Progressively De-Differentiated Pancreatic Cancer Cells Shift from Glycolysis to Oxidative Metabolism and Gain a Quiescent Stem State
Abstract
:1. Background
2. Materials and Methods
2.1. Drugs and Chemicals
2.2. Cell Lines
2.3. RNA Extraction and qPCR
2.4. Cell Proliferation Assay
2.5. Clonogenic Assay
2.6. Soft Agar Colony Formation Assay
2.7. Cell Cycle Analysis
2.8. Senescence Analysis
2.9. Omic Analyses
2.10. Oxygen Consumption Rate (OCR)
2.11. Zebrafish
2.12. Statistical Analysis
3. Results
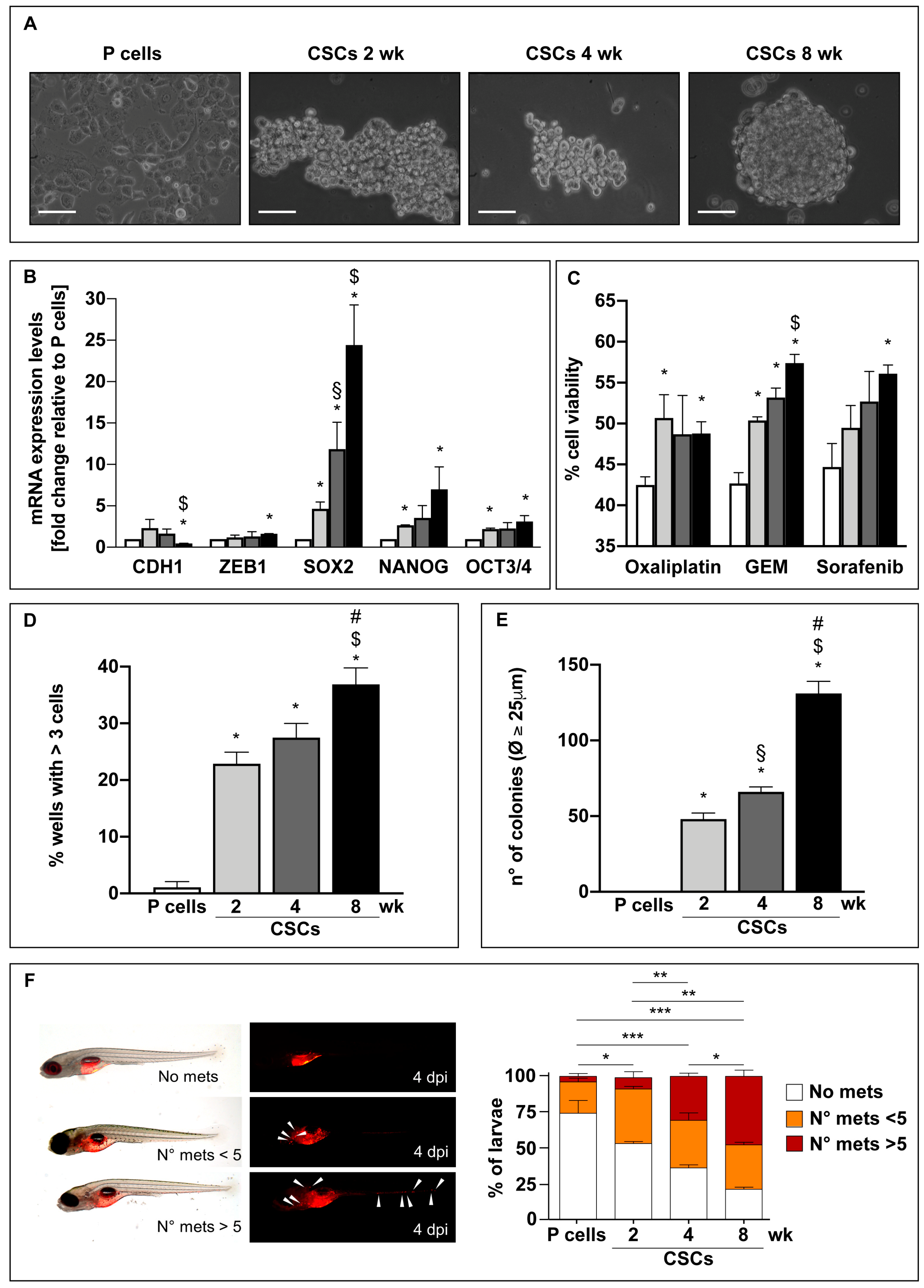
3.1. Long-Term Culture in the Stem-Specific Medium Elicits the Stem and Mesenchymal Properties in PDAC Cells
3.2. Long-Term Cultured CSCs Enter a Quiescent State
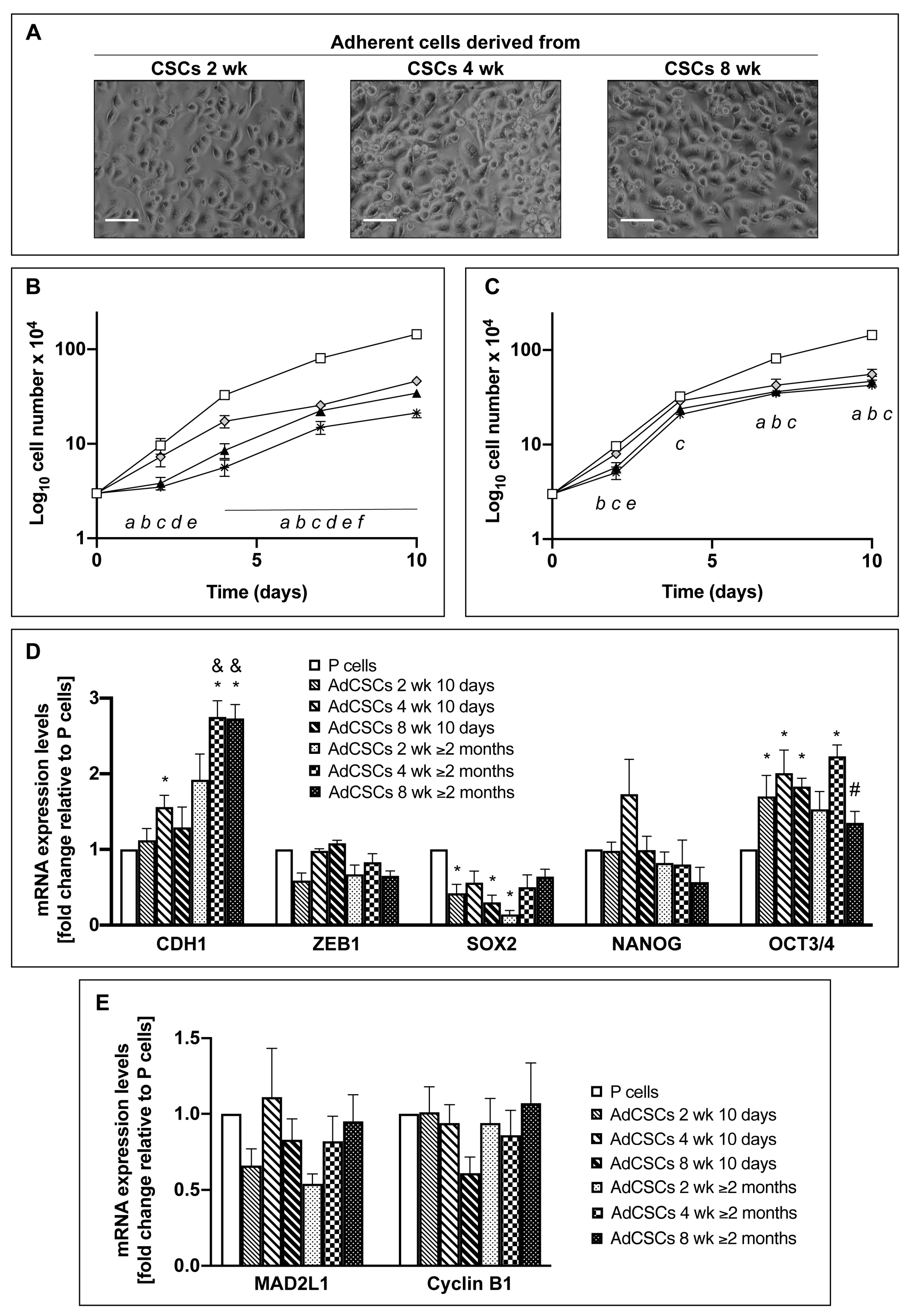
3.3. CSCs Cultured in the Differentiated-Cell Medium Re-Acquire Epithelial Features and Exit from the Quiescent State
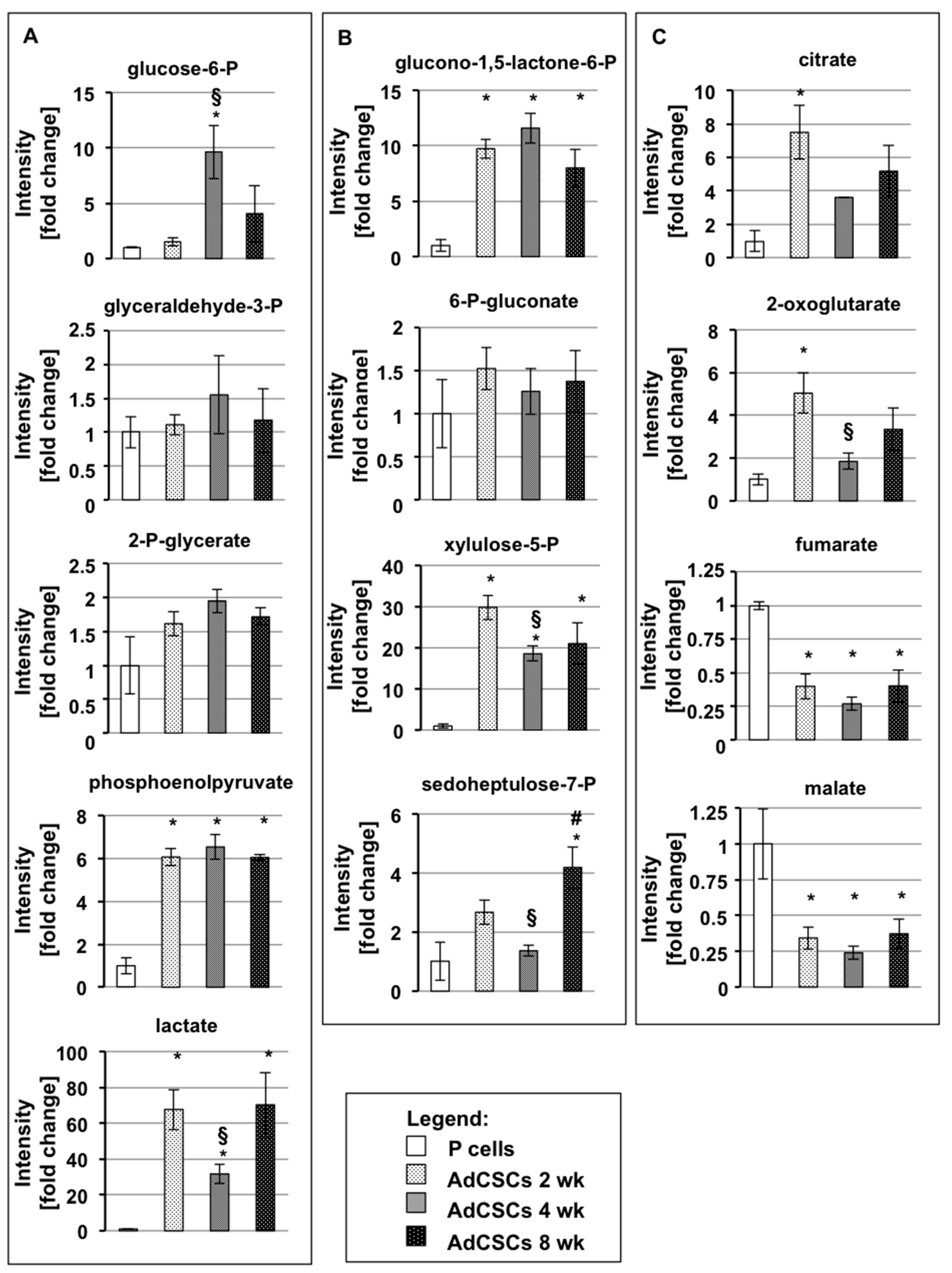
3.4. CSCs Modify Their Metabolic Settings Towards the Acquisition of Quiescence Properties
3.5. CSCs Demonstrate an Upregulation of Glycolysis and the PPP, and Downregulation of Oxidative Phosphorylation, at 2 Weeks of Culture
3.6. CSCs Demonstrate an Upregulation of Oxidative Phosphorylation at 4 Weeks of Culture
3.7. Week-Quiescent CSCs Downregulate Metabolic Processes
3.8. Metabolic Phenotypes Confer Targetable
3.9. Adherent Cells Derived from CSCs Escape the Quiescent State and Become Metabolically Active
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbruzzese, J.L.; Hess, K.R. New option for the initial management of metastatic pancreatic cancer? J. Clin. Oncol. 2014, 32, 2405–2407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neoptolemos, J.P.; Stocken, D.D.; Friess, H.; Bassi, C.; Dunn, J.A.; Hickey, H.; Beger, H.; Fernandez-Cruz, L.; Dervenis, C.; Lacaine, F.; et al. A randomized trial of chemoradiotherapy and chemotherapy after resection of pancreatic cancer. N. Engl. J. Med. 2004, 350, 1200–1210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasheed, Z.A.; Matsui, W. Biological and clinical relevance of stem cells in pancreatic adenocarcinoma. J. Gastroenterol. Hepatol. 2012, 27 (Suppl. 2), 15–18. [Google Scholar] [CrossRef] [Green Version]
