Checkpoint Inhibitors and Engineered Cells: New Weapons for Natural Killer Cell Arsenal Against Hematological Malignancies
Abstract
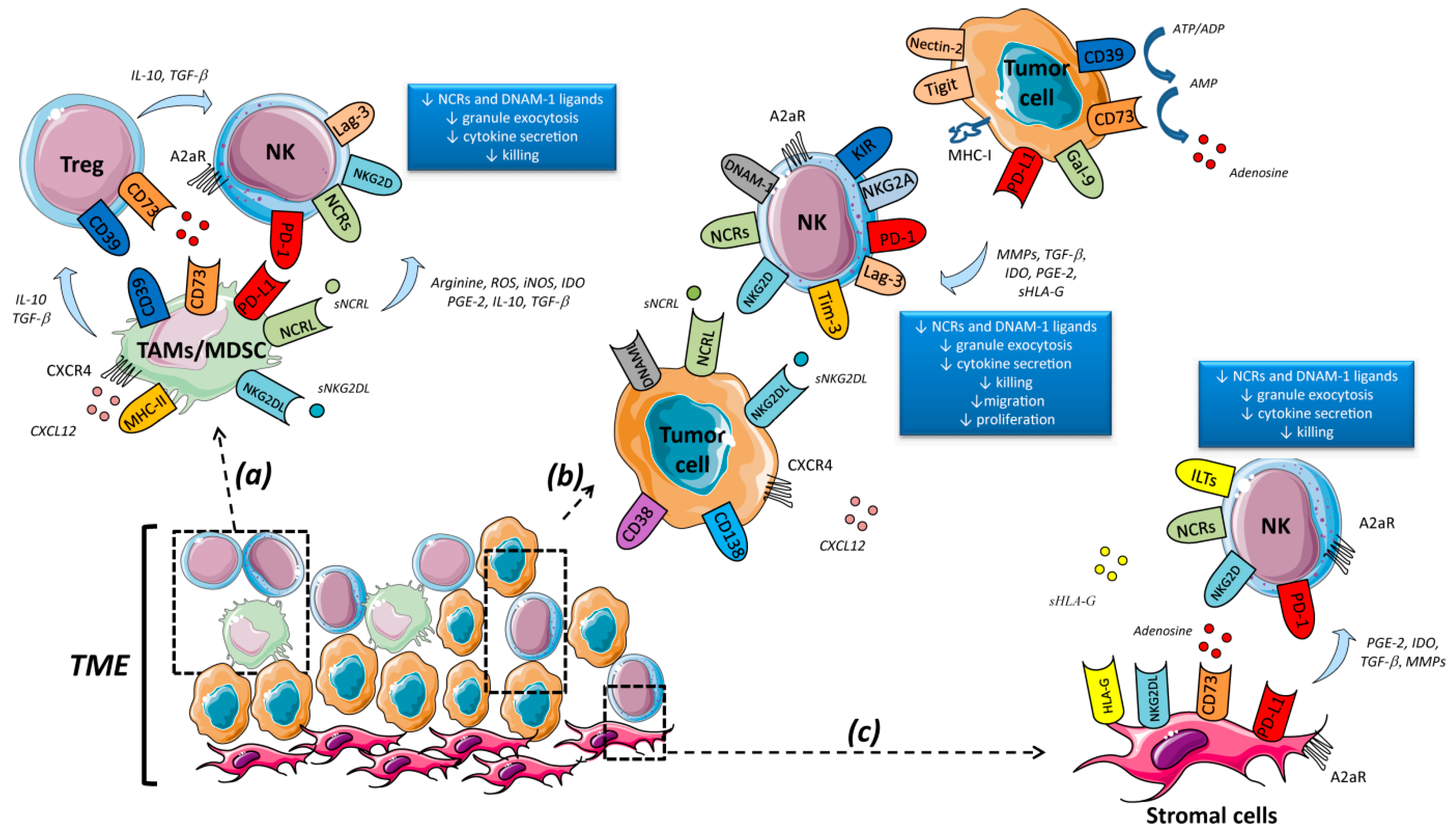
1. Tumor-Mediated NK Cell Exhaustion in Hematological Malignancies
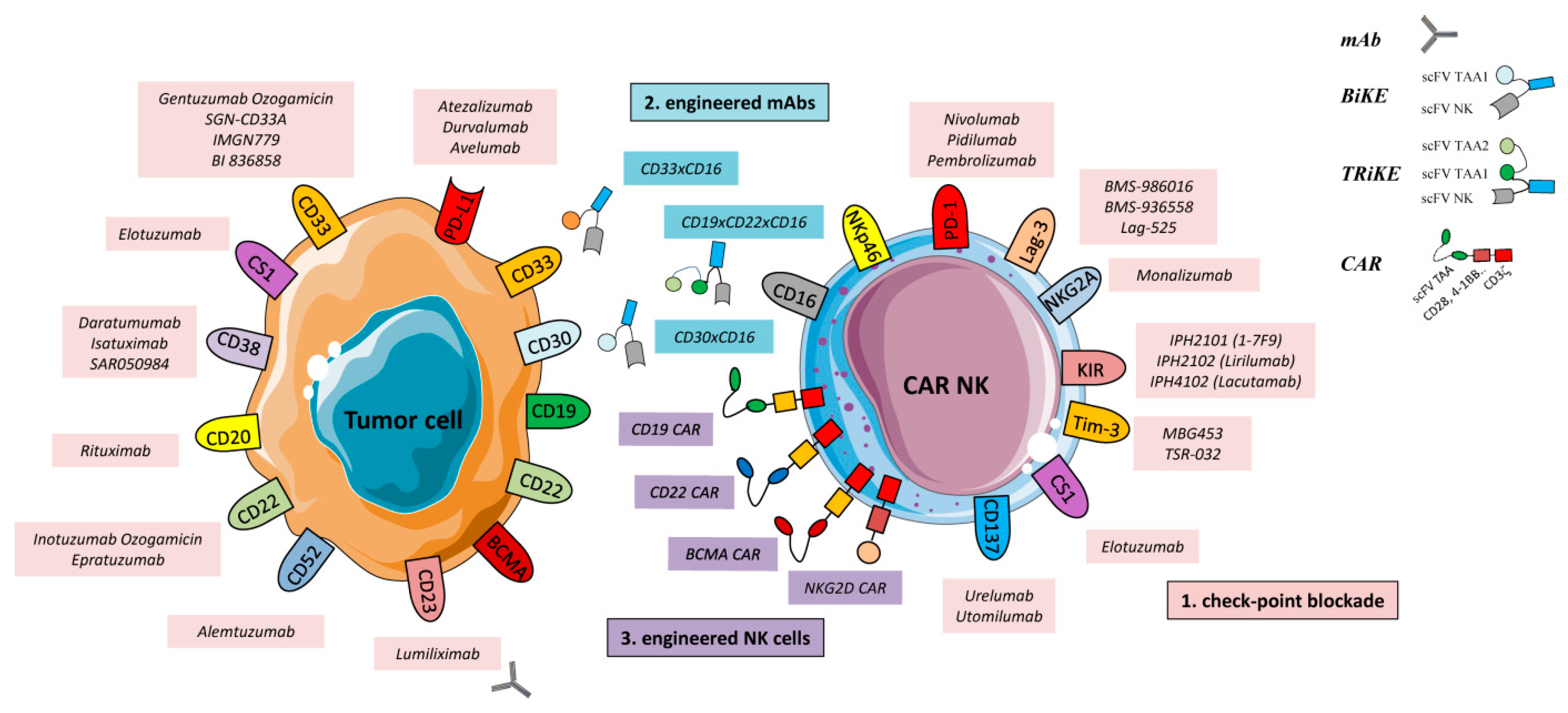
2. Current Advanced Therapy in Hematological Malignancies
2.1. Monoclonal Antibodies (mAbs)
- - Adenosine, CD39, and CD73
- - Other Molecular Targets for NK Cell Immunotherapy
2.2. Checkpoint Inhibitors
- - Anti-KIRs and Anti-NKG2A mAbs
- - HLA-G
- - Lymphocyte-activation gene-3 (LAG-3), TIM-3, PD-1, and T cell immunoglobulin and ITIM domain (TIGIT)
- - Other molecular Targets for NK Cell-Mediated Immunotherapy
2.3. Engineered mAbs
2.3.1. Bi-specific T cell Engagers, Bi-Specific Killer Engagers, and Tri-Specific Killer Engagers (BITEs, BIKEs, and TRIKEs)
2.3.2. Dual-Affinity Re-Targeting T cells (DARTs)
3. Engineered Effector Cells
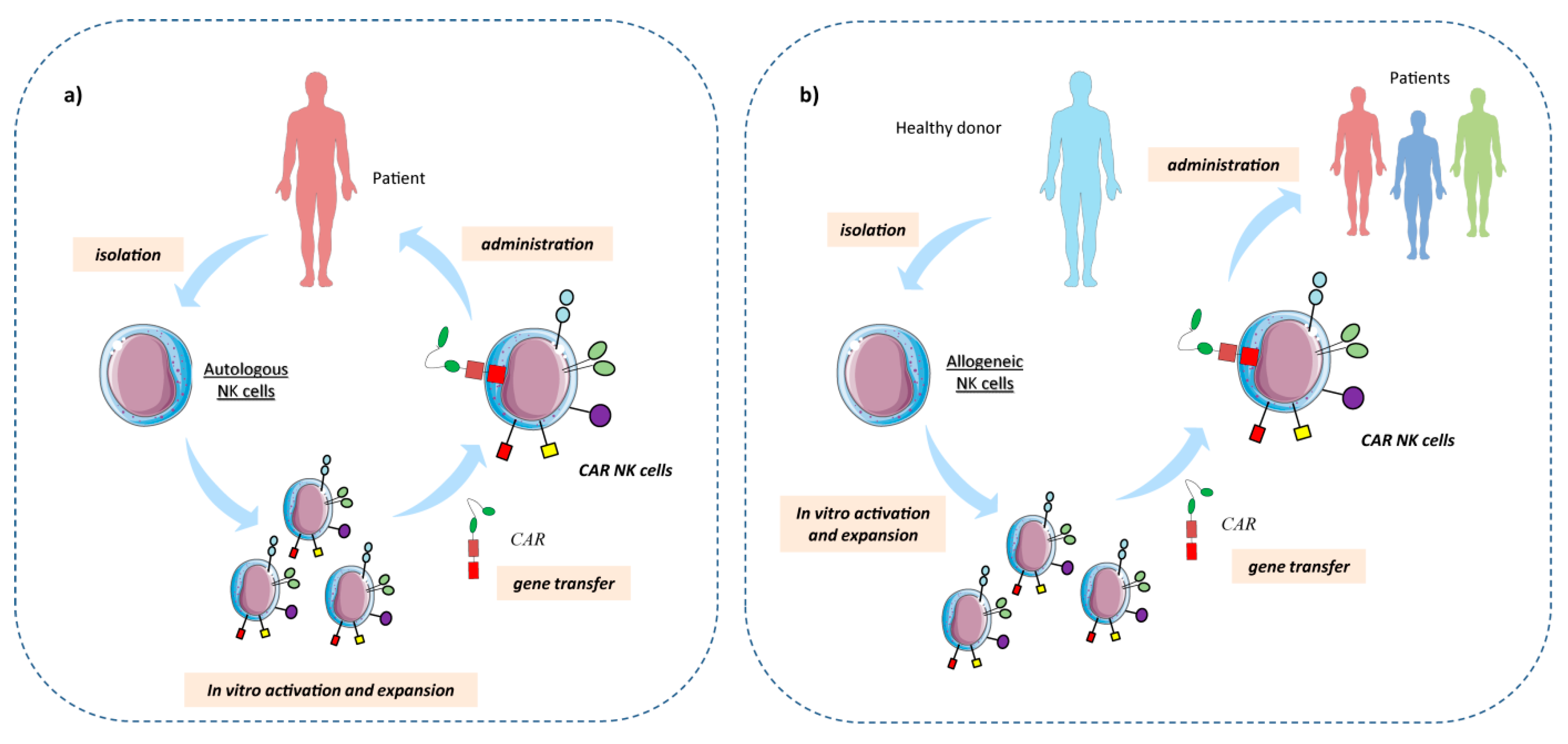
3.1. Chimeric Antigen Receptor (CAR) NK Cells
3.2. T-Cell Redirected for Universal Cytokine-Mediated Killing (TRUCKs)
4. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Lanier, L.L. NK cell recognition. Annu. Rev. Immunol. 2005, 23, 225–274. [Google Scholar] [CrossRef]
- Ljunggren, H.G.; Karre, K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol. Today 1990, 11, 237–244. [Google Scholar] [CrossRef]
- Moretta, A.; Bottino, C.; Vitale, M.; Pende, D.; Cantoni, C.; Mingari, M.C.; Biassoni, R.; Moretta, L. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu. Rev. Immunol. 2001, 19, 197–223. [Google Scholar] [CrossRef] [PubMed]
- Moretta, L.; Moretta, A. Unravelling natural killer cell function: Triggering and inhibitory human NK receptors. Embo. J. 2004, 23, 255–259. [Google Scholar] [CrossRef]
- Vivier, E.; Tomasello, E.; Baratin, M.; Walzer, T.; Ugolini, S. Functions of natural killer cells. Nat. Immunol. 2008, 9, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Pende, D.; Falco, M.; Vitale, M.; Cantoni, C.; Vitale, C.; Munari, E.; Bertaina, A.; Moretta, F.; Del Zotto, G.; Pietra, G.; et al. Killer Ig-Like Receptors (KIRs): Their Role in NK Cell Modulation and Developments Leading to Their Clinical Exploitation. Front. Immunol. 2019, 10, 1179. [Google Scholar] [CrossRef] [PubMed]
- Barrow, A.D.; Colonna, M. Tailoring Natural Killer cell immunotherapy to the tumour microenvironment. Semin. Immunol. 2017, 31, 30–36. [Google Scholar] [CrossRef]
- Bassani, B.; Baci, D.; Gallazzi, M.; Poggi, A.; Bruno, A.; Mortara, L. Natural Killer Cells as Key Players of Tumor Progression and Angiogenesis: Old and Novel Tools to Divert Their Pro-Tumor Activities into Potent Anti-Tumor Effects. Cancers 2019, 11, 461. [Google Scholar] [CrossRef]
- Nicholas, N.S.; Apollonio, B.; Ramsay, A.G. Tumor microenvironment (TME)-driven immune suppression in B cell malignancy. Biochim. Biophys. Acta. 2016, 1863, 471–482. [Google Scholar] [CrossRef]
- Poggi, A.; Giuliani, M. Mesenchymal Stromal Cells Can Regulate the Immune Response in the Tumor Microenvironment. Vaccines 2016, 4, 41. [Google Scholar] [CrossRef] [PubMed]
- Scott, D.W.; Gascoyne, R.D. The tumour microenvironment in B cell lymphomas. Nat. Rev. Cancer 2014, 14, 517–534. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Correa, B.; Gayoso, I.; Bergua, J.M.; Casado, J.G.; Morgado, S.; Solana, R.; Tarazona, R. Decreased expression of DNAM-1 on NK cells from acute myeloid leukemia patients. Immunol. Cell Biol. 2012, 90, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Spear, P.; Wu, M.R.; Sentman, M.L.; Sentman, C.L. NKG2D ligands as therapeutic targets. Cancer Immun. 2013, 13, 8. [Google Scholar]
- Carlsten, M.; Jaras, M. Natural Killer Cells in Myeloid Malignancies: Immune Surveillance, NK Cell Dysfunction, and Pharmacological Opportunities to Bolster the Endogenous NK Cells. Front. Immunol. 2019, 10, 2357. [Google Scholar] [CrossRef] [PubMed]
- Dulphy, N.; Chretien, A.S.; Khaznadar, Z.; Fauriat, C.; Nanbakhsh, A.; Caignard, A.; Chouaib, S.; Olive, D.; Toubert, A. Underground Adaptation to a Hostile Environment: Acute Myeloid Leukemia vs. Natural Killer Cells. Front. Immunol. 2016, 7, 94. [Google Scholar] [CrossRef] [PubMed]
- Farnault, L.; Sanchez, C.; Baier, C.; Le Treut, T.; Costello, R.T. Hematological malignancies escape from NK cell innate immune surveillance: Mechanisms and therapeutic implications. Clin. Dev. Immunol. 2012, 2012, 421702. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Rodriguez, A.P.; Villa-Alvarez, M.; Sordo-Bahamonde, C.; Lorenzo-Herrero, S.; Gonzalez, S. NK Cells in the Treatment of Hematological Malignancies. J. Clin. Med. 2019, 8, 1557. [Google Scholar] [CrossRef]
- Hofland, T.; Eldering, E.; Kater, A.P.; Tonino, S.H. Engaging Cytotoxic T and NK Cells for Immunotherapy in Chronic Lymphocytic Leukemia. Int. J. Mol. Sci. 2019, 20, 4315. [Google Scholar] [CrossRef]
- Parry, H.M.; Stevens, T.; Oldreive, C.; Zadran, B.; McSkeane, T.; Rudzki, Z.; Paneesha, S.; Chadwick, C.; Stankovic, T.; Pratt, G.; et al. NK cell function is markedly impaired in patients with chronic lymphocytic leukaemia but is preserved in patients with small lymphocytic lymphoma. Oncotarget 2016, 7, 68513–68526. [Google Scholar] [CrossRef]
- El-Sherbiny, Y.M.; Meade, J.L.; Holmes, T.D.; McGonagle, D.; Mackie, S.L.; Morgan, A.W.; Cook, G.; Feyler, S.; Richards, S.J.; Davies, F.E.; et al. The requirement for DNAM-1, NKG2D, and NKp46 in the natural killer cell-mediated killing of myeloma cells. Cancer Res. 2007, 67, 8444–8449. [Google Scholar] [CrossRef]
- Godfrey, J.; Benson, D.M., Jr. The role of natural killer cells in immunity against multiple myeloma. Leuk. Lymphoma 2012, 53, 1666–1676. [Google Scholar] [CrossRef] [PubMed]
- Khaznadar, Z.; Boissel, N.; Agaugue, S.; Henry, G.; Cheok, M.; Vignon, M.; Geromin, D.; Cayuela, J.M.; Castaigne, S.; Pautas, C.; et al. Defective NK Cells in Acute Myeloid Leukemia Patients at Diagnosis Are Associated with Blast Transcriptional Signatures of Immune Evasion. J. Immunol. 2015, 195, 2580–2590. [Google Scholar] [CrossRef]
- Lion, E.; Willemen, Y.; Berneman, Z.N.; Van Tendeloo, V.F.; Smits, E.L. Natural killer cell immune escape in acute myeloid leukemia. Leukemia 2012, 26, 2019–2026. [Google Scholar] [CrossRef] [PubMed]
- Stringaris, K.; Sekine, T.; Khoder, A.; Alsuliman, A.; Razzaghi, B.; Sargeant, R.; Pavlu, J.; Brisley, G.; de Lavallade, H.; Sarvaria, A.; et al. Leukemia-induced phenotypic and functional defects in natural killer cells predict failure to achieve remission in acute myeloid leukemia. Haematologica 2014, 99, 836–847. [Google Scholar] [CrossRef]
- Khaznadar, Z.; Henry, G.; Setterblad, N.; Agaugue, S.; Raffoux, E.; Boissel, N.; Dombret, H.; Toubert, A.; Dulphy, N. Acute myeloid leukemia impairs natural killer cells through the formation of a deficient cytotoxic immunological synapse. Eur. J. Immunol 2014, 44, 3068–3080. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, A.G.; Johnson, A.J.; Lee, A.M.; Gorgun, G.; Le Dieu, R.; Blum, W.; Byrd, J.C.; Gribben, J.G. Chronic lymphocytic leukemia T cells show impaired immunological synapse formation that can be reversed with an immunomodulating drug. J. Clin. Investig. 2008, 118, 2427–2437. [Google Scholar] [CrossRef] [PubMed]
- Giuliani, M.; Janji, B.; Berchem, G. Activation of NK cells and disruption of PD-L1/PD-1 axis: Two different ways for lenalidomide to block myeloma progression. Oncotarget 2017, 8, 24031–24044. [Google Scholar] [CrossRef]
- Konjevic, G.; Vuletic, A.; Mirjacic Martinovic, K.; Colovic, N.; Colovic, M.; Jurisic, V. Decreased CD161 activating and increased CD158a inhibitory receptor expression on NK cells underlies impaired NK cell cytotoxicity in patients with multiple myeloma. J. Clin. Pathol. 2016, 69, 1009–1016. [Google Scholar] [CrossRef]
- MacFarlane, A.W.t.; Jillab, M.; Smith, M.R.; Alpaugh, R.K.; Cole, M.E.; Litwin, S.; Millenson, M.M.; Al-Saleem, T.; Cohen, A.D.; Campbell, K.S. NK cell dysfunction in chronic lymphocytic leukemia is associated with loss of the mature cells expressing inhibitory killer cell Ig-like receptors. Oncoimmunology 2017, 6, e1330235. [Google Scholar] [CrossRef]
- Sandoval-Borrego, D.; Moreno-Lafont, M.C.; Vazquez-Sanchez, E.A.; Gutierrez-Hoya, A.; Lopez-Santiago, R.; Montiel-Cervantes, L.A.; Ramirez-Saldana, M.; Vela-Ojeda, J. Overexpression of CD158 and NKG2A Inhibitory Receptors and Underexpression of NKG2D and NKp46 Activating Receptors on NK Cells in Acute Myeloid Leukemia. Arch. Med. Res. 2016, 47, 55–64. [Google Scholar] [CrossRef]
- Hadadi, L.; Hafezi, M.; Amirzargar, A.A.; Sharifian, R.A.; Abediankenari, S.; Asgarian-Omran, H. Dysregulated Expression of Tim-3 and NKp30 Receptors on NK Cells of Patients with Chronic Lymphocytic Leukemia. Oncol. Res. Treat. 2019, 42, 202–208. [Google Scholar] [CrossRef]
- Chiossone, L.; Vienne, M.; Kerdiles, Y.M.; Vivier, E. Natural killer cell immunotherapies against cancer: Checkpoint inhibitors and more. Semin. Immunol. 2017, 31, 55–63. [Google Scholar] [CrossRef]
- Dempke, W.C.M.; Fenchel, K.; Uciechowski, P.; Dale, S.P. Second- and third-generation drugs for immuno-oncology treatment-The more the better? Eur J. Cancer 2017, 74, 55–72. [Google Scholar] [CrossRef] [PubMed]
- Marin-Acevedo, J.A.; Dholaria, B.; Soyano, A.E.; Knutson, K.L.; Chumsri, S.; Lou, Y. Next generation of immune checkpoint therapy in cancer: New developments and challenges. J. Hematol. Oncol. 2018, 11, 39. [Google Scholar] [CrossRef] [PubMed]
- Ok, C.Y.; Young, K.H. Checkpoint inhibitors in hematological malignancies. J. Hematol. Oncol. 2017, 10, 103. [Google Scholar] [CrossRef]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Marshall, M.J.E.; Stopforth, R.J.; Cragg, M.S. Therapeutic Antibodies: What Have We Learnt from Targeting CD20 and Where Are We Going? Front. Immunol. 2017, 8, 1245. [Google Scholar] [CrossRef] [PubMed]
- Salles, G.; Barrett, M.; Foa, R.; Maurer, J.; O’Brien, S.; Valente, N.; Wenger, M.; Maloney, D.G. Rituximab in B-Cell Hematologic Malignancies: A Review of 20 Years of Clinical Experience. Adv. Ther. 2017, 34, 2232–2273. [Google Scholar] [CrossRef]
- Wei, G.; Wang, J.; Huang, H.; Zhao, Y. Novel immunotherapies for adult patients with B-lineage acute lymphoblastic leukemia. J. Hematol. Oncol. 2017, 10, 150. [Google Scholar] [CrossRef]
