Effects of Mutations in the Drosophila melanogaster Rif1 Gene on the Replication and Underreplication of Pericentromeric Heterochromatin in Salivary Gland Polytene Chromosomes
Abstract
1. Introduction
2. Materials and Methods
2.1. Fly Stocks
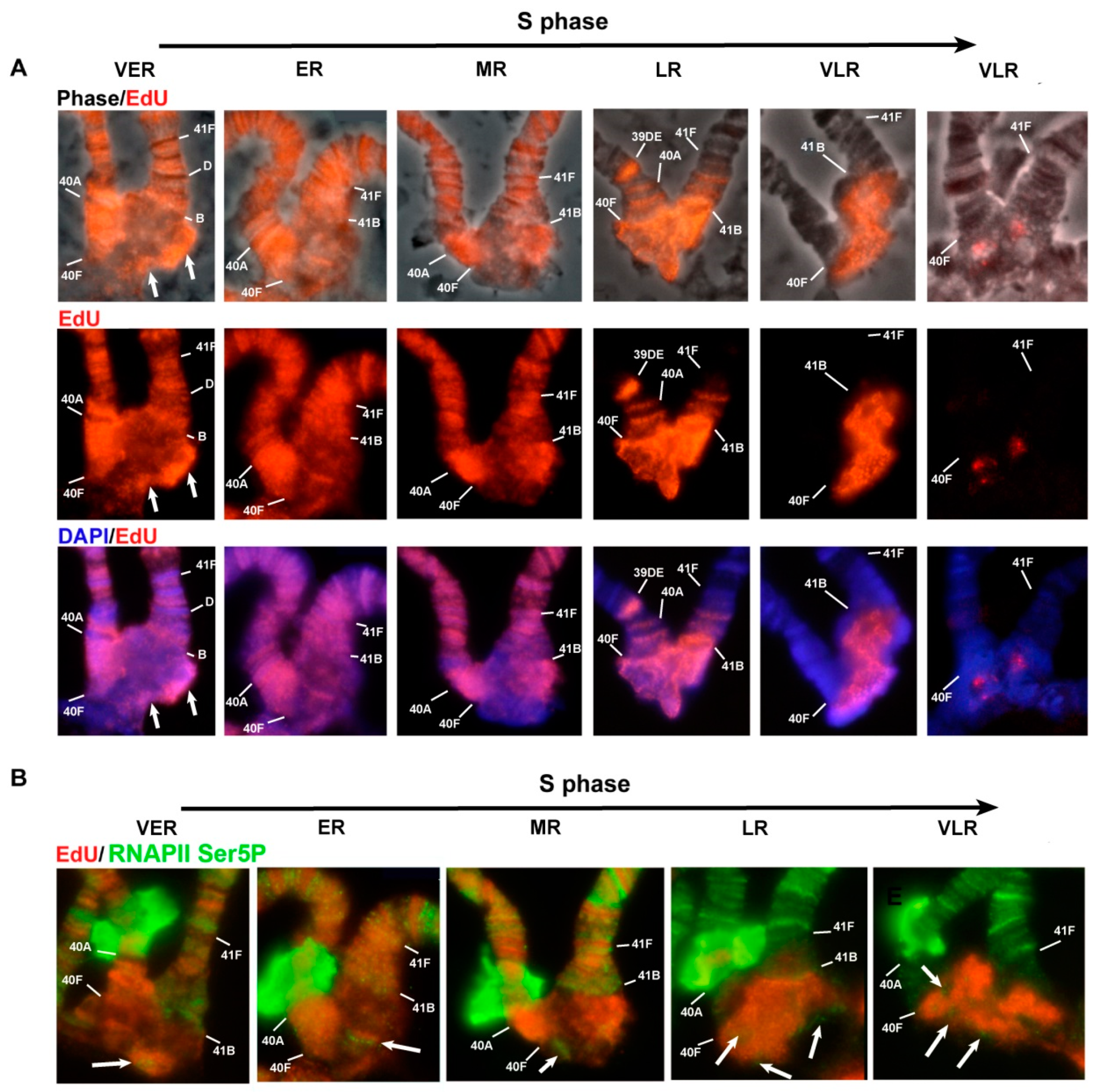
2.2. Fluorescence in Situ Hybridization (FISH)
2.3. Indirect Immunofluorescent Staining
2.4. 5-Ethynyl-2′-Deoxyuridine (EdU) Incorporation and Detection
2.5. Morphological Analyses and Microscopy of Polytene Chromosomes