- Bünger, S.; Barow, M.; Thorns, C.; Freitag-Wolf, S.; Danner, S.; Tiede, S.; Pries, R.; Görg, S.; Bruch, H.P.; Roblick, U.J.; et al. Pancreatic carcinoma cell lines reflect frequency and variability of cancer stem cell markers in clinical tissue. Eur. Surg. Res. 2012, 49, 88–98. [Google Scholar] [CrossRef]
- Dalla Pozza, E.; Dando, I.; Biondani, G.; Brandi, J.; Costanzo, C.; Zoratti, E.; Fassan, M.; Boschi, F.; Melisi, D.; Cecconi, D.; et al. Pancreatic ductal adenocarcinoma cell lines display a plastic ability to bidirectionally convert into cancer stem cells. Int. J. Oncol. 2015, 46, 1099–1108. [Google Scholar] [CrossRef] [Green Version]
- Sancho, P.; Alcala, S.; Usachov, V.; Hermann, P.C.; Sainz, B., Jr. The ever-changing landscape of pancreatic cancer stem cells. Pancreatology 2016, 16, 489–496. [Google Scholar] [CrossRef] [Green Version]
- Rhim, A.D.; Mirek, E.T.; Aiello, N.M.; Maitra, A.; Bailey, J.M.; McAllister, F.; Reichert, M.; Beatty, G.L.; Rustgi, A.K.; Vonderheide, R.H.; et al. EMT and dissemination precede pancreatic tumor formation. Cell 2012, 148, 349–361. [Google Scholar] [CrossRef] [Green Version]
- Shibue, T.; Weinberg, R.A. EMT, CSCs, and drug resistance: The mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 2017, 14, 611–629. [Google Scholar] [CrossRef] [Green Version]
- Phi, L.T.; Sari, I.N.; Yang, Y.G.; Lee, S.H.; Jun, N.; Kim, K.S.; Lee, Y.K.; Kwon, H.Y. Cancer Stem Cells (CSCs) in Drug Resistance and their Therapeutic Implications in Cancer Treatment. Stem Cells Int. 2018, 2018, 5416923. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.J.; Wechsler-Reya, R.J. Hit’em where they live: Targeting the cancer stem cell niche. Cancer Cell 2007, 11, 3–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Carlo, C.; Brandi, J.; Cecconi, D. Pancreatic cancer stem cells: Perspectives on potential therapeutic approaches of pancreatic ductal adenocarcinoma. World J. Stem Cells 2018, 10, 172–182. [Google Scholar] [CrossRef] [PubMed]