- Dahlberg, C.I.; Sarhan, D.; Chrobok, M.; Duru, A.D.; Alici, E. Natural Killer Cell-Based Therapies Targeting Cancer: Possible Strategies to Gain and Sustain Anti-Tumor Activity. Front. Immunol. 2015, 6, 605. [Google Scholar] [CrossRef]
- Kohrt, H.E.; Thielens, A.; Marabelle, A.; Sagiv-Barfi, I.; Sola, C.; Chanuc, F.; Fuseri, N.; Bonnafous, C.; Czerwinski, D.; Rajapaksa, A.; et al. Anti-KIR antibody enhancement of anti-lymphoma activity of natural killer cells as monotherapy and in combination with anti-CD20 antibodies. Blood 2014, 123, 678–686. [Google Scholar] [CrossRef]
- Le Garff-Tavernier, M.; Decocq, J.; de Romeuf, C.; Parizot, C.; Dutertre, C.A.; Chapiro, E.; Davi, F.; Debre, P.; Prost, J.F.; Teillaud, J.L.; et al. Analysis of CD16+CD56dim NK cells from CLL patients: Evidence supporting a therapeutic strategy with optimized anti-CD20 monoclonal antibodies. Leukemia 2011, 25, 101–109. [Google Scholar] [CrossRef]
- Le Garff-Tavernier, M.; Herbi, L.; de Romeuf, C.; Azar, N.; Roos-Weil, D.; Bonnemye, P.; Urbain, R.; Leblond, V.; Merle-Beral, H.; Vieillard, V. The optimized anti-CD20 monoclonal antibody ublituximab bypasses natural killer phenotypic features in Waldenstrom macroglobulinemia. Haematologica 2015, 100, e147–e151. [Google Scholar] [CrossRef][Green Version]
- Le Garff-Tavernier, M.; Herbi, L.; de Romeuf, C.; Prost, J.-F.; Debré, P.; Urbain, R.M.; Leblond, V.R.; Vieillard, V.; Merle-Béral, H.L.N. Ublituximab, an Optimized Anti-CD20 Monoclonal Antibody, Demonstrates Greater NK-Mediated ADCC Than Rituximab in Waldenstrom’s Macroglobulinemia Patients Supporting a Therapeutic Strategy with Ublituximab. Blood 2012, 120, 1654. [Google Scholar] [CrossRef]
- Werlenius, O.; Aurelius, J.; Hallner, A.; Akhiani, A.A.; Simpanen, M.; Martner, A.; Andersson, P.O.; Hellstrand, K.; Thoren, F.B. Reactive oxygen species induced by therapeutic CD20 antibodies inhibit natural killer cell-mediated antibody-dependent cellular cytotoxicity against primary CLL cells. Oncotarget 2016, 7, 32046–32053. [Google Scholar] [CrossRef]
- Chen, W.C.; Kanate, A.S.; Craig, M.; Petros, W.P.; Hazlehurst, L.A. Emerging combination therapies for the management of multiple myeloma: The role of elotuzumab. Cancer Manag Res. 2017, 9, 307–314. [Google Scholar] [CrossRef]
- Collins, S.M.; Bakan, C.E.; Swartzel, G.D.; Hofmeister, C.C.; Efebera, Y.A.; Kwon, H.; Starling, G.C.; Ciarlariello, D.; Bhaskar, S.; Briercheck, E.L.; et al. Elotuzumab directly enhances NK cell cytotoxicity against myeloma via CS1 ligation: Evidence for augmented NK cell function complementing ADCC. Cancer Immunol. Immunother. 2013, 62, 1841–1849. [Google Scholar] [CrossRef]
- Hsi, E.D.; Steinle, R.; Balasa, B.; Szmania, S.; Draksharapu, A.; Shum, B.P.; Huseni, M.; Powers, D.; Nanisetti, A.; Zhang, Y.; et al. CS1, a potential new therapeutic antibody target for the treatment of multiple myeloma. Clin. Cancer Res. 2008, 14, 2775–2784. [Google Scholar] [CrossRef]
- Liu, Y.C.; Szmania, S.; van Rhee, F. Profile of elotuzumab and its potential in the treatment of multiple myeloma. Blood Lymphat Cancer 2014, 2014, 15–27. [Google Scholar]
- Pazina, T.; James, A.M.; MacFarlane, A.W.t.; Bezman, N.A.; Henning, K.A.; Bee, C.; Graziano, R.F.; Robbins, M.D.; Cohen, A.D.; Campbell, K.S. The anti-SLAMF7 antibody elotuzumab mediates NK cell activation through both CD16-dependent and -independent mechanisms. Oncoimmunology 2017, 6, e1339853. [Google Scholar] [CrossRef]
- Tai, Y.T.; Dillon, M.; Song, W.; Leiba, M.; Li, X.F.; Burger, P.; Lee, A.I.; Podar, K.; Hideshima, T.; Rice, A.G.; et al. Anti-CS1 humanized monoclonal antibody HuLuc63 inhibits myeloma cell adhesion and induces antibody-dependent cellular cytotoxicity in the bone marrow milieu. Blood 2008, 112, 1329–1337. [Google Scholar] [CrossRef] [PubMed]
- Afifi, S.; Michael, A.; Lesokhin, A. Immunotherapy: A New Approach to Treating Multiple Myeloma with Daratumumab and Elotuzumab. Ann. Pharm. 2016, 50, 555–568. [Google Scholar] [CrossRef] [PubMed]
- Lonial, S.; Dimopoulos, M.; Palumbo, A.; White, D.; Grosicki, S.; Spicka, I.; Walter-Croneck, A.; Moreau, P.; Mateos, M.V.; Magen, H.; et al. Elotuzumab Therapy for Relapsed or Refractory Multiple Myeloma. N. Engl. J. Med. 2015, 373, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Sherbenou, D.W.; Mark, T.M.; Forsberg, P. Monoclonal Antibodies in Multiple Myeloma: A New Wave of the Future. Clin. Lymphomamyeloma Leuk. 2017, 17, 545–554. [Google Scholar] [CrossRef]
- Liu, L.; Zhao, N.; Xu, W.; Sheng, Z.; Wang, L. Pooled analysis of the reports of carfilzomib, panobinostat, and elotuzumab combinations in patients with refractory/relapsed multiple myeloma. J. Hematol. Oncol. 2016, 9, 54. [Google Scholar] [CrossRef]
- Jakubowiak, A.; Offidani, M.; Pegourie, B.; De La Rubia, J.; Garderet, L.; Laribi, K.; Bosi, A.; Marasca, R.; Laubach, J.; Mohrbacher, A.; et al. Randomized phase 2 study: Elotuzumab plus bortezomib/dexamethasone vs bortezomib/dexamethasone for relapsed/refractory MM. Blood 2016, 127, 2833–2840. [Google Scholar] [CrossRef]
- van Rhee, F.; Szmania, S.M.; Dillon, M.; van Abbema, A.M.; Li, X.; Stone, M.K.; Garg, T.K.; Shi, J.; Moreno-Bost, A.M.; Yun, R.; et al. Combinatorial efficacy of anti-CS1 monoclonal antibody elotuzumab (HuLuc63) and bortezomib against multiple myeloma. Mol. Cancer 2009, 8, 2616–2624. [Google Scholar] [CrossRef]
- Gormley, N.J.; Ko, C.W.; Deisseroth, A.; Nie, L.; Kaminskas, E.; Kormanik, N.; Goldberg, K.B.; Farrell, A.T.; Pazdur, R. FDA Drug Approval: Elotuzumab in Combination with Lenalidomide and Dexamethasone for the Treatment of Relapsed or Refractory Multiple Myeloma. Clin. Cancer Res. 2017, 23, 6759–6763. [Google Scholar] [CrossRef]
- Mateos, M.V.; Granell, M.; Oriol, A.; Martinez-Lopez, J.; Blade, J.; Hernandez, M.T.; Martin, J.; Gironella, M.; Lynch, M.; Bleickardt, E.; et al. Elotuzumab in combination with thalidomide and low-dose dexamethasone: A phase 2 single-arm safety study in patients with relapsed/refractory multiple myeloma. Br. J. Haematol. 2016, 175, 448–456. [Google Scholar] [CrossRef]
- Richardson, P.G.; Jagannath, S.; Moreau, P.; Jakubowiak, A.J.; Raab, M.S.; Facon, T.; Vij, R.; White, D.; Reece, D.E.; Benboubker, L.; et al. Elotuzumab in combination with lenalidomide and dexamethasone in patients with relapsed multiple myeloma: Final phase 2 results from the randomised, open-label, phase 1b-2 dose-escalation study. Lancet Haematol. 2015, 2, e516–e527. [Google Scholar] [CrossRef]
- Fang, F.; Xiao, W.; Tian, Z. NK cell-based immunotherapy for cancer. Semin Immunol 2017, 31, 37–54. [Google Scholar] [CrossRef]
- Makkouk, A.; Chester, C.; Kohrt, H.E. Rationale for anti-CD137 cancer immunotherapy. Eur. J. Cancer 2016, 54, 112–119. [Google Scholar] [CrossRef]
- Weigelin, B.; Bolanos, E.; Teijeira, A.; Martinez-Forero, I.; Labiano, S.; Azpilikueta, A.; Morales-Kastresana, A.; Quetglas, J.I.; Wagena, E.; Sanchez-Paulete, A.R.; et al. Focusing and sustaining the antitumor CTL effector killer response by agonist anti-CD137 mAb. Proc. Natl. Acad. Sci. USA 2015, 112, 7551–7556. [Google Scholar] [CrossRef] [PubMed]
- Chester, C.; Sanmamed, M.F.; Wang, J.; Melero, I. Immunotherapy targeting 4-1BB: Mechanistic rationale, clinical results, and future strategies. Blood 2018, 131, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Mahoney, K.M.; Rennert, P.D.; Freeman, G.J. Combination cancer immunotherapy and new immunomodulatory targets. Nat. Rev. Drug Discov. 2015, 14, 561–584. [Google Scholar] [CrossRef] [PubMed]
- Muntasell, A.; Ochoa, M.C.; Cordeiro, L.; Berraondo, P.; Lopez-Diaz de Cerio, A.; Cabo, M.; Lopez-Botet, M.; Melero, I. Targeting NK-cell checkpoints for cancer immunotherapy. Curr. Opin. Immunol. 2017, 45, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Yonezawa, A.; Dutt, S.; Chester, C.; Kim, J.; Kohrt, H.E. Boosting Cancer Immunotherapy with Anti-CD137 Antibody Therapy. Clin. Cancer Res. 2015, 21, 3113–3120. [Google Scholar] [CrossRef]
- Allard, B.; Longhi, M.S.; Robson, S.C.; Stagg, J. The ectonucleotidases CD39 and CD73: Novel checkpoint inhibitor targets. Immunol. Rev. 2017, 276, 121–144. [Google Scholar] [CrossRef]
- Whiteside, T.L. Targeting adenosine in cancer immunotherapy: A review of recent progress. Expert Rev. Anticancer 2017, 17, 527–535. [Google Scholar] [CrossRef]
- Antonioli, L.; Blandizzi, C.; Malavasi, F.; Ferrari, D.; Hasko, G. Anti-CD73 immunotherapy: A viable way to reprogram the tumor microenvironment. Oncoimmunology 2016, 5, e1216292. [Google Scholar] [CrossRef]
- Leone, R.D.; Lo, Y.C.; Powell, J.D. A2aR antagonists: Next generation checkpoint blockade for cancer immunotherapy. Comput. Struct. Biotechnol. J. 2015, 13, 265–272. [Google Scholar] [CrossRef]
- Beavis, P.A.; Divisekera, U.; Paget, C.; Chow, M.T.; John, L.B.; Devaud, C.; Dwyer, K.; Stagg, J.; Smyth, M.J.; Darcy, P.K. Blockade of A2A receptors potently suppresses the metastasis of CD73+ tumors. Proc. Natl. Acad. Sci. USA 2013, 110, 14711–14716. [Google Scholar] [CrossRef]
- Chambers, A.M.; Matosevic, S. Immunemetabolic Dysfunction of Natural Killer Cells Mediated by the Hypoxia-CD73 Axis in Solid Tumors. Front. Mol. Biosci. 2019, 6, 60. [Google Scholar] [CrossRef]
- Vigano, S.; Alatzoglou, D.; Irving, M.; Menetrier-Caux, C.; Caux, C.; Romero, P.; Coukos, G. Targeting Adenosine in Cancer Immunotherapy to Enhance T-Cell Function. Front. Immunol. 2019, 10, 925. [Google Scholar] [CrossRef]
- Neo, S.Y.; Yang, Y.; Record, J.; Ma, R.; Chen, X.; Chen, Z.; Tobin, N.P.; Blake, E.; Seitz, C.; Thomas, R.; et al. CD73 immune checkpoint defines regulatory NK cells within the tumor microenvironment. J. Clin. Investig. 2020, 130, 1185–1198. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, T.; Song, Z.; Li, L.; Zhang, X.; Liu, J.; Liu, X.; Qiu, L.; Qian, Z.; Zhou, S.; et al. Tumor CD73/A2aR adenosine immunosuppressive axis and tumor-infiltrating lymphocytes in diffuse large B-cell lymphoma: Correlations with clinicopathological characteristics and clinical outcome. Int. J. Cancer 2019, 145, 1414–1422. [Google Scholar] [CrossRef]
- Cai, Y.; Feng, L.; Wang, X. Targeting the tumor promoting effects of adenosine in chronic lymphocytic leukemia. Crit. Rev. Oncol. Hematol. 2018, 126, 24–31. [Google Scholar] [CrossRef]
