required for AML1-ETO activity in Drosophila hemocytes | Hemocytes | [79] | ||
| - In vivo RNAi in Drosophila expressing human AML1-ETO identifies Pontin/RUVBL1 as a gene responsible for AML1-ETO-induced lethality and blood cell proliferation | Hemocytes | [111] | ||
| - Expression in majority of circulating hemocytes using (hml-Gal4) increased hemocytes count and along with expansion of hemocytes progenitors | Hemocytes | [80] | ||
| Human MLF1 | - Drosophila mlf appeared to play a role in RUNX1-ETO stabilization - Human MLF1 expressed under the control of lz-Gal4 reversed mlf-associated crystal cell defects | Crystal cell lineage | [114] | |
| Human NUP98-HOXA9 (NA9) | - Expression of NA9 in Drosophila cortical zone of lymph gland and circulating hemocytes results in increased cellular proliferation and enlargement of posterior signaling center | Lymph gland and hemocytes | [88] | |
| ATL | HTLV-1Tax transactivator (Tax-1) | - Eye defects and increased circulating hemocytes - Knockdown of Relish of the IMD pathway reversed the rough eye phenotype through an RNAi-based screen | Eye imaginal discs and hemocytes | [123] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al Outa, A.; Abubaker, D.; Madi, J.; Nasr, R.; Shirinian, M. The Leukemic Fly: Promises and Challenges. Cells 2020, 9, 1737. https://doi.org/10.3390/cells9071737
Al Outa A, Abubaker D, Madi J, Nasr R, Shirinian M. The Leukemic Fly: Promises and Challenges. Cells. 2020; 9(7):1737. https://doi.org/10.3390/cells9071737
Chicago/Turabian StyleAl Outa, Amani, Dana Abubaker, Joelle Madi, Rihab Nasr, and Margret Shirinian. 2020. "The Leukemic Fly: Promises and Challenges" Cells 9, no. 7: 1737. https://doi.org/10.3390/cells9071737
APA StyleAl Outa, A., Abubaker, D., Madi, J., Nasr, R., & Shirinian, M. (2020). The Leukemic Fly: Promises and Challenges. Cells, 9(7), 1737. https://doi.org/10.3390/cells9071737