- da Veiga Moreira, J.; Hamraz, M.; Abolhassani, M.; Schwartz, L.; Jolicoeur, M.; Peres, S. Metabolic therapies inhibit tumor growth in vivo and in silico. Sci. Rep. 2019, 9, 3153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viale, A.; Pettazzoni, P.; Lyssiotis, C.A.; Ying, H.; Sánchez, N.; Marchesini, M.; Carugo, A.; Green, T.; Seth, S.; Giuliani, V.; et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 2014, 514, 628–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dando, I.; Dalla Pozza, E.; Biondani, G.; Cordani, M.; Palmieri, M.; Donadelli, M. The metabolic landscape of cancer stem cells. IUBMB Life 2015, 67, 687–693. [Google Scholar] [CrossRef] [Green Version]
- Peiris-Pages, M.; Martinez-Outschoorn, U.E.; Pestell, R.G.; Sotgia, F.; Lisanti, M.P. Cancer stem cell metabolism. Breast Cancer Res. 2016, 18, 55. [Google Scholar] [CrossRef]
- Sancho, P.; Burgos-Ramos, E.; Tavera, A.; Kheir, T.B.; Jagust, P.; Schoenhals, M.; Barneda, D.; Sellers, K.; Campos-Olivas, R.; Graña, O.; et al. MYC/PGC-1alpha Balance Determines the Metabolic Phenotype and Plasticity of Pancreatic Cancer Stem Cells. Cell Metab. 2015, 22, 590–605. [Google Scholar] [CrossRef] [Green Version]
- Dobbin, Z.C.; Landen, C.N. Isolation and characterization of potential cancer stem cells from solid human tumors--potential applications. Curr. Protoc. Pharmacol. 2013, 63, 14–28. [Google Scholar] [CrossRef]
- Franco, S.S.; Szczesna, K.; Iliou, M.S.; Al-Qahtani, M.; Mobasheri, A.; Kobolák, J.; Dinnyés, A. In vitro models of cancer stem cells and clinical applications. BMC Cancer 2016, 16 (Suppl. 2), 738. [Google Scholar] [CrossRef] [Green Version]
- Brandi, J.; Dando, I.; Dalla Pozza, E.; Biondani, G.; Jenkins, R.; Elliott, V.; Park, K.; Fanelli, G.; Zolla, L.; Costello, E.; et al. Proteomic analysis of pancreatic cancer stem cells: Functional role of fatty acid synthesis and mevalonate pathways. J. Proteom. 2017, 150, 310–322. [Google Scholar] [CrossRef]
- Lemma, S.; Avnet, S.; Salerno, M.; Chano, T.; Baldini, N. Identification and Validation of Housekeeping Genes for Gene Expression Analysis of Cancer Stem Cells. PLoS ONE 2016, 11, e0149481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brandi, J.; Di Carlo, C.; Manfredi, M.; Federici, F.; Bazaj, A.; Rizzi, E.; Cornaglia, G.; Manna, L.; Marengo, E.; Cecconi, D. Investigating the Proteomic Profile of HT-29 Colon Cancer Cells After Lactobacillus kefiri SGL 13 Exposure Using the SWATH Method. J. Am. Soc. Mass Spectrom. 2019, 30, 1690–1699. [Google Scholar] [CrossRef]