- Kicova, M.; Michalova, Z.; Coma, M.; Gabzdilova, J.; Dedinska, K.; Guman, T.; Bernatova, S.; Hajikova, M.; Giertlova, M.; Veselinyova, D.; et al. The expression of CD73 on pathological B-cells is associated with shorter overall survival of patients with CLL. Neoplasma 2020. [Google Scholar] [CrossRef]
- Vaisitti, T.; Arruga, F.; Guerra, G.; Deaglio, S. Ectonucleotidases in Blood Malignancies: A Tale of Surface Markers and Therapeutic Targets. Front. Immunol. 2019, 10, 2301. [Google Scholar] [CrossRef]
- Saldanha-Araujo, F.; Ferreira, F.I.; Palma, P.V.; Araujo, A.G.; Queiroz, R.H.; Covas, D.T.; Zago, M.A.; Panepucci, R.A. Mesenchymal stromal cells up-regulate CD39 and increase adenosine production to suppress activated T-lymphocytes. Stem Cell Res. 2011, 7, 66–74. [Google Scholar] [CrossRef]
- Chatterjee, D.; Tufa, D.M.; Baehre, H.; Hass, R.; Schmidt, R.E.; Jacobs, R. Natural killer cells acquire CD73 expression upon exposure to mesenchymal stem cells. Blood 2014, 123, 594–595. [Google Scholar] [CrossRef]
- Vijayan, D.; Young, A.; Teng, M.W.L.; Smyth, M.J. Targeting immunosuppressive adenosine in cancer. Nat. Rev. Cancer 2017, 17, 709–724. [Google Scholar] [CrossRef]
- Young, A.; Ngiow, S.F.; Gao, Y.; Patch, A.M.; Barkauskas, D.S.; Messaoudene, M.; Lin, G.; Coudert, J.D.; Stannard, K.A.; Zitvogel, L.; et al. A2AR Adenosine Signaling Suppresses Natural Killer Cell Maturation in the Tumor Microenvironment. Cancer Res. 2018, 78, 1003–1016. [Google Scholar] [CrossRef]
- Atanackovic, D.; Steinbach, M.; Radhakrishnan, S.V.; Luetkens, T. Immunotherapies targeting CD38 in Multiple Myeloma. Oncoimmunology 2016, 5, e1217374. [Google Scholar] [CrossRef]
- Costa, F.; Dalla Palma, B.; Giuliani, N. CD38 Expression by Myeloma Cells and Its Role in the Context of Bone Marrow Microenvironment: Modulation by Therapeutic Agents. Cells 2019, 8, 1632. [Google Scholar] [CrossRef]
- Nijhof, I.S.; Lammerts van Bueren, J.J.; van Kessel, B.; Andre, P.; Morel, Y.; Lokhorst, H.M.; van de Donk, N.W.; Parren, P.W.; Mutis, T. Daratumumab-mediated lysis of primary multiple myeloma cells is enhanced in combination with the human anti-KIR antibody IPH2102 and lenalidomide. Haematologica 2015, 100, 263–268. [Google Scholar] [CrossRef]
- Sanchez, L.; Wang, Y.; Siegel, D.S.; Wang, M.L. Daratumumab: A first-in-class CD38 monoclonal antibody for the treatment of multiple myeloma. J. Hematol. Oncol. 2016, 9, 51. [Google Scholar] [CrossRef]
- Chari, A.; Suvannasankha, A.; Fay, J.W.; Arnulf, B.; Kaufman, J.L.; Ifthikharuddin, J.J.; Weiss, B.M.; Krishnan, A.; Lentzsch, S.; Comenzo, R.; et al. Daratumumab plus pomalidomide and dexamethasone in relapsed and/or refractory multiple myeloma. Blood 2017, 130, 974–981. [Google Scholar] [CrossRef]
- Chung, C. Role of Immunotherapy in Targeting the Bone Marrow Microenvironment in Multiple Myeloma: An Evolving Therapeutic Strategy. Pharmacotherapy 2017, 37, 129–143. [Google Scholar] [CrossRef]
- Petrucci, M.T.; Vozella, F. The Anti-CD38 Antibody Therapy in Multiple Myeloma. Cells 2019, 8, 1629. [Google Scholar] [CrossRef]
- van de Donk, N.; Usmani, S.Z. CD38 Antibodies in Multiple Myeloma: Mechanisms of Action and Modes of Resistance. Front. Immunol. 2018, 9, 2134. [Google Scholar] [CrossRef] [PubMed]
- Soekojo, C.Y.; Ooi, M.; de Mel, S.; Chng, W.J. Immunotherapy in Multiple Myeloma. Cells 2020, 9, 601. [Google Scholar] [CrossRef] [PubMed]
- Martin, T.; Baz, R.; Benson, D.M.; Lendvai, N.; Wolf, J.; Munster, P.; Lesokhin, A.M.; Wack, C.; Charpentier, E.; Campana, F.; et al. A phase 1b study of isatuximab plus lenalidomide and dexamethasone for relapsed/refractory multiple myeloma. Blood 2017, 129, 3294–3303. [Google Scholar] [CrossRef] [PubMed]
- Mikhael, J.; Richardson, P.; Usmani, S.Z.; Raje, N.; Bensinger, W.; Karanes, C.; Campana, F.; Kanagavel, D.; Dubin, F.; Liu, Q.; et al. A phase 1b study of isatuximab plus pomalidomide/dexamethasone in relapsed/refractory multiple myeloma. Blood 2019, 134, 123–133. [Google Scholar] [CrossRef]
- Moreno, L.; Perez, C.; Zabaleta, A.; Manrique, I.; Alignani, D.; Ajona, D.; Blanco, L.; Lasa, M.; Maiso, P.; Rodriguez, I.; et al. The Mechanism of Action of the Anti-CD38 Monoclonal Antibody Isatuximab in Multiple Myeloma. Clin. Cancer Res. 2019, 25, 3176–3187. [Google Scholar] [CrossRef]
- Assi, R.; Kantarjian, H.; Ravandi, F.; Daver, N. Immune therapies in acute myeloid leukemia: A focus on monoclonal antibodies and immune checkpoint inhibitors. Curr. Opin. Hematol. 2018, 25, 136–145. [Google Scholar] [CrossRef]
- Daver, N.; Kantarjian, H.; Ravandi, F.; Estey, E.; Wang, X.; Garcia-Manero, G.; Jabbour, E.; Konopleva, M.; O’Brien, S.; Verstovsek, S.; et al. A phase II study of decitabine and gemtuzumab ozogamicin in newly diagnosed and relapsed acute myeloid leukemia and high-risk myelodysplastic syndrome. Leukemia 2016, 30, 268–273. [Google Scholar] [CrossRef]
- Vasu, S.; He, S.; Cheney, C.; Gopalakrishnan, B.; Mani, R.; Lozanski, G.; Mo, X.; Groh, V.; Whitman, S.P.; Konopitzky, R.; et al. Decitabine enhances anti-CD33 monoclonal antibody BI 836858-mediated natural killer ADCC against AML blasts. Blood 2016, 127, 2879–2889. [Google Scholar] [CrossRef]
- Chao, M.P.; Takimoto, C.H.; Feng, D.D.; McKenna, K.; Gip, P.; Liu, J.; Volkmer, J.P.; Weissman, I.L.; Majeti, R. Therapeutic Targeting of the Macrophage Immune Checkpoint CD47 in Myeloid Malignancies. Front. Oncol. 2019, 9, 1380. [Google Scholar] [CrossRef]
- Matlung, H.L.; Szilagyi, K.; Barclay, N.A.; van den Berg, T.K. The CD47-SIRPalpha signaling axis as an innate immune checkpoint in cancer. Immunol. Rev. 2017, 276, 145–164. [Google Scholar] [CrossRef]
- Weiskopf, K. Cancer immunotherapy targeting the CD47/SIRPalpha axis. Eur J. Cancer 2017, 76, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.M., Jr.; Bakan, C.E.; Zhang, S.; Collins, S.M.; Liang, J.; Srivastava, S.; Hofmeister, C.C.; Efebera, Y.; Andre, P.; Romagne, F.; et al. IPH2101, a novel anti-inhibitory KIR antibody, and lenalidomide combine to enhance the natural killer cell versus multiple myeloma effect. Blood 2011, 118, 6387–6391. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.M., Jr.; Cohen, A.D.; Jagannath, S.; Munshi, N.C.; Spitzer, G.; Hofmeister, C.C.; Efebera, Y.A.; Andre, P.; Zerbib, R.; Caligiuri, M.A. A Phase I Trial of the Anti-KIR Antibody IPH2101 and Lenalidomide in Patients with Relapsed/Refractory Multiple Myeloma. Clin. Cancer Res. 2015, 21, 4055–4061. [Google Scholar] [CrossRef]
- Benson, D.M., Jr.; Hofmeister, C.C.; Padmanabhan, S.; Suvannasankha, A.; Jagannath, S.; Abonour, R.; Bakan, C.; Andre, P.; Efebera, Y.; Tiollier, J.; et al. A phase 1 trial of the anti-KIR antibody IPH2101 in patients with relapsed/refractory multiple myeloma. Blood 2012, 120, 4324–4333. [Google Scholar] [CrossRef]
- Carlsten, M.; Korde, N.; Kotecha, R.; Reger, R.; Bor, S.; Kazandjian, D.; Landgren, O.; Childs, R.W. Checkpoint Inhibition of KIR2D with the Monoclonal Antibody IPH2101 Induces Contraction and Hyporesponsiveness of NK Cells in Patients with Myeloma. Clin. Cancer Res. 2016, 22, 5211–5222. [Google Scholar] [CrossRef]
- Romagne, F.; Andre, P.; Spee, P.; Zahn, S.; Anfossi, N.; Gauthier, L.; Capanni, M.; Ruggeri, L.; Benson, D.M., Jr.; Blaser, B.W.; et al. Preclinical characterization of 1-7F9, a novel human anti-KIR receptor therapeutic antibody that augments natural killer-mediated killing of tumor cells. Blood 2009, 114, 2667–2677. [Google Scholar] [CrossRef]
- Vey, N.; Bourhis, J.H.; Boissel, N.; Bordessoule, D.; Prebet, T.; Charbonnier, A.; Etienne, A.; Andre, P.; Romagne, F.; Benson, D.; et al. A phase 1 trial of the anti-inhibitory KIR mAb IPH2101 for AML in complete remission. Blood 2012, 120, 4317–4323. [Google Scholar] [CrossRef]
- Ansell, S.M.; Lesokhin, A.M.; Borrello, I.; Halwani, A.; Scott, E.C.; Gutierrez, M.; Schuster, S.J.; Millenson, M.M.; Cattry, D.; Freeman, G.J.; et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N. Engl. J. Med. 2015, 372, 311–319. [Google Scholar] [CrossRef]
- Pianko, M.J.; Liu, Y.; Bagchi, S.; Lesokhin, A.M. Immune checkpoint blockade for hematologic malignancies: A review. Stem Cell Investig. 2017, 4, 32. [Google Scholar] [CrossRef]
- Sarkar, S.; van Gelder, M.; Noort, W.; Xu, Y.; Rouschop, K.M.; Groen, R.; Schouten, H.C.; Tilanus, M.G.; Germeraad, W.T.; Martens, A.C.; et al. Optimal selection of natural killer cells to kill myeloma: The role of HLA-E and NKG2A. Cancer Immunol. Immunother. 2015, 64, 951–963. [Google Scholar] [CrossRef]
- Mahaweni, N.M.; Ehlers, F.A.I.; Bos, G.M.J.; Wieten, L. Tuning Natural Killer Cell Anti-multiple Myeloma Reactivity by Targeting Inhibitory Signaling via KIR and NKG2A. Front. Immunol. 2018, 9, 2848. [Google Scholar] [CrossRef]
- McWilliams, E.M.; Mele, J.M.; Cheney, C.; Timmerman, E.A.; Fiazuddin, F.; Strattan, E.J.; Mo, X.; Byrd, J.C.; Muthusamy, N.; Awan, F.T. Therapeutic CD94/NKG2A blockade improves natural killer cell dysfunction in chronic lymphocytic leukemia. Oncoimmunology 2016, 5, e1226720. [Google Scholar] [CrossRef] [PubMed]
- van Hall, T.; Andre, P.; Horowitz, A.; Ruan, D.F.; Borst, L.; Zerbib, R.; Narni-Mancinelli, E.; van der Burg, S.H.; Vivier, E. Monalizumab: Inhibiting the novel immune checkpoint NKG2A. J. Immunother. Cancer 2019, 7, 263. [Google Scholar] [CrossRef] [PubMed]
- Godal, R.; Bachanova, V.; Gleason, M.; McCullar, V.; Yun, G.H.; Cooley, S.; Verneris, M.R.; McGlave, P.B.; Miller, J.S. Natural killer cell killing of acute myelogenous leukemia and acute lymphoblastic leukemia blasts by killer cell immunoglobulin-like receptor-negative natural killer cells after NKG2A and LIR-1 blockade. Biol Blood Marrow Transpl. 2010, 16, 612–621. [Google Scholar] [CrossRef]
- Rouas-Freiss, N.; Moreau, P.; LeMaoult, J.; Carosella, E.D. The dual role of HLA-G in cancer. J. Immunol. Res. 2014, 2014, 359748. [Google Scholar] [CrossRef]
- Arruga, F.; Gyau, B.B.; Iannello, A.; Vitale, N.; Vaisitti, T.; Deaglio, S. Immune Response Dysfunction in Chronic Lymphocytic Leukemia: Dissecting Molecular Mechanisms and Microenvironmental Conditions. Int. J. Mol. Sci. 2020, 21, 1825. [Google Scholar] [CrossRef]
- Villa-Alvarez, M.; Sordo-Bahamonde, C.; Lorenzo-Herrero, S.; Gonzalez-Rodriguez, A.P.; Payer, A.R.; Gonzalez-Garcia, E.; Villa-Alvarez, M.C.; Lopez-Soto, A.; Gonzalez, S. Ig-Like Transcript 2 (ILT2) Blockade and Lenalidomide Restore NK Cell Function in Chronic Lymphocytic Leukemia. Front. Immunol. 2018, 9, 2917. [Google Scholar] [CrossRef]