2.6. Microdissection
2.7. Data Processing
3. Results
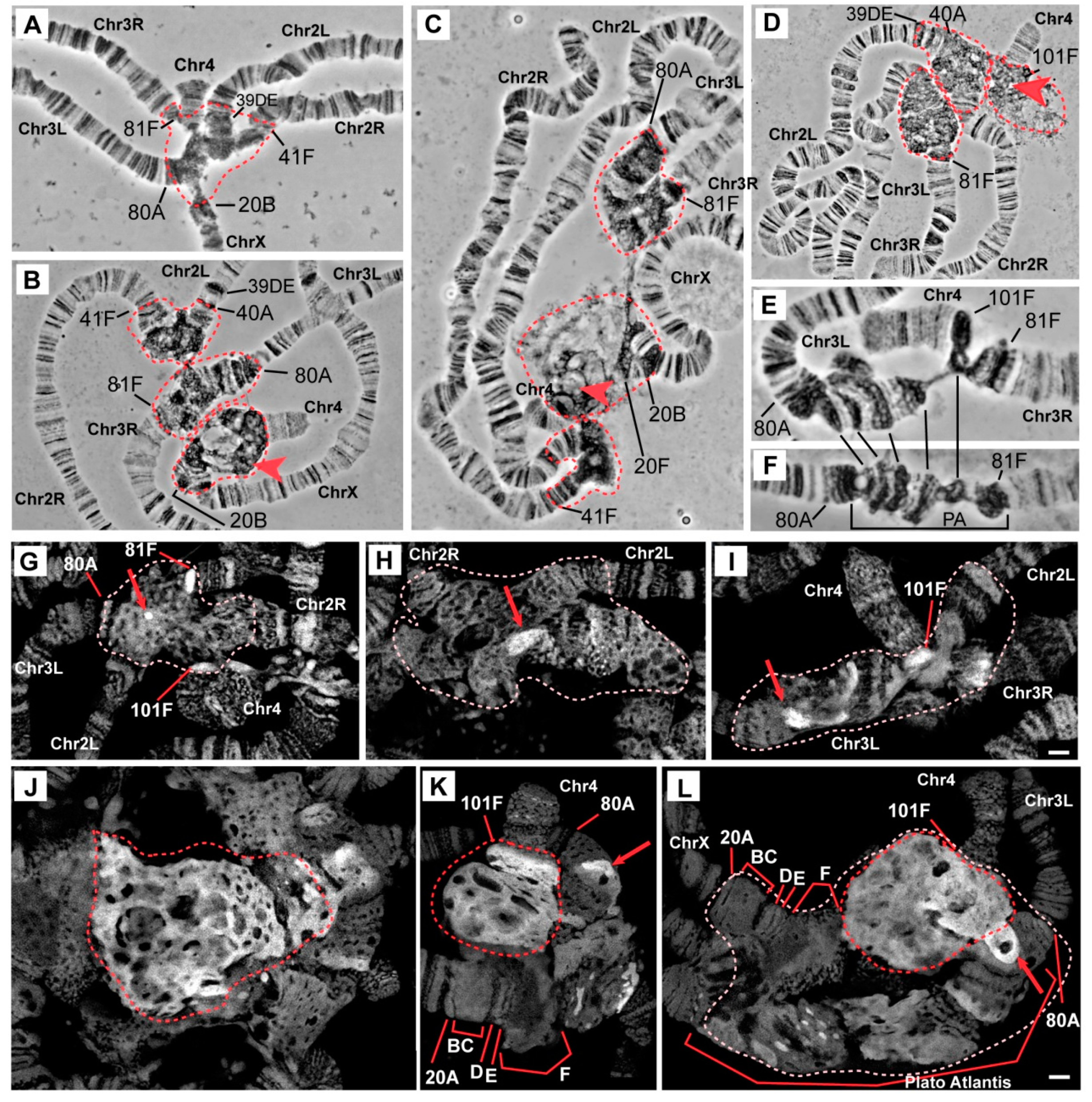
3.1. The Effect of Mutations in the Rif1 Gene on the Morphology of Pericentromeric Regions in Salivary Gland Polytene Chromosomes
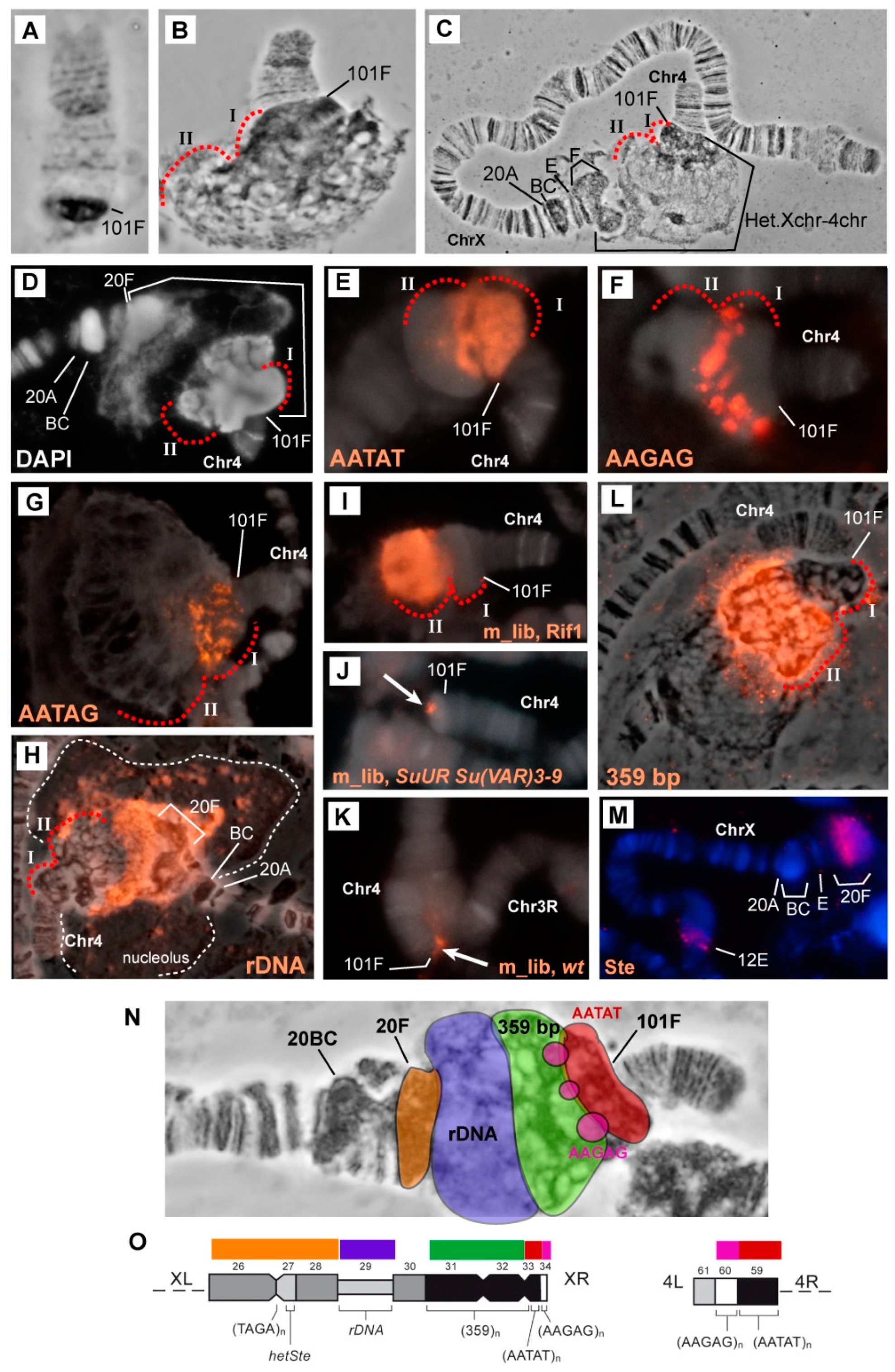
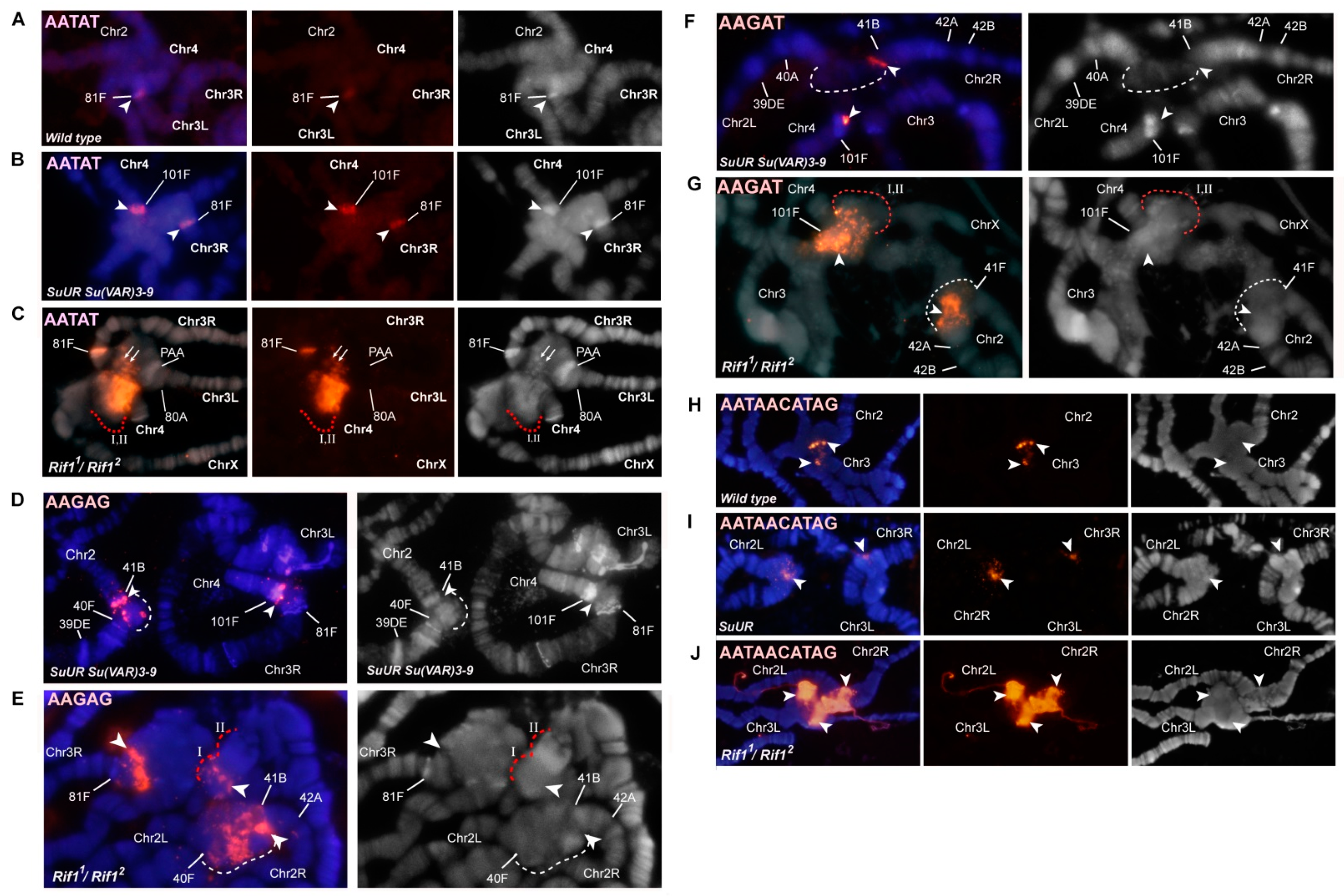
3.2. Mutations in the Rif1 Gene Result in Polytenization of the Sequences of Proximal Heterochromatin on Chromosome X and Heterochromatin on Chromosome 4