- Masgras, I.; Ciscato, F.; Brunati, A.M.; Tibaldi, E.; Indraccolo, S.; Curtarello, M.; Chiara, F.; Cannino, G.; Papaleo, E.; Lambrughi, M.; et al. Absence of Neurofibromin Induces an Oncogenic Metabolic Switch via Mitochondrial ERK-Mediated Phosphorylation of the Chaperone TRAP1. Cell Rep. 2017, 18, 659–672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lister, J.A.; Robertson, C.P.; Lepage, T.; Johnson, S.L.; Raible, D.W. nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development 1999, 126, 3757–3767. [Google Scholar] [PubMed]
- Rajendran, V.; Jain, M.V. In Vitro Tumorigenic Assay: Colony Forming Assay for Cancer Stem Cells. Methods Mol. Biol. 2018, 1692, 89–95. [Google Scholar]
- Borowicz, S.; Van Scoyk, M.; Avasarala, S.; Rathinam, M.K.; Tauler, J.; Bikkavilli, R.K.; Winn, R.A. The soft agar colony formation assay. J. Vis. Exp. 2014, e51998. [Google Scholar] [CrossRef] [Green Version]
- Pang, M.F.; Georgoudaki, A.M.; Lambut, L.; Johansson, J.; Tabor, V.; Hagikura, K.; Jin, Y.; Jansson, M.; Alexander, J.S.; Nelson, C.M.; et al. TGF-beta1-induced EMT promotes targeted migration of breast cancer cells through the lymphatic system by the activation of CCR7/CCL21-mediated chemotaxis. Oncogene 2016, 35, 748–760. [Google Scholar] [CrossRef]
- Hwang, K.L.; Goessling, W. Baiting for Cancer: Using the Zebrafish as a Model in Liver and Pancreatic Cancer. Adv. Exp. Med. Biol. 2016, 916, 391–410. [Google Scholar]
- Parasido, E.; Avetian, G.S.; Naeem, A.; Graham, G.; Pishvaian, M.; Glasgow, E.; Mudambi, S.; Lee, Y.; Ihemelandu, C.; Choudhry, M.; et al. The Sustained Induction of c-MYC Drives Nab-Paclitaxel Resistance in Primary Pancreatic Ductal Carcinoma Cells. Mol. Cancer Res. 2019, 17, 1815–1827. [Google Scholar] [CrossRef]
- Teng, Y.; Xie, X.; Walker, S.; White, D.T.; Mumm, J.S.; Cowell, J.K. Evaluating human cancer cell metastasis in zebrafish. BMC Cancer 2013, 13, 453. [Google Scholar] [CrossRef] [Green Version]