- Long, L.; Zhang, X.; Chen, F.; Pan, Q.; Phiphatwatchara, P.; Zeng, Y.; Chen, H. The promising immune checkpoint LAG-3: From tumor microenvironment to cancer immunotherapy. Genes Cancer 2018, 9, 176–189. [Google Scholar]
- Anderson, A.C.; Joller, N.; Kuchroo, V.K. Lag-3, Tim-3, and TIGIT: Co-inhibitory Receptors with Specialized Functions in Immune Regulation. Immunity 2016, 44, 989–1004. [Google Scholar] [CrossRef]
- Grosso, J.F.; Goldberg, M.V.; Getnet, D.; Bruno, T.C.; Yen, H.R.; Pyle, K.J.; Hipkiss, E.; Vignali, D.A.; Pardoll, D.M.; Drake, C.G. Functionally distinct LAG-3 and PD-1 subsets on activated and chronically stimulated CD8 T cells. J. Immunol. 2009, 182, 6659–6669. [Google Scholar] [CrossRef]
- Woo, S.R.; Turnis, M.E.; Goldberg, M.V.; Bankoti, J.; Selby, M.; Nirschl, C.J.; Bettini, M.L.; Gravano, D.M.; Vogel, P.; Liu, C.L.; et al. Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T-cell function to promote tumoral immune escape. Cancer Res. 2012, 72, 917–927. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.Z.; Kim, H.J.; Villasboas, J.C.; Chen, Y.P.; Price-Troska, T.; Jalali, S.; Wilson, M.; Novak, A.J.; Ansell, S.M. Expression of LAG-3 defines exhaustion of intratumoral PD-1(+) T cells and correlates with poor outcome in follicular lymphoma. Oncotarget. 2017, 8, 61425–61439. [Google Scholar] [CrossRef] [PubMed]
- Keane, C.; Law, S.C.; Gould, C.; Birch, S.; Sabdia, M.B.; Merida de Long, L.; Thillaiyampalam, G.; Abro, E.; Tobin, J.W.; Tan, X.; et al. LAG3: A novel immune checkpoint expressed by multiple lymphocyte subsets in diffuse large B-cell lymphoma. Blood Adv. 2020, 4, 1367–1377. [Google Scholar] [CrossRef] [PubMed]
- Gleason, M.K.; Lenvik, T.R.; McCullar, V.; Felices, M.; O’Brien, M.S.; Cooley, S.A.; Verneris, M.R.; Cichocki, F.; Holman, C.J.; Panoskaltsis-Mortari, A.; et al. Tim-3 is an inducible human natural killer cell receptor that enhances interferon gamma production in response to galectin-9. Blood 2012, 119, 3064–3072. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.; Rangachari, M.; Kuchroo, V.K. Tim-3: A co-receptor with diverse roles in T cell exhaustion and tolerance. Semin. Immunol. 2019, 42, 101302. [Google Scholar] [CrossRef]
- Folgiero, V.; Cifaldi, L.; Li Pira, G.; Goffredo, B.M.; Vinti, L.; Locatelli, F. TIM-3/Gal-9 interaction induces IFNgamma-dependent IDO1 expression in acute myeloid leukemia blast cells. J. Hematol. Oncol. 2015, 8, 36. [Google Scholar] [CrossRef]
- Gallois, A.; Silva, I.; Osman, I.; Bhardwaj, N. Reversal of natural killer cell exhaustion by TIM-3 blockade. Oncoimmunology 2014, 3, e946365. [Google Scholar] [CrossRef]
- Ndhlovu, L.C.; Lopez-Verges, S.; Barbour, J.D.; Jones, R.B.; Jha, A.R.; Long, B.R.; Schoeffler, E.C.; Fujita, T.; Nixon, D.F.; Lanier, L.L. Tim-3 marks human natural killer cell maturation and suppresses cell-mediated cytotoxicity. Blood 2012, 119, 3734–3743. [Google Scholar] [CrossRef]
- Przespolewski, A.; Szeles, A.; Wang, E.S. Advances in immunotherapy for acute myeloid leukemia. Future Oncol. 2018, 14, 963–978. [Google Scholar] [CrossRef]
- Asayama, T.; Tamura, H.; Ishibashi, M.; Kuribayashi-Hamada, Y.; Onodera-Kondo, A.; Okuyama, N.; Yamada, A.; Shimizu, M.; Moriya, K.; Takahashi, H.; et al. Functional expression of Tim-3 on blasts and clinical impact of its ligand galectin-9 in myelodysplastic syndromes. Oncotarget. 2017, 8, 88904–88917. [Google Scholar] [CrossRef]
- Dong, H.; Strome, S.E.; Salomao, D.R.; Tamura, H.; Hirano, F.; Flies, D.B.; Roche, P.C.; Lu, J.; Zhu, G.; Tamada, K.; et al. Tumor-associated B7-H1 promotes T-cell apoptosis: A potential mechanism of immune evasion. Nat. Med. 2002, 8, 793–800. [Google Scholar] [CrossRef] [PubMed]
- Francisco, L.M.; Sage, P.T.; Sharpe, A.H. The PD-1 pathway in tolerance and autoimmunity. Immunol. Rev. 2010, 236, 219–242. [Google Scholar] [CrossRef] [PubMed]
- Daver, N.; Ravandi, F. Enhancing cytotoxicity of immunotoxins in AML. Blood 2016, 127, 2787–2788. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sehgal, A.; Whiteside, T.L.; Boyiadzis, M. Programmed death-1 checkpoint blockade in acute myeloid leukemia. Expert Opin. Biol. 2015, 15, 1191–1203. [Google Scholar] [CrossRef]
- Atanackovic, D.; Luetkens, T.; Radhakrishnan, S.; Kroger, N. Coinhibitory Molecule PD-1 as a Therapeutic Target in the Microenvironment of Multiple Myeloma. Curr. Cancer Drug Targets 2017, 17, 839–845. [Google Scholar] [CrossRef]
- Paiva, B.; Azpilikueta, A.; Puig, N.; Ocio, E.M.; Sharma, R.; Oyajobi, B.O.; Labiano, S.; San-Segundo, L.; Rodriguez, A.; Aires-Mejia, I.; et al. PD-L1/PD-1 presence in the tumor microenvironment and activity of PD-1 blockade in multiple myeloma. Leukemia 2015, 29, 2110–2113. [Google Scholar] [CrossRef]
- Tremblay-LeMay, R.; Rastgoo, N.; Chang, H. Modulating PD-L1 expression in multiple myeloma: An alternative strategy to target the PD-1/PD-L1 pathway. J. Hematol. Oncol. 2018, 11, 46. [Google Scholar] [CrossRef]
- Andorsky, D.J.; Yamada, R.E.; Said, J.; Pinkus, G.S.; Betting, D.J.; Timmerman, J.M. Programmed death ligand 1 is expressed by non-hodgkin lymphomas and inhibits the activity of tumor-associated T cells. Clin. Cancer Res. 2011, 17, 4232–4244. [Google Scholar] [CrossRef]
- Goodman, A.; Patel, S.P.; Kurzrock, R. PD-1-PD-L1 immune-checkpoint blockade in B-cell lymphomas. Nat. Rev. Clin. Oncol. 2017, 14, 203–220. [Google Scholar] [CrossRef]
- Batlevi, C.L.; Matsuki, E.; Brentjens, R.J.; Younes, A. Novel immunotherapies in lymphoid malignancies. Nat. Rev. Clin. Oncol. 2016, 13, 25–40. [Google Scholar] [CrossRef]
- de Charette, M.; Houot, R. Hide or defend, the two strategies of lymphoma immune evasion: Potential implications for immunotherapy. Haematologica 2018, 103, 1256–1268. [Google Scholar] [CrossRef] [PubMed]
- Grzywnowicz, M.; Zaleska, J.; Mertens, D.; Tomczak, W.; Wlasiuk, P.; Kosior, K.; Piechnik, A.; Bojarska-Junak, A.; Dmoszynska, A.; Giannopoulos, K. Programmed death-1 and its ligand are novel immunotolerant molecules expressed on leukemic B cells in chronic lymphocytic leukemia. PLoS ONE 2012, 7, e35178. [Google Scholar] [CrossRef] [PubMed]
- Carreras, J.; Lopez-Guillermo, A.; Roncador, G.; Villamor, N.; Colomo, L.; Martinez, A.; Hamoudi, R.; Howat, W.J.; Montserrat, E.; Campo, E. High numbers of tumor-infiltrating programmed cell death 1-positive regulatory lymphocytes are associated with improved overall survival in follicular lymphoma. J. Clin. Oncol. 2009, 27, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Myklebust, J.H.; Irish, J.M.; Brody, J.; Czerwinski, D.K.; Houot, R.; Kohrt, H.E.; Timmerman, J.; Said, J.; Green, M.R.; Delabie, J.; et al. High PD-1 expression and suppressed cytokine signaling distinguish T cells infiltrating follicular lymphoma tumors from peripheral T cells. Blood 2013, 121, 1367–1376. [Google Scholar] [CrossRef]
- Xie, M.; Huang, X.; Ye, X.; Qian, W. Prognostic and clinicopathological significance of PD-1/PD-L1 expression in the tumor microenvironment and neoplastic cells for lymphoma. Int. Immunopharmacol. 2019, 77, 105999. [Google Scholar] [CrossRef]
- Yang, Z.Z.; Grote, D.M.; Ziesmer, S.C.; Xiu, B.; Novak, A.J.; Ansell, S.M. PD-1 expression defines two distinct T-cell sub-populations in follicular lymphoma that differentially impact patient survival. Blood Cancer J. 2015, 5, e281. [Google Scholar] [CrossRef]
- Della Chiesa, M.; Pesce, S.; Muccio, L.; Carlomagno, S.; Sivori, S.; Moretta, A.; Marcenaro, E. Features of Memory-Like and PD-1(+) Human NK Cell Subsets. Front. Immunol. 2016, 7, 351. [Google Scholar] [CrossRef]
- Pesce, S.; Greppi, M.; Tabellini, G.; Rampinelli, F.; Parolini, S.; Olive, D.; Moretta, L.; Moretta, A.; Marcenaro, E. Identification of a subset of human natural killer cells expressing high levels of programmed death 1: A phenotypic and functional characterization. J. Allergy Clin. Immunol. 2017, 139, 335–346.e333. [Google Scholar] [CrossRef]
- Danhof, S.; Schreder, M.; Knop, S.; Rasche, L.; Strifler, S.; Loffler, C.; Gogishvili, T.; Einsele, H.; Hudecek, M. Expression of programmed death-1 on lymphocytes in myeloma patients is lowered during lenalidomide maintenance. Haematologica 2018, 103, e126–e129. [Google Scholar] [CrossRef]
- Cornell, R.F.; Kassim, A.A. Evolving paradigms in the treatment of relapsed/refractory multiple myeloma: Increased options and increased complexity. Bone Marrow Transpl. 2016, 51, 479–491. [Google Scholar] [CrossRef]
- Mateos, M.V.; Blacklock, H.; Schjesvold, F.; Oriol, A.; Simpson, D.; George, A.; Goldschmidt, H.; Larocca, A.; Chanan-Khan, A.; Sherbenou, D.; et al. Pembrolizumab plus pomalidomide and dexamethasone for patients with relapsed or refractory multiple myeloma (KEYNOTE-183): A randomised, open-label, phase 3 trial. Lancet. Haematol. 2019, 6, e459–e469. [Google Scholar] [CrossRef]
- San Miguel, J.; Mateos, M.-V.; Shah, J.J.; Ocio, E.M.; Rodriguez-Otero, P.; Reece, D.; Munshi, N.C.; Avigan, D.; Ge, Y.; Balakumaran, A.; et al. Pembrolizumab in Combination with Lenalidomide and Low-Dose Dexamethasone for Relapsed/Refractory Multiple Myeloma (RRMM): Keynote-023. Blood 2015, 126, 505. [Google Scholar] [CrossRef]
- Daver, N.G.; Garcia-Manero, G.; Konopleva, M.Y.; Alfayez, M.; Pemmaraju, N.; Kadia, T.M.; DiNardo, C.D.; Cortes, J.E.; Ravandi, F.; Abbas, H.; et al. Azacitidine (AZA) with Nivolumab (Nivo), and AZA with Nivo + Ipilimumab (Ipi) in Relapsed/Refractory Acute Myeloid Leukemia: A Non-Randomized, Prospective, Phase 2 Study. Blood 2019, 134, 830. [Google Scholar] [CrossRef]
- Garcia-Manero, G.; Sasaki, K.; Montalban-Bravo, G.; Daver, N.G.; Jabbour, E.J.; Alvarado, Y.; DiNardo, C.D.; Ravandi, F.; Borthakur, G.; Bose, P.; et al. A Phase II Study of Nivolumab or Ipilimumab with or without Azacitidine for Patients with Myelodysplastic Syndrome (MDS). Blood 2018, 132, 465. [Google Scholar] [CrossRef]
- Westin, J.R.; Chu, F.; Zhang, M.; Fayad, L.E.; Kwak, L.W.; Fowler, N.; Romaguera, J.; Hagemeister, F.; Fanale, M.; Samaniego, F.; et al. Safety and activity of PD1 blockade by pidilizumab in combination with rituximab in patients with relapsed follicular lymphoma: A single group, open-label, phase 2 trial. Lancet Oncol. 2014, 15, 69–77. [Google Scholar] [CrossRef]