3.3. In the Rif1 Mutants, Chromosome Y is Visible in Salivary Gland Polytene Chromosomes
3.4. The effect of Rif1 on the Heterochromatin of Chromosome 2
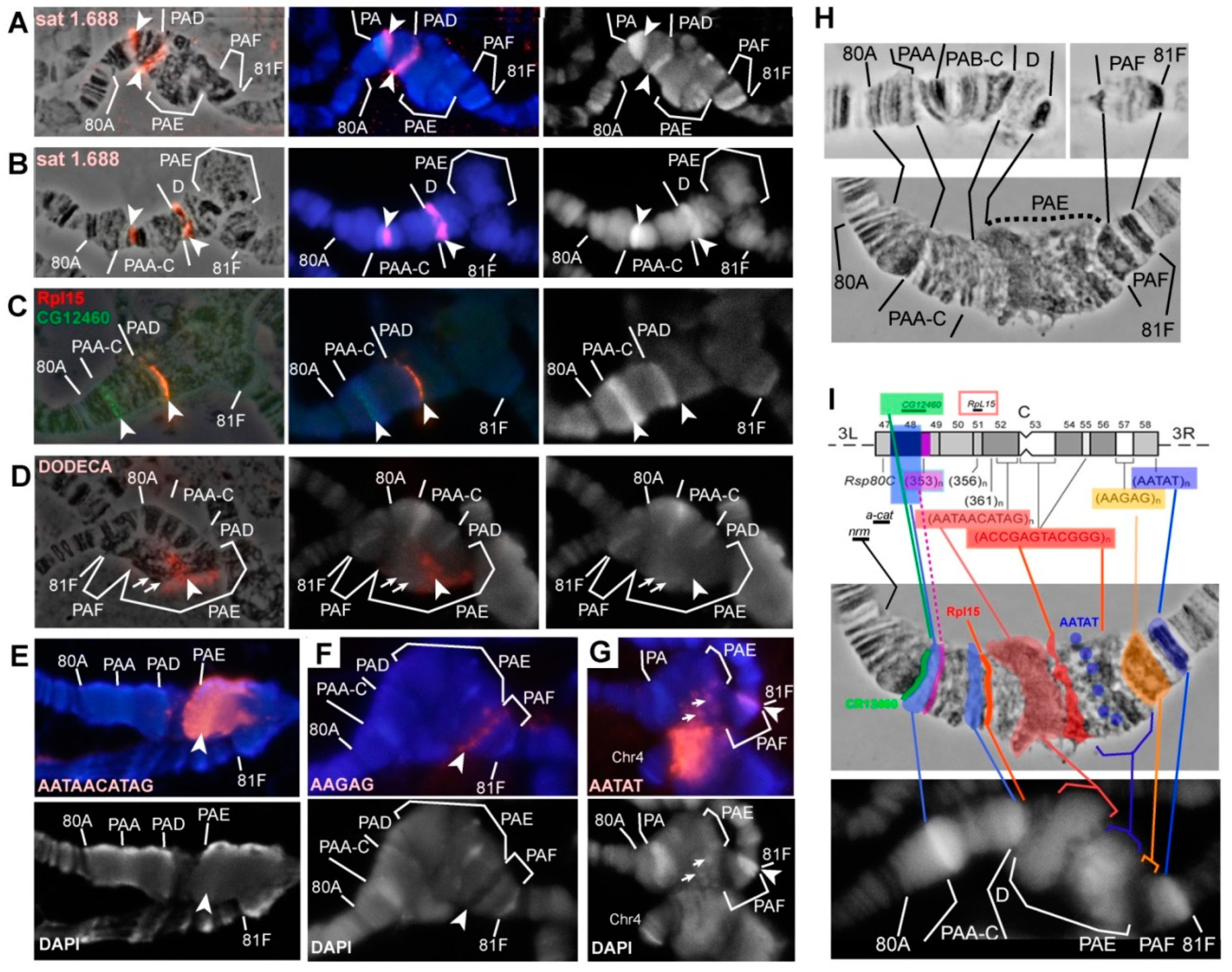
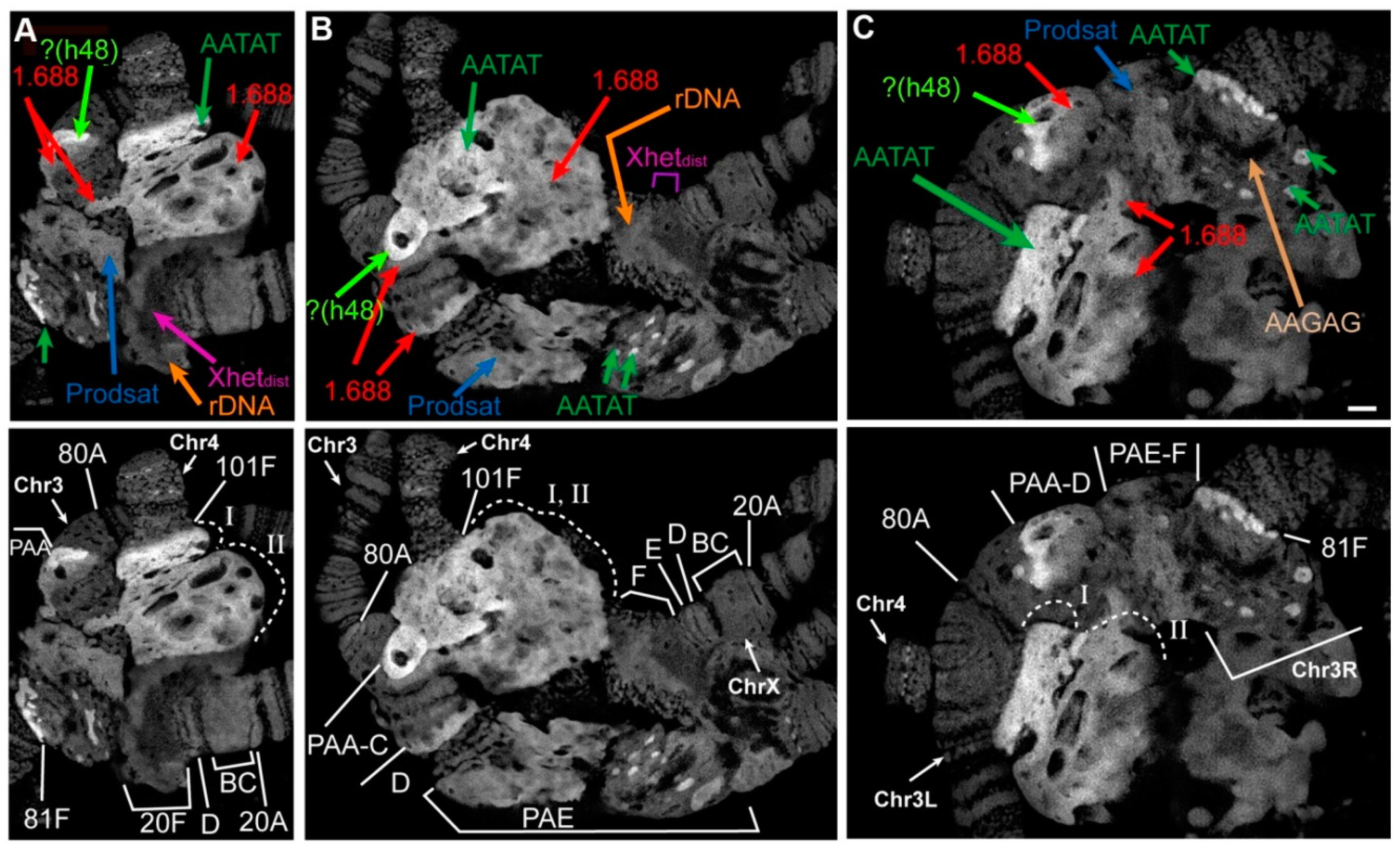
3.5. Mutations in the Rif1 Gene lead to the Polytenization of Satellite Sequences Surrounding the Centromere of Chromosome 3
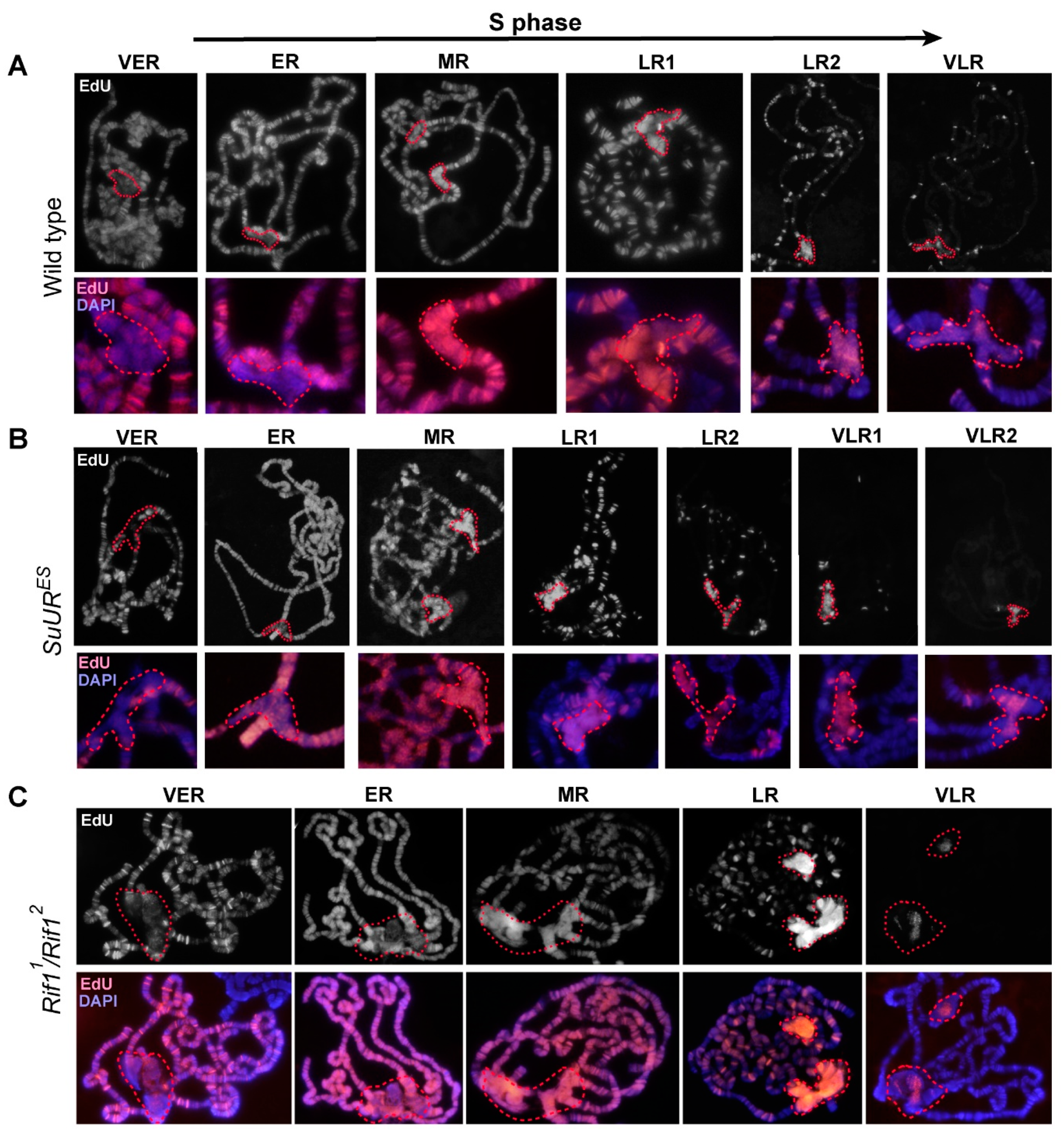
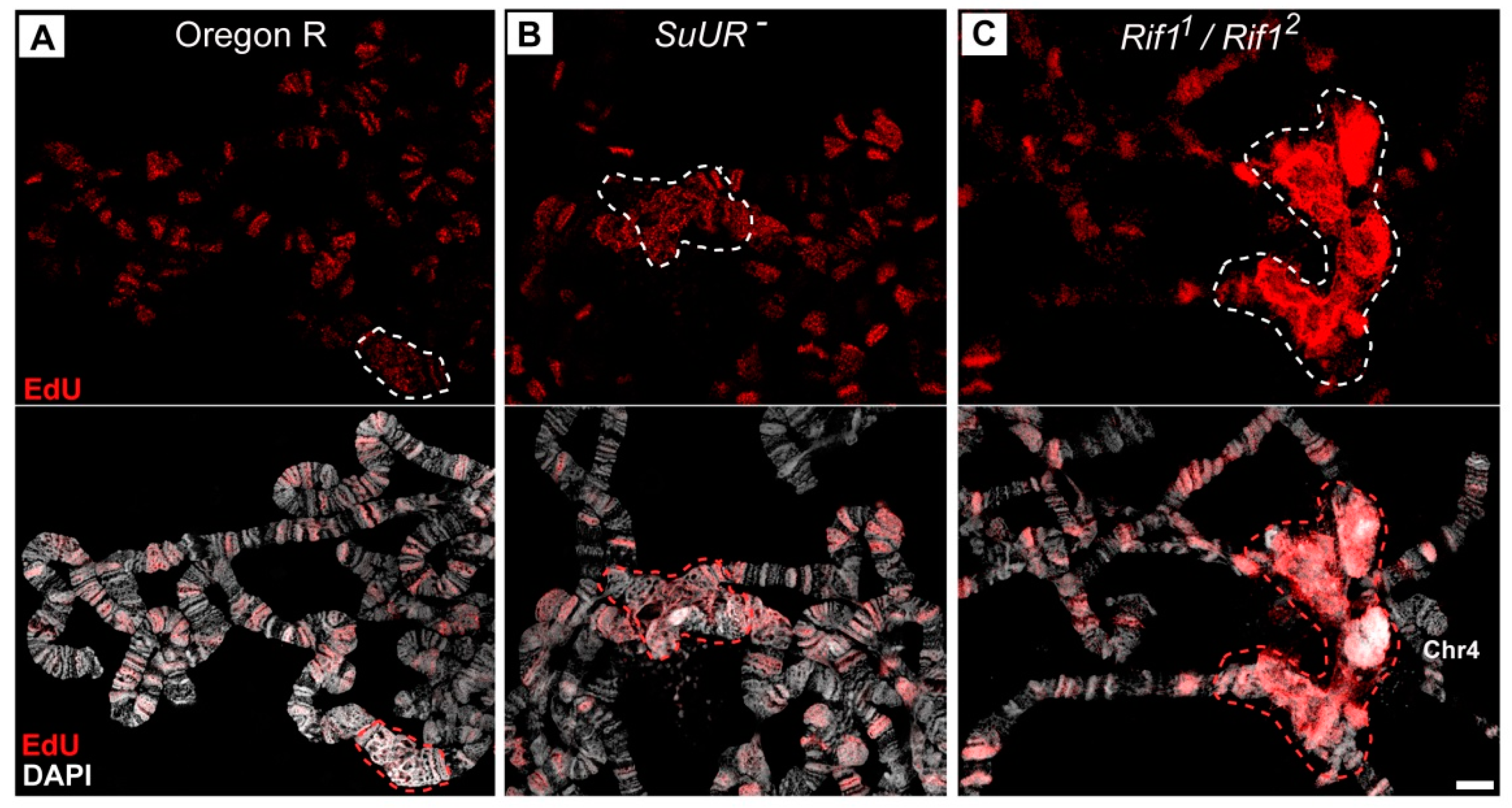
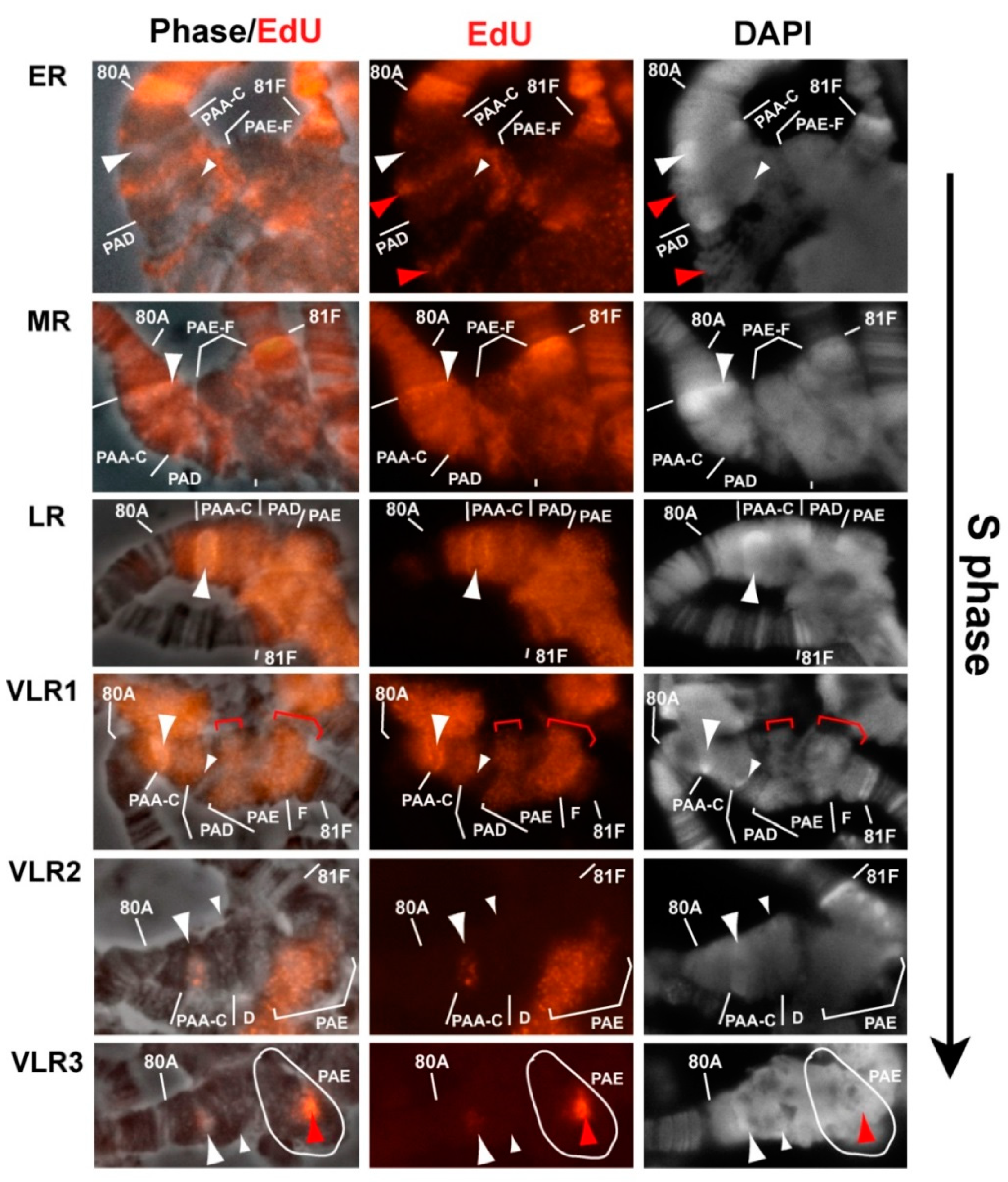