- Bao, B.; Ahmad, A.; Azmi, A.S.; Ali, S.; Sarkar, F.H. Overview of cancer stem cells (CSCs) and mechanisms of their regulation: Implications for cancer therapy. Curr. Protoc. Pharmacol. 2013, 61, 14–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lonardo, E.; Cioffi, M.; Sancho, P.; Sanchez-Ripoll, Y.; Trabulo, S.M.; Dorado, J.; Balic, A.; Hidalgo, M.; Heeschen, C. Metformin targets the metabolic achilles heel of human pancreatic cancer stem cells. PLoS ONE 2013, 8, e76518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mellor, H.R.; Ferguson, D.J.; Callaghan, R. A model of quiescent tumour microregions for evaluating multicellular resistance to chemotherapeutic drugs. Br. J. Cancer 2005, 93, 302–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, I.J.; Lui, P.P.; Obajdin, J.; Riccio, F.; Stroukov, W.; Willis, T.L.; Spagnoli, F.; Watt, F.M. Mechanisms, Hallmarks, and Implications of Stem Cell Quiescence. Stem Cell Rep. 2019, 12, 1190–1200. [Google Scholar] [CrossRef] [Green Version]
- Calcinotto, A.; Kohli, J.; Zagato, E.; Pellegrini, L.; Demaria, M.; Alimonti, A. Cellular Senescence: Aging, Cancer, and Injury. Physiol. Rev. 2019, 99, 1047–1078. [Google Scholar] [CrossRef]
- Coller, H.A.; Sang, L.; Roberts, J.M. A new description of cellular quiescence. PLoS Biol. 2006, 4, e83. [Google Scholar] [CrossRef] [Green Version]
- Moore, N.; Lyle, S. Quiescent, slow-cycling stem cell populations in cancer: A review of the evidence and discussion of significance. J. Oncol. 2011, 2011, 396076. [Google Scholar] [CrossRef]
- Marcon, B.H.; Holetz, F.B.; Eastman, G.; Origa-Alves, A.C.; Amorós, M.A.; de Aguiar, A.M.; Rebelatto, C.K.; Brofman, P.R.; Sotelo-Silveira, J.; Dallagiovanna, B. Downregulation of the protein synthesis machinery is a major regulatory event during early adipogenic differentiation of human adipose-derived stromal cells. Stem Cell Res. 2017, 25, 191–201. [Google Scholar] [CrossRef]
- Signer, R.A.; Magee, J.A.; Salic, A.; Morrison, S.J. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 2014, 509, 49–54. [Google Scholar] [CrossRef]