- Ansell, S.; Gutierrez, M.E.; Shipp, M.A.; Gladstone, D.; Moskowitz, A.; Borello, I.; Popa-Mckiver, M.; Farsaci, B.; Zhu, L.; Lesokhin, A.M.; et al. A Phase 1 Study of Nivolumab in Combination with Ipilimumab for Relapsed or Refractory Hematologic Malignancies (CheckMate 039). Blood 2016, 128, 183. [Google Scholar] [CrossRef]
- Ray, A.; Das, D.S.; Song, Y.; Hideshima, T.; Tai, Y.T.; Chauhan, D.; Anderson, K.C. Combination of a novel HDAC6 inhibitor ACY-241 and anti-PD-L1 antibody enhances anti-tumor immunity and cytotoxicity in multiple myeloma. Leukemia 2018, 32, 843–846. [Google Scholar] [CrossRef] [PubMed]
- Tamura, H.; Ishibashi, M.; Yamashita, T.; Tanosaki, S.; Okuyama, N.; Kondo, A.; Hyodo, H.; Shinya, E.; Takahashi, H.; Dong, H.; et al. Marrow stromal cells induce B7-H1 expression on myeloma cells, generating aggressive characteristics in multiple myeloma. Leukemia 2013, 27, 464–472. [Google Scholar] [CrossRef]
- Chen, L.; Han, X. Anti-PD-1/PD-L1 therapy of human cancer: Past, present, and future. J. Clin. Investig. 2015, 125, 3384–3391. [Google Scholar] [CrossRef]
- Georgiou, K.; Chen, L.; Berglund, M.; Ren, W.; de Miranda, N.F.C.C.; Lisboa, S.; Fangazio, M.; Zhu, S.; Hou, Y.; Wu, K.; et al. Genetic basis of PD-L1 overexpression in diffuse large B-cell lymphomas. Blood 2016, 127, 3026–3034. [Google Scholar] [CrossRef]
- Laurent, C.; Charmpi, K.; Gravelle, P.; Tosolini, M.; Franchet, C.; Ysebaert, L.; Brousset, P.; Bidaut, A.; Ycart, B.; Fournie, J.J. Several immune escape patterns in non-Hodgkin’s lymphomas. Oncoimmunology 2015, 4, e1026530. [Google Scholar] [CrossRef] [PubMed]
- Roemer, M.G.; Advani, R.H.; Ligon, A.H.; Natkunam, Y.; Redd, R.A.; Homer, H.; Connelly, C.F.; Sun, H.H.; Daadi, S.E.; Freeman, G.J.; et al. PD-L1 and PD-L2 Genetic Alterations Define Classical Hodgkin Lymphoma and Predict Outcome. J. Clin. Oncol. 2016, 34, 2690–2697. [Google Scholar] [CrossRef] [PubMed]
- Yousef, S.; Marvin, J.; Steinbach, M.; Langemo, A.; Kovacsovics, T.; Binder, M.; Kroger, N.; Luetkens, T.; Atanackovic, D. Immunomodulatory molecule PD-L1 is expressed on malignant plasma cells and myeloma-propagating pre-plasma cells in the bone marrow of multiple myeloma patients. Blood Cancer J. 2015, 5, e285. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.M., Jr.; Bakan, C.E.; Mishra, A.; Hofmeister, C.C.; Efebera, Y.; Becknell, B.; Baiocchi, R.A.; Zhang, J.; Yu, J.; Smith, M.K.; et al. The PD-1/PD-L1 axis modulates the natural killer cell versus multiple myeloma effect: A therapeutic target for CT-011, a novel monoclonal anti-PD-1 antibody. Blood 2010, 116, 2286–2294. [Google Scholar] [CrossRef]
- Guo, Y.; Feng, X.; Jiang, Y.; Shi, X.; Xing, X.; Liu, X.; Li, N.; Fadeel, B.; Zheng, C. PD1 blockade enhances cytotoxicity of in vitro expanded natural killer cells towards myeloma cells. Oncotarget 2016, 7, 48360–48374. [Google Scholar] [CrossRef]
- Hallett, W.H.; Jing, W.; Drobyski, W.R.; Johnson, B.D. Immunosuppressive effects of multiple myeloma are overcome by PD-L1 blockade. Biol. Blood Marrow Transpl. 2011, 17, 1133–1145. [Google Scholar] [CrossRef]
- Rosenblatt, J.; Glotzbecker, B.; Mills, H.; Vasir, B.; Tzachanis, D.; Levine, J.D.; Joyce, R.M.; Wellenstein, K.; Keefe, W.; Schickler, M.; et al. PD-1 blockade by CT-011, anti-PD-1 antibody, enhances ex vivo T-cell responses to autologous dendritic cell/myeloma fusion vaccine. J. Immunother. 2011, 34, 409–418. [Google Scholar] [CrossRef]
- Whitehill, G.D.; Chivers, A.; Danielson, B.; Tessier, J.S.; Hernandez Duran, G.; Bender, G.L.; Sborov, D.W.; Hofmeister, C.C.; Bradner, J.; Hideshima, T.; et al. Immunomodulatory Effects of HDACi in Combination with Checkpoint Blockade and Lenalidomide in the Immunosuppressive Multiple Myeloma Bone Marrow Microenvironment. Blood 2017, 130, 4422. [Google Scholar]
- Ilcus, C.; Bagacean, C.; Tempescul, A.; Popescu, C.; Parvu, A.; Cenariu, M.; Bocsan, C.; Zdrenghea, M. Immune checkpoint blockade: The role of PD-1-PD-L axis in lymphoid malignancies. Onco. Targets 2017, 10, 2349–2363. [Google Scholar] [CrossRef]
- Xu-Monette, Z.Y.; Zhou, J.; Young, K.H. PD-1 expression and clinical PD-1 blockade in B-cell lymphomas. Blood 2018, 131, 68–83. [Google Scholar] [CrossRef]
- Armand, P. Immune checkpoint blockade in hematologic malignancies. Blood 2015, 125, 3393–3400. [Google Scholar] [CrossRef] [PubMed]
- Korkmaz, S.; Erdem, S.; Akay, E.; Tasdemir, E.A.; Karaman, H.; Keklik, M. Do PD-1 and PD-L2 expressions have prognostic impact in hematologic malignancies? Turk. J. Med. Sci 2019, 49, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Panjwani, P.K.; Charu, V.; DeLisser, M.; Molina-Kirsch, H.; Natkunam, Y.; Zhao, S. Programmed death-1 ligands PD-L1 and PD-L2 show distinctive and restricted patterns of expression in lymphoma subtypes. Hum. Pathol. 2018, 71, 91–99. [Google Scholar] [CrossRef]
- Rozali, E.N.; Hato, S.V.; Robinson, B.W.; Lake, R.A.; Lesterhuis, W.J. Programmed death ligand 2 in cancer-induced immune suppression. Clin. Dev. Immunol. 2012, 2012, 656340. [Google Scholar] [CrossRef]
- Tanaka, Y.; Maeshima, A.M.; Nomoto, J.; Makita, S.; Fukuhara, S.; Munakata, W.; Maruyama, D.; Tobinai, K.; Kobayashi, Y. Expression pattern of PD-L1 and PD-L2 in classical Hodgkin lymphoma, primary mediastinal large B-cell lymphoma, and gray zone lymphoma. Eur. J. Haematol 2018, 100, 511–517. [Google Scholar] [CrossRef]
- Johnston, R.J.; Comps-Agrar, L.; Hackney, J.; Yu, X.; Huseni, M.; Yang, Y.; Park, S.; Javinal, V.; Chiu, H.; Irving, B.; et al. The immunoreceptor TIGIT regulates antitumor and antiviral CD8(+) T cell effector function. Cancer Cell 2014, 26, 923–937. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Arooj, S.; Wang, H. NK Cell-Based Immune Checkpoint Inhibition. Front. Immunol. 2020, 11, 167. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.J.; Martinet, L.; Gilfillan, S.; Souza-Fonseca-Guimaraes, F.; Chow, M.T.; Town, L.; Ritchie, D.S.; Colonna, M.; Andrews, D.M.; Smyth, M.J. The receptors CD96 and CD226 oppose each other in the regulation of natural killer cell functions. Nat. Immunol. 2014, 15, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Zheng, Q.; Xin, N.; Wang, W.; Zhao, C. CD155, an onco-immunologic molecule in human tumors. Cancer Sci. 2017, 108, 1934–1938. [Google Scholar] [CrossRef]
- Martinet, L.; Smyth, M.J. Balancing natural killer cell activation through paired receptors. Nat. Rev. Immunol 2015, 15, 243–254. [Google Scholar] [CrossRef]
- Stanietsky, N.; Simic, H.; Arapovic, J.; Toporik, A.; Levy, O.; Novik, A.; Levine, Z.; Beiman, M.; Dassa, L.; Achdout, H.; et al. The interaction of TIGIT with PVR and PVRL2 inhibits human NK cell cytotoxicity. Proc. Natl. Acad. Sci. USA 2009, 106, 17858–17863. [Google Scholar] [CrossRef] [PubMed]
- Kucan Brlic, P.; Lenac Rovis, T.; Cinamon, G.; Tsukerman, P.; Mandelboim, O.; Jonjic, S. Targeting PVR (CD155) and its receptors in anti-tumor therapy. Cell Mol. Immunol. 2019, 16, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Stengel, K.F.; Harden-Bowles, K.; Yu, X.; Rouge, L.; Yin, J.; Comps-Agrar, L.; Wiesmann, C.; Bazan, J.F.; Eaton, D.L.; Grogan, J.L. Structure of TIGIT immunoreceptor bound to poliovirus receptor reveals a cell-cell adhesion and signaling mechanism that requires cis-trans receptor clustering. Proc. Natl. Acad. Sci. USA 2012, 109, 5399–5404. [Google Scholar] [CrossRef] [PubMed]
- Blake, S.J.; Dougall, W.C.; Miles, J.J.; Teng, M.W.; Smyth, M.J. Molecular Pathways: Targeting CD96 and TIGIT for Cancer Immunotherapy. Clin. Cancer Res. 2016, 22, 5183–5188. [Google Scholar] [CrossRef]
- Dougall, W.C.; Kurtulus, S.; Smyth, M.J.; Anderson, A.C. TIGIT and CD96: New checkpoint receptor targets for cancer immunotherapy. Immunol. Rev. 2017, 276, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Chauvin, J.M.; Pagliano, O.; Fourcade, J.; Sun, Z.; Wang, H.; Sander, C.; Kirkwood, J.M.; Chen, T.H.; Maurer, M.; Korman, A.J.; et al. TIGIT and PD-1 impair tumor antigen-specific CD8(+) T cells in melanoma patients. J. Clin. Investig. 2015, 125, 2046–2058. [Google Scholar] [CrossRef]
- Sanchez-Correa, B.; Valhondo, I.; Hassouneh, F.; Lopez-Sejas, N.; Pera, A.; Bergua, J.M.; Arcos, M.J.; Banas, H.; Casas-Aviles, I.; Duran, E.; et al. DNAM-1 and the TIGIT/PVRIG/TACTILE Axis: Novel Immune Checkpoints for Natural Killer Cell-Based Cancer Immunotherapy. Cancers 2019, 11, 877. [Google Scholar] [CrossRef]
- Asimakopoulos, F. TIGIT checkpoint inhibition for myeloma. Blood 2018, 132, 1629–1630. [Google Scholar] [CrossRef]
- Wang, M.; Bu, J.; Zhou, M.; Sido, J.; Lin, Y.; Liu, G.; Lin, Q.; Xu, X.; Leavenworth, J.W.; Shen, E. CD8(+)T cells expressing both PD-1 and TIGIT but not CD226 are dysfunctional in acute myeloid leukemia (AML) patients. Clin. Immunol. 2018, 190, 64–73. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, H.; Chen, L.; Feng, Z.; Gao, L.; Li, Q. TIGIT expression is upregulated in T cells and causes T cell dysfunction independent of PD-1 and Tim-3 in adult B lineage acute lymphoblastic leukemia. Cell Immunol. 2019, 344, 103958. [Google Scholar] [CrossRef]
- Friedrich, M.; Henn, A.; Raum, T.; Bajtus, M.; Matthes, K.; Hendrich, L.; Wahl, J.; Hoffmann, P.; Kischel, R.; Kvesic, M.; et al. Preclinical characterization of AMG 330, a CD3/CD33-bispecific T-cell-engaging antibody with potential for treatment of acute myelogenous leukemia. Mol. Cancer 2014, 13, 1549–1557. [Google Scholar] [CrossRef] [PubMed]
- Stamm, H.; Klingler, F.; Grossjohann, E.M.; Muschhammer, J.; Vettorazzi, E.; Heuser, M.; Mock, U.; Thol, F.; Vohwinkel, G.; Latuske, E.; et al. Immune checkpoints PVR and PVRL2 are prognostic markers in AML and their blockade represents a new therapeutic option. Oncogene 2018, 37, 5269–5280. [Google Scholar] [CrossRef] [PubMed]
- Daly, J.; Carlsten, M.; O’Dwyer, M. Sugar Free: Novel Immunotherapeutic Approaches Targeting Siglecs and Sialic Acids to Enhance Natural Killer Cell Cytotoxicity Against Cancer. Front. Immunol. 2019, 10, 1047. [Google Scholar] [CrossRef] [PubMed]
- Daly, J.; Duggan, T.; Hu, J.; Natoni, A.; Sarkar, S.; Kirkham-McCarthy, L.; McEllistrim, C.; Krawczyk, J.; O’Dwyer, M. Targeting Siglec-7: A Novel Immunotherapeutic Approach to Potentiate the Cytotoxic Functions of Natural Killer Cells Against Multiple Myeloma. Blood 2017, 130, 1799. [Google Scholar]