3.6. Replication of Pericentromeric Heterochromatin in the Polytene Chromosomes of the Rif1 Mutants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rudkin, G.T. Nonreplicating DNA in giant chromosomes. Nonreplicating DNA Giant Chromosom. 1965, 52, 470. [Google Scholar]
- Rudkin, G.T. Non replicating DNA in Drosophila. Genetics 1969, 61, 227–238. [Google Scholar]
- Gall, J.G.; Cohen, E.H.; Polan, M.L. Repetitive DNA sequences in Drosophila. Chromosoma 1971, 33, 319–344. [Google Scholar] [CrossRef] [PubMed]
- Zhimulev, I.F. Polytene Chromosomes, Heterochromatin, and Position Effect Variegation. In Advances in Genetics; Hall, J.C., Dunlap, J.C., Friedmann, T., Giannelli, F., Eds.; Academic Press: Cambridge, MA, USA, 1998; Volume 37, pp. 1–555. [Google Scholar]
- Belyaeva, E.S.; Zhimulev, I.F.; Volkova, E.I.; Alekseyenko, A.A.; Moshkin, Y.M.; Koryakov, D.E. Su(UR)ES: A gene suppressing DNA underreplication in intercalary and pericentric heterochromatin of Drosophila melanogaster polytene chromosomes. Proc. Natl. Acad. Sci. USA 1998, 95, 7532–7537. [Google Scholar] [CrossRef] [PubMed]
- Yarosh, W.; Spradling, A.C. Incomplete replication generates somatic DNA alterations within Drosophila polytene salivary gland cells. Genes Dev. 2014, 28, 1840–1855. [Google Scholar] [CrossRef]
- Lilly, M.A.; Spradling, A.C. The Drosophila endocycle is controlled by Cyclin E and lacks a checkpoint ensuring S-phase