- Chen, W.; Dong, J.; Haiech, J.; Kilhoffer, M.C.; Zeniou, M. Cancer Stem Cell Quiescence and Plasticity as Major Challenges in Cancer Therapy. Stem Cells Int. 2016, 2016, 1740936. [Google Scholar] [CrossRef] [Green Version]
- Wellen, K.E.; Hatzivassiliou, G.; Sachdeva, U.M.; Bui, T.V.; Cross, J.R.; Thompson, C.B. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 2009, 324, 1076–1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, C.C.; Qian, Y.; Yu, J. Interplay between epigenetics and metabolism in oncogenesis: Mechanisms and therapeutic approaches. Oncogene 2017, 36, 3359–3374. [Google Scholar] [CrossRef] [PubMed]
- Curtis, N.J.; Mooney, L.; Hopcroft, L.; Michopoulos, F.; Whalley, N.; Zhong, H.; Murray, C.; Logie, A.; Revill, M.; Byth, K.F.; et al. Pre-clinical pharmacology of AZD3965, a selective inhibitor of MCT1: DLBCL, NHL and Burkitt’s lymphoma anti-tumor activity. Oncotarget 2017, 8, 69219–69236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, S.; Kildegaard, H.F.; Andersen, M.R. Impact of CHO Metabolism on Cell Growth and Protein Production: An Overview of Toxic and Inhibiting Metabolites and Nutrients. Biotechnol. J. 2018, 13, e1700499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shono, Y.; Kamouchi, M.; Kitazono, T.; Kuroda, J.; Nakamura, K.; Hagiwara, N.; Ooboshi, H.; Ibayashi, S.; Iida, M. Change in intracellular pH causes the toxic Ca2+ entry via NCX1 in neuron- and glia-derived cells. Cell Mol. Neurobiol. 2010, 30, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Ayob, A.Z.; Ramasamy, T.S. Cancer stem cells as key drivers of tumour progression. J. Biomed. Sci. 2018, 25, 20. [Google Scholar] [CrossRef]
- Valcourt, J.R.; Lemons, J.M.; Haley, E.M.; Kojima, M.; Demuren, O.O.; Coller, H.A. Staying alive: Metabolic adaptations to quiescence. Cell Cycle. 2012, 11, 1680–1696. [Google Scholar] [CrossRef] [Green Version]
- Yumoto, K.; Eber, M.R.; Berry, J.E.; Taichman, R.S.; Shiozawa, Y. Molecular pathways: Niches in metastatic dormancy. Clin. Cancer Res. 2014, 20, 3384–3389. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, K.M.; Dewhirst, M.W. Tumor metabolism of lactate: The influence and therapeutic potential for MCT and CD147 regulation. Future Oncol. 2010, 6, 127–148. [Google Scholar] [CrossRef] [Green Version]