- Shao, J.Y.; Yin, W.W.; Zhang, Q.F.; Liu, Q.; Peng, M.L.; Hu, H.D.; Hu, P.; Ren, H.; Zhang, D.Z. Siglec-7 Defines a Highly Functional Natural Killer Cell Subset and Inhibits Cell-Mediated Activities. Scand. J. Immunol. 2016, 84, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, Y.; Ito, A.; Withers, D.A.; Taima, T.; Kakoi, N.; Saito, S.; Arai, Y. Ganglioside DSGb5, preferred ligand for Siglec-7, inhibits NK cell cytotoxicity against renal cell carcinoma cells. Glycobiology 2010, 20, 1373–1379. [Google Scholar] [CrossRef]
- Nicoll, G.; Avril, T.; Lock, K.; Furukawa, K.; Bovin, N.; Crocker, P.R. Ganglioside GD3 expression on target cells can modulate NK cell cytotoxicity via siglec-7-dependent and -independent mechanisms. Eur J. Immunol. 2003, 33, 1642–1648. [Google Scholar] [CrossRef]
- Crocker, P.R.; Paulson, J.C.; Varki, A. Siglecs and their roles in the immune system. Nat. Rev. Immunol. 2007, 7, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Jandus, C.; Boligan, K.F.; Chijioke, O.; Liu, H.; Dahlhaus, M.; Demoulins, T.; Schneider, C.; Wehrli, M.; Hunger, R.E.; Baerlocher, G.M.; et al. Interactions between Siglec-7/9 receptors and ligands influence NK cell-dependent tumor immunosurveillance. J. Clin. Investig. 2014, 124, 1810–1820. [Google Scholar] [CrossRef] [PubMed]
- Chapoval, A.I.; Ni, J.; Lau, J.S.; Wilcox, R.A.; Flies, D.B.; Liu, D.; Dong, H.; Sica, G.L.; Zhu, G.; Tamada, K.; et al. B7-H3: A costimulatory molecule for T cell activation and IFN-gamma production. Nat. Immunol. 2001, 2, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Loos, M.; Hedderich, D.M.; Friess, H.; Kleeff, J. B7-h3 and its role in antitumor immunity. Clin. Dev. Immunol. 2010, 2010, 683875. [Google Scholar] [CrossRef] [PubMed]
- Picarda, E.; Ohaegbulam, K.C.; Zang, X. Molecular Pathways: Targeting B7-H3 (CD276) for Human Cancer Immunotherapy. Clin. Cancer Res. 2016, 22, 3425–3431. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Kang, F.B.; Shan, B.E. B7-H3-mediated tumor immunology: Friend or foe? Int. J. Cancer 2014, 134, 2764–2771. [Google Scholar] [CrossRef] [PubMed]
- Yonesaka, K.; Haratani, K.; Takamura, S.; Sakai, H.; Kato, R.; Takegawa, N.; Takahama, T.; Tanaka, K.; Hayashi, H.; Takeda, M.; et al. B7-H3 Negatively Modulates CTL-Mediated Cancer Immunity. Clin. Cancer Res. 2018, 24, 2653–2664. [Google Scholar] [CrossRef] [PubMed]
- Guery, T.; Roumier, C.; Berthon, C.; Renneville, A.; Preudhomme, C.; Quesnel, B. B7-H3 protein expression in acute myeloid leukemia. Cancer Med. 2015, 4, 1879–1883. [Google Scholar] [CrossRef]
- Hu, Y.; Lv, X.; Wu, Y.; Xu, J.; Wang, L.; Chen, W.; Zhang, W.; Li, J.; Zhang, S.; Qiu, H. Expression of costimulatory molecule B7-H3 and its prognostic implications in human acute leukemia. Hematology 2015, 20, 187–195. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, Y.; Wang, J.; Dong, F.; Zhu, M.; Wan, W.; Li, H.; Wu, F.; Yan, X.; Ke, X. B7-H3 silencing inhibits tumor progression of mantle cell lymphoma and enhances chemosensitivity. Int. J. Oncol. 2015, 46, 2562–2572. [Google Scholar] [CrossRef][Green Version]
- Sun, X.; Yu, Y.; Ma, L.; Xue, X.; Gao, Z.; Ma, J.; Zhang, M. T cell cytotoxicity toward hematologic malignancy via B7-H3 targeting. Invest. New Drugs 2019, 38, 722–732. [Google Scholar] [CrossRef]
- Germain, C.; Guillaudeux, T.; Galsgaard, E.D.; Hervouet, C.; Tekaya, N.; Gallouet, A.S.; Fassy, J.; Bihl, F.; Poupon, G.; Lazzari, A.; et al. Lectin-like transcript 1 is a marker of germinal center-derived B-cell non-Hodgkin’s lymphomas dampening natural killer cell functions. Oncoimmunology 2015, 4, e1026503. [Google Scholar] [CrossRef]
- Sivori, S.; Meazza, R.; Quintarelli, C.; Carlomagno, S.; Della Chiesa, M.; Falco, M.; Moretta, L.; Locatelli, F.; Pende, D. NK Cell-Based Immunotherapy for Hematological Malignancies. J. Clin. Med. 2019, 8, 1702. [Google Scholar] [CrossRef] [PubMed]
- Bialoszewska, A.; Malejczyk, J. Biological and Clinical Significance of Human NKRP1A/LLT1 Receptor/Ligand Interactions. Crit Rev. Immunol. 2018, 38, 479–489. [Google Scholar] [CrossRef]
- Deeks, E.D. Polatuzumab Vedotin: First Global Approval. Drugs 2019, 79, 1467–1475. [Google Scholar] [CrossRef]
- Sehn, L.H.; Herrera, A.F.; Flowers, C.R.; Kamdar, M.K.; McMillan, A.; Hertzberg, M.; Assouline, S.; Kim, T.M.; Kim, W.S.; Ozcan, M.; et al. Polatuzumab Vedotin in Relapsed or Refractory Diffuse Large B-Cell Lymphoma. J. Clin. Oncol. 2020, 38, 155–165. [Google Scholar] [CrossRef]
- Tilly, H.; Morschhauser, F.; Bartlett, N.L.; Mehta, A.; Salles, G.; Haioun, C.; Munoz, J.; Chen, A.I.; Kolibaba, K.; Lu, D.; et al. Polatuzumab vedotin in combination with immunochemotherapy in patients with previously untreated diffuse large B-cell lymphoma: An open-label, non-randomised, phase 1b-2 study. Lancet. Oncol. 2019, 20, 998–1010. [Google Scholar] [CrossRef]
- Morschhauser, F.; Flinn, I.W.; Advani, R.; Sehn, L.H.; Diefenbach, C.; Kolibaba, K.; Press, O.W.; Salles, G.; Tilly, H.; Chen, A.I.; et al. Polatuzumab vedotin or pinatuzumab vedotin plus rituximab in patients with relapsed or refractory non-Hodgkin lymphoma: Final results from a phase 2 randomised study (ROMULUS). Lancet. Haematol. 2019, 6, e254–e265. [Google Scholar] [CrossRef]
- Barrett, D.M.; Singh, N.; Porter, D.L.; Grupp, S.A.; June, C.H. Chimeric antigen receptor therapy for cancer. Annu. Rev. Med. 2014, 65, 333–347. [Google Scholar] [CrossRef] [PubMed]
- Goebeler, M.E.; Bargou, R.C. T cell-engaging therapies—BiTEs and beyond. Nat. Rev. Clin. Oncol. 2020, 17, 418–434. [Google Scholar] [CrossRef]
- Nelson, M.H.; Paulos, C.M. Novel immunotherapies for hematologic malignancies. Immunol. Rev. 2015, 263, 90–105. [Google Scholar] [CrossRef] [PubMed]
- Strohl, W.R.; Naso, M. Bispecific T-Cell Redirection versus Chimeric Antigen Receptor (CAR)-T Cells as Approaches to Kill Cancer Cells. Antibodies 2019, 8, 41. [Google Scholar] [CrossRef]
- Suryadevara, C.M.; Gedeon, P.C.; Sanchez-Perez, L.; Verla, T.; Alvarez-Breckenridge, C.; Choi, B.D.; Fecci, P.E.; Sampson, J.H. Are BiTEs the “missing link” in cancer therapy? Oncoimmunology 2015, 4, e1008339. [Google Scholar] [CrossRef]
- Aldoss, I.; Bargou, R.C.; Nagorsen, D.; Friberg, G.R.; Baeuerle, P.A.; Forman, S.J. Redirecting T cells to eradicate B-cell acute lymphoblastic leukemia: Bispecific T-cell engagers and chimeric antigen receptors. Leukemia 2017, 31, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Felices, M.; Lenvik, T.R.; Davis, Z.B.; Miller, J.S.; Vallera, D.A. Generation of BiKEs and TriKEs to Improve NK Cell-Mediated Targeting of Tumor Cells. Methods Mol. Biol. 2016, 1441, 333–346. [Google Scholar] [PubMed]
- Tay, S.S.; Carol, H.; Biro, M. TriKEs and BiKEs join CARs on the cancer immunotherapy highway. Hum. Vaccines Immunother. 2016, 12, 2790–2796. [Google Scholar] [CrossRef] [PubMed]
- Gleason, M.K.; Verneris, M.R.; Todhunter, D.A.; Zhang, B.; McCullar, V.; Zhou, S.X.; Panoskaltsis-Mortari, A.; Weiner, L.M.; Vallera, D.A.; Miller, J.S. Bispecific and trispecific killer cell engagers directly activate human NK cells through CD16 signaling and induce cytotoxicity and cytokine production. Mol. Cancer 2012, 11, 2674–2684. [Google Scholar] [CrossRef]
- Gleason, M.K.; Ross, J.A.; Warlick, E.D.; Lund, T.C.; Verneris, M.R.; Wiernik, A.; Spellman, S.; Haagenson, M.D.; Lenvik, A.J.; Litzow, M.R.; et al. CD16 × CD33 bispecific killer cell engager (BiKE) activates NK cells against primary MDS and MDSC CD33+ targets. Blood 2014, 123, 3016–3026. [Google Scholar] [CrossRef]
- Portner, L.M.; Schonberg, K.; Hejazi, M.; Brunnert, D.; Neumann, F.; Galonska, L.; Reusch, U.; Little, M.; Haas, R.; Uhrberg, M. T and NK cells of B cell NHL patients exert cytotoxicity against lymphoma cells following binding of bispecific tetravalent antibody CD19 × CD3 or CD19 × CD16. Cancer Immunol. Immunother. 2012, 61, 1869–1875. [Google Scholar] [CrossRef]
- Reiners, K.S.; Kessler, J.; Sauer, M.; Rothe, A.; Hansen, H.P.; Reusch, U.; Hucke, C.; Kohl, U.; Durkop, H.; Engert, A.; et al. Rescue of impaired NK cell activity in hodgkin lymphoma with bispecific antibodies in vitro and in patients. Mol. Ther. 2013, 21, 895–903. [Google Scholar] [CrossRef]
- Reusch, U.; Burkhardt, C.; Fucek, I.; Le Gall, F.; Le Gall, M.; Hoffmann, K.; Knackmuss, S.H.; Kiprijanov, S.; Little, M.; Zhukovsky, E.A. A novel tetravalent bispecific TandAb (CD30/CD16A) efficiently recruits NK cells for the lysis of CD30+ tumor cells. MAbs 2014, 6, 728–739. [Google Scholar] [CrossRef]
- Wu, J.; Fu, J.; Zhang, M.; Liu, D. AFM13: A first-in-class tetravalent bispecific anti-CD30/CD16A antibody for NK cell-mediated immunotherapy. J. Hematol. Oncol. 2015, 8, 96. [Google Scholar] [CrossRef]
- Pahl, J.H.W.; Koch, J.; Gotz, J.J.; Arnold, A.; Reusch, U.; Gantke, T.; Rajkovic, E.; Treder, M.; Cerwenka, A. CD16A Activation of NK Cells Promotes NK Cell Proliferation and Memory-Like Cytotoxicity against Cancer Cells. Cancer Immunol. Res. 2018, 6, 517–527. [Google Scholar] [CrossRef]
- Glorius, P.; Baerenwaldt, A.; Kellner, C.; Staudinger, M.; Dechant, M.; Stauch, M.; Beurskens, F.J.; Parren, P.W.; Winkel, J.G.; Valerius, T.; et al. The novel tribody [(CD20)(2) × CD16] efficiently triggers effector cell-mediated lysis of malignant B cells. Leukemia 2013, 27, 190–201. [Google Scholar] [CrossRef]
- Berrien-Elliott, M.M.; Wagner, J.A.; Fehniger, T.A. Human Cytokine-Induced Memory-Like Natural Killer Cells. J. Innate. Immun. 2015, 7, 563–571. [Google Scholar] [CrossRef]
- Childs, R.W.; Carlsten, M. Therapeutic approaches to enhance natural killer cell cytotoxicity against cancer: The force awakens. Nat. Rev. Drug Discov. 2015, 14, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Davis, Z.B.; Vallera, D.A.; Miller, J.S.; Felices, M. Natural killer cells unleashed: Checkpoint receptor blockade and BiKE/TriKE utilization in NK-mediated anti-tumor immunotherapy. Semin. Immunol. 2017, 31, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Del Bano, J.; Chames, P.; Baty, D.; Kerfelec, B. Taking up Cancer Immunotherapy Challenges: Bispecific Antibodies, the Path Forward? Antibodies 2015, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Rothe, A.; Sasse, S.; Topp, M.S.; Eichenauer, D.A.; Hummel, H.; Reiners, K.S.; Dietlein, M.; Kuhnert, G.; Kessler, J.; Buerkle, C.; et al. A phase 1 study of the bispecific anti-CD30/CD16A antibody construct AFM13 in patients with relapsed or refractory Hodgkin lymphoma. Blood 2015, 125, 4024–4031. [Google Scholar] [CrossRef]