| - | Doubling Time (Day) | ||
|---|---|---|---|
| - | Standard culture conditions | Adherent cells derived from CSCs cultured in DM for 10 days | Adherent cells derived from CSCs cultured in DM for ≥ 2 months |
| P cells | 2 ± 0.25 | / | / |
| CSCs 2 wk | 3.6 ± 0.05 * | 2.9 ± 0.31 *,£ | 2.9 ± 0.04 *,£ |
| CSCs 4 wk | 3.5 ± 0.02 * | 2.8 ± 0.34 *,£ | 2.6 ± 0.09 *,£ |
| CSCs 8 wk | 7.2 ± 0.25 *$,# | 3.5 ± 0.46 *,£ | 2.6 ± 0.10 *,£,& |
| Doubling Time (Day) | |||
| Standard culture conditions | Adherent cells derived from CSCs cultured in DM for 10 days | Adherent cells derived from CSCs cultured in DM for ≥ 2 months | |
| P cells | 2 ± 0.25 | / | / |
| CSCs 2 wk | 3.6 ± 0.05 * | 2.9 ± 0.31 *,£ | 2.9 ± 0.04 *,£ |
| CSCs 4 wk | 3.5 ± 0.02 * | 2.8 ± 0.34 *,£ | 2.6 ± 0.09 *,£ |
| CSCs 8 wk | 7.2 ± 0.25 *$,# | 3.5 ± 0.46 0,*£ | 2.6 ± 0.10 *,£,& |
| Doubling Time (Day) | |||
| Standard culture conditions | Adherent cells derived from CSCs cultured in DM for 10 days | Adherent cells derived from CSCs cultured in DM for ≥ 2 months | |
| P cells | 2 ± 0.25 | / | / |
| CSCs 2 wk | 3.6 ± 0.05 * | 2.9 ± 0.31 *,£ | 2.9 ± 0.04 *,£ |
| CSCs 4 wk | 3.5 ± 0.02 * | 2.8 ± 0.34 *,£ | 2.6 ± 0.09 *,£ |
| CSCs 8 wk | 7.2 ± 0.25 *,$,# | 3.5 ± 0.46 *,£ | 2.6 ± 0.10 *,£,& |
| CDH1 (fold change) | ZEB1 (fold change) | SOX2 (fold change) | NANOG (fold change) | OCT3/4 (fold change) | MAD2L1 (fold change) | cyclinB1 (fold change) | |
|---|---|---|---|---|---|---|---|
| P | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| CSCs 2wk | 2.32 ± 1.05 | 1.19 ± 0.28 | 4.64 ± 0.82 | 2.67 ± 0.04 | 2.20 ± 0.10 | 0.65 ± 0.33 | 0.31 ± 0.08 |
| CSCs 4wk | 1.65 ± 0.54 | 1.30 ± 0.58 | 11.85± 3.23 | 3.54 ± 1.50 | 2.28 ± 0.69 | 0.61 ± 0.12 | 0.69 ± 0.14 |
| CSCs 8wk | 0.17 ± 0.01 | 1.64 ± 0.02 | 24.42± 6.84 | 6.99 ± 2.71 | 3.12 ± 0.70 | 0.16 ± 0.02 | 0.14 ± 0.07 |
| AdCSC 2w 10 days | 1.12 ± 0.15 | 0.59± 0.09 | 0.42 ± 0.11 | 0.98 ± 0.11 | 1.70 ± 0.28 | 0.66 ± 0.11 | 1.01 ± 0.16 |
| AdCSC 4w 10 days | 1.56 ± 0.15 | 0.98 ± 0.03 | 0.56 ± 0.15 | 1.73± 0.46 | 2.01 ± 0.30 | 1.11 ± 0.32 | 0.94 ± 0.12 |
| AdCSC 8w 10 days | 1.29 ± 0.27 | 1.08 ± 0.04 | 0.30 ± 0.09 | 0.99 ± 0.18 | 1.83 ± 0.11 | 0.83 ± 0.13 | 0.61 ± 0.11 |
| AdCSC 2w ≥2 months | 1.92 ± 0.34 | 0.67 ± 0.12 | 0.14 ± 0.05 | 0.82 ± 0.14 | 1.53 ± 0.24 | 0.54 ± 0.06 | 0.94 ± 0.16 |
| AdCSC 4w ≥2 months | 2.75 ± 0.21 | 0.83 ± 0.11 | 0.50 ± 0.16 | 0.80 ± 0.32 | 2.23 ± 0.15 | 0.82 ± 0.16 | 0.86 ± 0.16 |
| AdCSC 8w ≥2 months | 2.73± 0.19 | 0.65 ± 0.06 | 0.64 ± 0.10 | 0.57 ± 0.19 | 1.35 ± 0.15 | 0.95 ± 0.17 | 1.07 ± 0.27 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ambrosini, G.; Dalla Pozza, E.; Fanelli, G.; Di Carlo, C.; Vettori, A.; Cannino, G.; Cavallini, C.; Carmona-Carmona, C.A.; Brandi, J.; Rinalducci, S.; et al. Progressively De-Differentiated Pancreatic Cancer Cells Shift from Glycolysis to Oxidative Metabolism and Gain a Quiescent Stem State. Cells 2020, 9, 1572. https://doi.org/10.3390/cells9071572
Ambrosini G, Dalla Pozza E, Fanelli G, Di Carlo C, Vettori A, Cannino G, Cavallini C, Carmona-Carmona CA, Brandi J, Rinalducci S, et al. Progressively De-Differentiated Pancreatic Cancer Cells Shift from Glycolysis to Oxidative Metabolism and Gain a Quiescent Stem State. Cells. 2020; 9(7):1572. https://doi.org/10.3390/cells9071572
Chicago/Turabian StyleAmbrosini, Giulia, Elisa Dalla Pozza, Giuseppina Fanelli, Claudia Di Carlo, Andrea Vettori, Giuseppe Cannino, Chiara Cavallini, Cristian Andres Carmona-Carmona, Jessica Brandi, Sara Rinalducci, and et al. 2020. "Progressively De-Differentiated Pancreatic Cancer Cells Shift from Glycolysis to Oxidative Metabolism and Gain a Quiescent Stem State" Cells 9, no. 7: 1572. https://doi.org/10.3390/cells9071572
APA StyleAmbrosini, G., Dalla Pozza, E., Fanelli, G., Di Carlo, C., Vettori, A., Cannino, G., Cavallini, C., Carmona-Carmona, C. A., Brandi, J., Rinalducci, S., Scupoli, M. T., Rasola, A., Cecconi, D., Palmieri, M., & Dando, I. (2020). Progressively De-Differentiated Pancreatic Cancer Cells Shift from Glycolysis to Oxidative Metabolism and Gain a Quiescent Stem State. Cells, 9(7), 1572. https://doi.org/10.3390/cells9071572