- Grandjenette, C.; Dicato, M.; Diederich, M. Bispecific Antibodies: An Innovative Arsenal to Hunt, Grab and Destroy Cancer Cells. Curr. Pharm. Biotechnol. 2015, 16, 670–683. [Google Scholar]
- Vallera, D.A.; Felices, M.; McElmurry, R.; McCullar, V.; Zhou, X.; Schmohl, J.U.; Zhang, B.; Lenvik, A.J.; Panoskaltsis-Mortari, A.; Verneris, M.R.; et al. IL15 Trispecific Killer Engagers (TriKE) Make Natural Killer Cells Specific to CD33+ Targets While Also Inducing Persistence, In Vivo Expansion, and Enhanced Function. Clin. Cancer Res. 2016, 22, 3440–3450. [Google Scholar] [CrossRef]
- Sarhan, D.; Brandt, L.; Felices, M.; Guldevall, K.; Lenvik, T.; Hinderlie, P.; Curtsinger, J.; Warlick, E.; Spellman, S.R.; Blazar, B.R.; et al. 161533 TriKE stimulates NK-cell function to overcome myeloid-derived suppressor cells in MDS. Blood Adv. 2018, 2, 1459–1469. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, L.; Morel, A.; Anceriz, N.; Rossi, B.; Blanchard-Alvarez, A.; Grondin, G.; Trichard, S.; Cesari, C.; Sapet, M.; Bosco, F.; et al. Multifunctional Natural Killer Cell Engagers Targeting NKp46 Trigger Protective Tumor Immunity. Cell 2019, 177, 1701–1713.e16. [Google Scholar] [CrossRef] [PubMed]
- Kloess, S.; Ede Valverde da Silva, A.; Oberschmidt, O.; Gardlowski, T.; Matthies, N.; Vyas, M.; Arseniev, L.; Heuser, M.; Pogge von Strandmann, E.; Kohl, U. Triplebody Mediates Increased Anti-Leukemic Reactivity of IL-2 Activated Donor Natural Killer (NK) Cells and Impairs Viability of Their CD33-Expressing NK Subset. Front. Immunol. 2017, 8, 1100. [Google Scholar] [CrossRef]
- Duell, J.; Lammers, P.E.; Djuretic, I.; Chunyk, A.G.; Alekar, S.; Jacobs, I.; Gill, S. Bispecific Antibodies in the Treatment of Hematologic Malignancies. Clin. Pharmacol. Ther. 2019, 106, 781–791. [Google Scholar] [CrossRef]
- Guy, D.G.; Uy, G.L. Bispecific Antibodies for the Treatment of Acute Myeloid Leukemia. Curr Hematol Malig Rep. 2018, 13, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.; Wang, Y.; Lu, X.; Han, W. Chimeric Antigen Receptors Modified T-Cells for Cancer Therapy. J. Natl. Cancer Inst. 2016, 108, djv439. [Google Scholar] [CrossRef]
- Maude, S.L.; Teachey, D.T.; Porter, D.L.; Grupp, S.A. CD19-targeted chimeric antigen receptor T-cell therapy for acute lymphoblastic leukemia. Blood 2015, 125, 4017–4023. [Google Scholar] [CrossRef] [PubMed]
- Maus, M.V.; June, C.H. Making Better Chimeric Antigen Receptors for Adoptive T-cell Therapy. Clin. Cancer Res. 2016, 22, 1875–1884. [Google Scholar] [CrossRef] [PubMed]
- Rezvani, K.; Rouce, R.; Liu, E.; Shpall, E. Engineering Natural Killer Cells for Cancer Immunotherapy. Mol. Ther. 2017, 25, 1769–1781. [Google Scholar] [CrossRef]
- Weber, E.W.; Maus, M.V.; Mackall, C.L. The Emerging Landscape of Immune Cell Therapies. Cell 2020, 181, 46–62. [Google Scholar] [CrossRef] [PubMed]
- Ye, B.; Stary, C.M.; Li, X.; Gao, Q.; Kang, C.; Xiong, X. Engineering chimeric antigen receptor-T cells for cancer treatment. Mol. Cancer 2018, 17, 32. [Google Scholar] [CrossRef]
- Oberschmidt, O.; Kloess, S.; Koehl, U. Redirected Primary Human Chimeric Antigen Receptor Natural Killer Cells As an “Off-the-Shelf Immunotherapy” for Improvement in Cancer Treatment. Front. Immunol. 2017, 8, 654. [Google Scholar] [CrossRef] [PubMed]
- Atanackovic, D.; Radhakrishnan, S.V.; Bhardwaj, N.; Luetkens, T. Chimeric Antigen Receptor (CAR) therapy for multiple myeloma. Br. J. Haematol. 2016, 172, 685–698. [Google Scholar] [CrossRef]
- Chu, J.; Deng, Y.; Benson, D.M.; He, S.; Hughes, T.; Zhang, J.; Peng, Y.; Mao, H.; Yi, L.; Ghoshal, K.; et al. CS1-specific chimeric antigen receptor (CAR)-engineered natural killer cells enhance in vitro and in vivo antitumor activity against human multiple myeloma. Leukemia 2014, 28, 917–927. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Zhang, W.; Shang, P.; Zhang, H.; Fu, W.; Ye, F.; Zeng, T.; Huang, H.; Zhang, X.; Sun, W.; et al. Transfection of chimeric anti-CD138 gene enhances natural killer cell activation and killing of multiple myeloma cells. Mol. Oncol. 2014, 8, 297–310. [Google Scholar] [CrossRef] [PubMed]
- Boissel, L.; Betancur-Boissel, M.; Lu, W.; Krause, D.S.; Van Etten, R.A.; Wels, W.S.; Klingemann, H. Retargeting NK-92 cells by means of CD19- and CD20-specific chimeric antigen receptors compares favorably with antibody-dependent cellular cytotoxicity. Oncoimmunology 2013, 2, e26527. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.; Hochberg, J.; Yahr, A.; Ayello, J.; van de Ven, C.; Barth, M.; Czuczman, M.; Cairo, M.S. Targeting CD20+ Aggressive B-cell Non-Hodgkin Lymphoma by Anti-CD20 CAR mRNA-Modified Expanded Natural Killer Cells In Vitro and in NSG Mice. Cancer Immunol. Res. 2015, 3, 333–344. [Google Scholar] [CrossRef]
- Kobayashi, E.; Kishi, H.; Ozawa, T.; Hamana, H.; Nakagawa, H.; Jin, A.; Lin, Z.; Muraguchi, A. A chimeric antigen receptor for TRAIL-receptor 1 induces apoptosis in various types of tumor cells. Biochem Biophys Res. Commun. 2014, 453, 798–803. [Google Scholar] [CrossRef] [PubMed]
- Caraccio, C.; Krishna, S.; Phillips, D.J.; Schurch, C.M. Bispecific Antibodies for Multiple Myeloma: A Review of Targets, Drugs, Clinical Trials, and Future Directions. Front. Immunol. 2020, 11, 501. [Google Scholar] [CrossRef]
- Lin, Q.; Zhao, J.; Song, Y.; Liu, D. Recent updates on CAR T clinical trials for multiple myeloma. Mol. Cancer 2019, 18, 154. [Google Scholar] [CrossRef]
- Timmers, M.; Roex, G.; Wang, Y.; Campillo-Davo, D.; Van Tendeloo, V.F.I.; Chu, Y.; Berneman, Z.N.; Luo, F.; Van Acker, H.H.; Anguille, S. Chimeric Antigen Receptor-Modified T Cell Therapy in Multiple Myeloma: Beyond B Cell Maturation Antigen. Front. Immunol. 2019, 10, 1613. [Google Scholar] [CrossRef]
- Wang, H.; Kaur, G.; Sankin, A.I.; Chen, F.; Guan, F.; Zang, X. Immune checkpoint blockade and CAR-T cell therapy in hematologic malignancies. J. Hematol. Oncol. 2019, 12, 59. [Google Scholar] [CrossRef]
- Sun, C.; Mahendravada, A.; Ballard, B.; Kale, B.; Ramos, C.; West, J.; Maguire, T.; McKay, K.; Lichtman, E.; Tuchman, S.; et al. Safety and efficacy of targeting CD138 with a chimeric antigen receptor for the treatment of multiple myeloma. Oncotarget 2019, 10, 2369–2383. [Google Scholar] [CrossRef] [PubMed]
- Demoulin, B.; Cook, W.J.; Murad, J.; Graber, D.J.; Sentman, M.L.; Lonez, C.; Gilham, D.E.; Sentman, C.L.; Agaugue, S. Exploiting natural killer group 2D receptors for CAR T-cell therapy. Future Oncol. 2017, 13, 1593–1605. [Google Scholar] [CrossRef]
- Glienke, W.; Esser, R.; Priesner, C.; Suerth, J.D.; Schambach, A.; Wels, W.S.; Grez, M.; Kloess, S.; Arseniev, L.; Koehl, U. Advantages and applications of CAR-expressing natural killer cells. Front. Pharm. 2015, 6, 21. [Google Scholar] [CrossRef] [PubMed]
- Sentman, C.L.; Meehan, K.R. NKG2D CARs as cell therapy for cancer. Cancer J. 2014, 20, 156–159. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.H.; Connolly, J.; Shimasaki, N.; Mimura, K.; Kono, K.; Campana, D. A chimeric receptor with NKG2D specificity enhances natural killer cell activation and killing of tumor cells. Cancer Res. 2013, 73, 1777–1786. [Google Scholar] [CrossRef]
- Leivas, A.; Rio, P.; Mateos, R.; Paciello, M.L.; Garcia-Ortiz, A.; Fernandez, L.; Perez-Martinez, A.; Lee, D.A.; Powell, D.J., Jr.; Valeri, A.; et al. NKG2D-CAR Transduced Primary Natural Killer Cells Efficiently Target Multiple Myeloma Cells. Blood 2018, 132, 590. [Google Scholar] [CrossRef]
- Wang, Z.; Guo, L.; Song, Y.; Zhang, Y.; Lin, D.; Hu, B.; Mei, Y.; Sandikin, D.; Liu, H. Augmented anti-tumor activity of NK-92 cells expressing chimeric receptors of TGF-betaR II and NKG2D. Cancer Immunol. Immunother. 2017, 66, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Di Stasi, A.; Tey, S.K.; Dotti, G.; Fujita, Y.; Kennedy-Nasser, A.; Martinez, C.; Straathof, K.; Liu, E.; Durett, A.G.; Grilley, B.; et al. Inducible apoptosis as a safety switch for adoptive cell therapy. N. Engl. J. Med. 2011, 365, 1673–1683. [Google Scholar] [CrossRef]
- Minagawa, K.; Al-Obaidi, M.; Di Stasi, A. Generation of Suicide Gene-Modified Chimeric Antigen Receptor-Redirected T-Cells for Cancer Immunotherapy. Methods Mol. Biol. 2019, 1895, 57–73. [Google Scholar] [PubMed]
- Liu, E.; Tong, Y.; Dotti, G.; Shaim, H.; Savoldo, B.; Mukherjee, M.; Orange, J.; Wan, X.; Lu, X.; Reynolds, A.; et al. Cord blood NK cells engineered to express IL-15 and a CD19-targeted CAR show long-term persistence and potent antitumor activity. Leukemia 2018, 32, 520–531. [Google Scholar] [CrossRef] [PubMed]
- Otahal, P.; Prukova, D.; Kral, V.; Fabry, M.; Vockova, P.; Lateckova, L.; Trneny, M.; Klener, P. Lenalidomide enhances antitumor functions of chimeric antigen receptor modified T cells. Oncoimmunology 2016, 5, e1115940. [Google Scholar] [CrossRef]
- Wang, X.; Walter, M.; Urak, R.; Weng, L.; Huynh, C.; Lim, L.; Wong, C.W.; Chang, W.C.; Thomas, S.H.; Sanchez, J.F.; et al. Lenalidomide Enhances the Function of CS1 Chimeric Antigen Receptor-Redirected T Cells Against Multiple Myeloma. Clin. Cancer Res. 2018, 24, 106–119. [Google Scholar] [CrossRef]
- Magen, H.; Muchtar, E. Elotuzumab: The first approved monoclonal antibody for multiple myeloma treatment. Ther. Adv. Hematol. 2016, 7, 187–195. [Google Scholar] [CrossRef]
- Cao, Y.; Lu, W.; Sun, R.; Jin, X.; Cheng, L.; He, X.; Wang, L.; Yuan, T.; Lyu, C.; Zhao, M. Anti-CD19 Chimeric Antigen Receptor T Cells in Combination With Nivolumab Are Safe and Effective Against Relapsed/Refractory B-Cell Non-hodgkin Lymphoma. Front. Oncol. 2019, 9, 767. [Google Scholar] [CrossRef]
- Petty, A.J.; Heyman, B.; Yang, Y. Chimeric Antigen Receptor Cell Therapy: Overcoming Obstacles to Battle Cancer. Cancers 2020, 12, 842. [Google Scholar] [CrossRef] [PubMed]
- Chong, E.A.; Melenhorst, J.J.; Svoboda, J.; Dwivedy Nasta, S.; Landsburg, D.J.; Mato, A.R.; Tian, L.; Parakandi, H.; Lacey, S.F.; June, C.H.; et al. Phase I/II Study of Pembrolizumab for Progressive Diffuse Large B Cell Lymphoma after Anti-CD19 Directed Chimeric Antigen Receptor Modified T Cell Therapy. Blood 2017, 130, 4121. [Google Scholar]
- Hodgins, J.J.; Khan, S.T.; Park, M.M.; Auer, R.C.; Ardolino, M. Killers 2.0: NK cell therapies at the forefront of cancer control. J. Clin. Investig. 2019, 129, 3499–3510. [Google Scholar] [CrossRef]
- Klingemann, H. Are natural killer cells superior CAR drivers? Oncoimmunology 2014, 3, e28147. [Google Scholar] [CrossRef]
- Kloess, S.; Kretschmer, A.; Stahl, L.; Fricke, S.; Koehl, U. CAR-Expressing Natural Killer Cells for Cancer Retargeting. Transfus. Med. Hemother. 2019, 46, 4–13. [Google Scholar] [CrossRef]
- Cheng, M.; Chen, Y.; Xiao, W.; Sun, R.; Tian, Z. NK cell-based immunotherapy for malignant diseases. Cell Mol. Immunol. 2013, 10, 230–252. [Google Scholar] [CrossRef]
- Hu, W.; Wang, G.; Huang, D.; Sui, M.; Xu, Y. Cancer Immunotherapy Based on Natural Killer Cells: Current Progress and New Opportunities. Front. Immunol. 2019, 10, 1205. [Google Scholar] [CrossRef] [PubMed]
- Saetersmoen, M.L.; Hammer, Q.; Valamehr, B.; Kaufman, D.S.; Malmberg, K.J. Off-the-shelf cell therapy with induced pluripotent stem cell-derived natural killer cells. Semin. Immunopathol. 2019, 41, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Hermanson, D.L.; Moriarity, B.S.; Kaufman, D.S. Human iPSC-Derived Natural Killer Cells Engineered with Chimeric Antigen Receptors Enhance Anti-tumor Activity. Cell Stem Cell 2018, 23, 181–192.e185. [Google Scholar] [CrossRef]
- Wang, W.N.; Zhou, G.Y.; Zhang, W.L. NK-92 cell, another ideal carrier for chimeric antigen receptor. Immunotherapy 2017, 9, 753–765. [Google Scholar] [CrossRef]
- Chmielewski, M.; Abken, H. CAR T cells transform to trucks: Chimeric antigen receptor-redirected T cells engineered to deliver inducible IL-12 modulate the tumour stroma to combat cancer. Cancer Immunol. Immunother 2012, 61, 1269–1277. [Google Scholar] [CrossRef]
- Chmielewski, M.; Abken, H. TRUCKs: The fourth generation of CARs. Expert Opin. Biol. 2015, 15, 1145–1154. [Google Scholar] [CrossRef]
- Krieg, S.; Ullrich, E. Novel immune modulators used in hematology: Impact on NK cells. Front. Immunol. 2012, 3, 388. [Google Scholar] [CrossRef] [PubMed]
- Kroesen, M.; Gielen, P.; Brok, I.C.; Armandari, I.; Hoogerbrugge, P.M.; Adema, G.J. HDAC inhibitors and immunotherapy; a double edged sword? Oncotarget. 2014, 5, 6558–6572. [Google Scholar] [CrossRef] [PubMed]
- Nijhof, I.S.; van de Donk, N.; Zweegman, S.; Lokhorst, H.M. Current and New Therapeutic Strategies for Relapsed and Refractory Multiple Myeloma: An Update. Drugs 2018, 78, 19–37. [Google Scholar] [CrossRef]
- Davies, F.; Baz, R. Lenalidomide mode of action: Linking bench and clinical findings. Blood Rev. 2010, 24 (Suppl. 1), S13–S19. [Google Scholar] [CrossRef]
- Gribben, J.G.; Fowler, N.; Morschhauser, F. Mechanisms of Action of Lenalidomide in B-Cell Non-Hodgkin Lymphoma. J. Clin. Oncol. 2015, 33, 2803–2811. [Google Scholar] [CrossRef]
- Quach, H.; Ritchie, D.; Stewart, A.K.; Neeson, P.; Harrison, S.; Smyth, M.J.; Prince, H.M. Mechanism of action of immunomodulatory drugs (IMiDS) in multiple myeloma. Leukemia 2010, 24, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Yang, L.; Li, Z.; Nalin, A.P.; Dai, H.; Xu, T.; Yin, J.; You, F.; Zhu, M.; Shen, W.; et al. First-in-man clinical trial of CAR NK-92 cells: Safety test of CD33-CAR NK-92 cells in patients with relapsed and refractory acute myeloid leukemia. Am. J. Cancer Res. 2018, 8, 1083–1089. [Google Scholar] [PubMed]
- Pfefferle, A.; Huntington, N.D. You Have Got a Fast CAR: Chimeric Antigen Receptor NK Cells in Cancer Therapy. Cancers 2020, 12, 706. [Google Scholar] [CrossRef] [PubMed]
| Target | mAb | Drug Combination | Disease | Trial Number (NCT) | Phase |
|---|---|---|---|---|---|
| CD38 | Daratumumab | Lenalidomide + Dexamethasone | MM | 02076009 | III |
| Bortezomib + Dexamethasone | MM (CASTOR) | 02136134 | III | ||
| MM (SIRIUS) | 01985126 | II | |||
| Carfilzomib, Lenalidomide, Dexamethasone | newly diagnosed MM | 03290950 | II | ||
| Bortezomib, Lenalidomide, Dexamethasone | untreated MM (PERSEUS) | 03710603 | III | ||
| Lenalidomide, Dexamethasone | MM r/r (POLLUX) | 02076009 | III | ||
| Bortezomib, Cyclophosphamide, Dexamethasone | MM r/r (LYRA) | 01951819 | II | ||
| Prednisone, Bortezomib, Melphalan vs Daratumumab alone | MM r/r (ALCYONE) | 02195479 | III | ||
| Lenalidomide, Dexamethasone | untreated MM (MAIA) | 02252172 | III | ||
| Bortezomib, Thalidomide, Dexamethasone | untreated MM (CASSEOPEIA) | 02541383 | III | ||
| Isatuximab (SAR650984) | Lenalidomide, Dexamethasone | MM | 01749969 | Ib | |
| Pomalidomide, Dexamethasone | MM r/r (ICARIA) | 02990338 | III | ||
| CD137 | Urelumab | Nivolumab | B-cell NHL | 02253992 | I/II |
| Elozutumab | MM | 02252263 | I | ||
| SLAM-F7 (CS1) | Elotuzumab | MM | 03003728 | II | |
| Nivolumab | MM r/r (Checkmate-602) | 02726581 | III | ||
| Lenalidomide, Dexamethasone | MM (ELOQUENT-2) | 01239797 | III | ||
| KIR2DL1/2/3 | IPH2102 (Lirilumab) | Nivolumab, 5-Azacytidine | Leukemia | 02599649 | II |
| Rituximab (anti-CD20) | High-risk Untreated and r/r CLL | 02481297 | II | ||
| Elotuzumab, Urelumab | MM | 02252263 | I | ||
| Lenalidomide | MM | 01217203 | I | ||
| IPH2101 (1-7F9) | SMM | 01222286 (KIRMONO) | II | ||
| MM | 00999830 (REMYKIR) | II | |||
| AML | 01256073 | I | |||
| MM | 00552396 | I | |||
| Lenalidomide | MM | 01217203 (KIRIMID) | I | ||
| MEDI4736 | 5-Azacytidine | Leukemia | 02399917 | II | |
| MEDI6469 | Tremelimumab (anti-CTLA4) or Rituximab or MEDI4736 | B-cell lymphoma, MDS | 02205333 | I/II | |
| KIR3DL2 | IPH4102 (Lacutamab) | Cutaneous T-cell lymphoma | 02593045 | I | |
| NKG2A | IPH2201 (Monalizumab) | Hematological cancers | 02921685 | I | |
| CLL | 03088059 | II | |||
| CLL | 02557516 | I/II | |||
| Sym-021 (anti-PD-1), Sym-022 (anti-Lag-3) | Lymphoma | 03311412 | I | ||
| MBG453 | Decitabine (hypomethylating agent) | AML, high risk MDS | 03066648 | I | |
| Lag-3 | Sym-022 | Lymphoma | 03489369 | I | |
| Sym-021 (anti-PD-1), Sym-023 (anti-Tim-3) | Lymphoma | 03311412 | I | ||
| BMS-986016 | Nivolumab (BMS-936558) | DLBCL r/r, HL r/r | 02061761 | I/II | |
| PD-1 | Pembrolizumab | cHL r/r | 01953692 (Keynote-013) | Ib | |
| cHL r/r post ASCT | 02458594 (Keynote-087) | II | |||
| Ibrutinib | NHL r/r | 02950220 | I | ||
| Brutoximab Vedotin (anti-CD30 mAb) | cHL r/r | 02684292 (Keynote-024) | III | ||
| Lenalidomide, Dexamethasone | MM | 02036502 (Keynote-023) | I | ||
| Pomalidomide, Dexamethasone | MM r/r | 02576977 (Keynote-183) | III | ||
| Nivolumab | cHL r/r (Checkmate-205) | 01592370 | II | ||
| Epacadostat (anti-IDO1 mAb) | DLBCL, HL | 02327078 | I/II | ||
| Lenalidomide | NHL, cHL r/r | 03015896 | I/II | ||
| FL r/r (Checkmate-140) | 02038946 | II | |||
| Lenalidomide, Rituximab | DLBCL | 03558750 | I | ||
| DLBCL r/r (CheckMate-139) | 02038933 | II | |||
| Cyclophosphamide, Prednisone, Doxorubicin Hydrochloride | DLCBL | 03704714 | I/II | ||
| Urelumab (anti-CD137 mAb) | NHL | 02253992 | I/II | ||
| Varlilumab (CDX-1127) (anti-CD27 mAb) | DLCBL | 03038672 | II | ||
| HL r/r (ANIMATE) | 03337919 | II | |||
| Lenalidomide, Dexamethasone | high risk SMM | 02903381 | II | ||
| Lenalidomide | MM r/r | 03333746 | II | ||
| Daratumumab with or without Cyclophosphamide | MM r/r | 03184194 | II | ||
| Daratumumab or Pomalidomide and Dexamethasone | Hematological cancers | 01592370 | I | ||
| Ipilimumab (anti-CTLA4) | high risk MM | 02681302 | I/II | ||
| Elotuzumab with or without Pomalidomide and Dexamethasone | MM r/r | 03227432 | II | ||
| Elotuzumab, Pomalidomide, Dexamethasone | MM r/r | 02726581 | III | ||
| AML | 02275533 | II | |||
| Dasatinib (tyrosine kinase receptor inhibitor) | CML | 02011945 | I | ||
| HL | 02181738 | II | |||
| Rituximab, Gemcitabine, Bendamustine (alkylating agent) | DLBCL r/r | 03259529 | I/II | ||
| Pidilizumab | Rituximab | FL r/r | 00904722 | II | |
| Lenalidomide | MM | 02077959 | I/II | ||
| PD-L1 | Atezolizumab | Obinutuzumab (anti-CD20) | DLBCL, FL r/r | 02220842 | I |
| Obinutuzumab, Polatuzumab vedotin (anti-CD79b mAb) | DLBCL, FL r/r | 02729896 | I | ||
| Obinutuzumab, Lenalidomide | FL r/r | 02631577 | I | ||
| Obinutuzumab, Ibrutinib | untreated, high risk or r/r CLL | 02846623 | II | ||
| Guadecitabine (hypomethylating agent) | AML, MDS, CML r/r | 02935361 | I/II | ||
| Daratumumab vs Daratumumab, IMiDs | MM | 02431208 | I | ||
| Avelumab | cHL r/r | 02603419 | Ib | ||
| Itolizumab (anti-CD6) vs Itolizumab, 5-Azacytidine vs Bendamustine, Rituximab | DLBCL r/r | 02951156 | Ib/II | ||
| 5-Azacytidine | AML r/r | 02953561 | I | ||
| Cetrelimab (JNJ-63723283) | Daratumumab | MM r/r | 03357952 | II/III | |
| Durvalumab | Daratumumab | MM r/r | 03000452 | II | |
| Lenalidomide, Dexamethasone | newly diagnosed MM | 02685826 | I | ||
| Pomalidomide, Dexamethasone | MM r/r | 02616640 | I | ||
| Rituximab, Lenalidomide with or without Ibrutinib | NHL, CLL | 02733042 | I/II |
| Sources of NK Cells | Disease | Receptor Target | Trial Number (NCT) | Phase |
|---|---|---|---|---|
| NK92 | Lymphoma and Leukemia | CD7 | 02742727 | I/II |
| Lymphoma and Leukemia | CD19 | 02892695 | I/II | |
| AML r/r | CD33 | 02944162 | I/II | |
| MM r/r | BCMA | 03940833 | I/II | |
| B-cell lymphoma r/r | CD19 | 03690310 | I | |
| CB-derived NK cells | ALL, CLL and NHL r/r | CD19 | 03056339 | I/II |
| B-cell lymphoma | CD19 | 03579927 | I/II | |
| unknown | B-cell lymphoma r/r | CD19+CD22 | 03824964 | I |
| PB NK cells | ALL | CD19 | 01974479 | I |
| ALL | CD19 | 00995137 | I | |
| unknown | B-cell lymphoma r/r | CD22 | 03692767 | I |
| iPSC-derived NK cells | B-cell lymphoma r/r | CD19 | 03824951 | I |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giuliani, M.; Poggi, A. Checkpoint Inhibitors and Engineered Cells: New Weapons for Natural Killer Cell Arsenal Against Hematological Malignancies. Cells 2020, 9, 1578. https://doi.org/10.3390/cells9071578
Giuliani M, Poggi A. Checkpoint Inhibitors and Engineered Cells: New Weapons for Natural Killer Cell Arsenal Against Hematological Malignancies. Cells. 2020; 9(7):1578. https://doi.org/10.3390/cells9071578
Chicago/Turabian StyleGiuliani, Massimo, and Alessandro Poggi. 2020. "Checkpoint Inhibitors and Engineered Cells: New Weapons for Natural Killer Cell Arsenal Against Hematological Malignancies" Cells 9, no. 7: 1578. https://doi.org/10.3390/cells9071578
APA StyleGiuliani, M., & Poggi, A. (2020). Checkpoint Inhibitors and Engineered Cells: New Weapons for Natural Killer Cell Arsenal Against Hematological Malignancies. Cells, 9(7), 1578. https://doi.org/10.3390/cells9071578