completion. Genes Dev. 1996, 10, 2514–2526. [Google Scholar] [CrossRef] [PubMed]
- Royzman, I.; Orr-Weaver, T.L. S phase and differential DNA replication during Drosophila oogenesis. Genes Cells Devoted Mol. Cell. Mech. 1998, 3, 767–776. [Google Scholar] [CrossRef]
- Doronkin, S.; Djagaeva, I.; Beckendorf, S.K. The COP9 signalosome promotes degradation of Cyclin E during early Drosophila oogenesis. Dev. Cell 2003, 4, 699–710. [Google Scholar] [CrossRef]
- Lee, H.O.; Davidson, J.M.; Duronio, R.J. Endoreplication: Polyploidy with purpose. Genes Dev. 2009, 23, 2461–2477. [Google Scholar] [CrossRef]
- Zielke, N.; Edgar, B.A.; DePamphilis, M.L. Endoreplication. Cold Spring Harb. Perspect. Biol. 2013, 5, a012948. [Google Scholar] [CrossRef]
- Edgar, B.A.; Zielke, N.; Gutierrez, C. Endocycles: A recurrent evolutionary innovation for post-mitotic cell growth. Nat. Rev. Mol. Cell Biol. 2014, 15, 197–210. [Google Scholar] [CrossRef] [PubMed]
- Shu, Z.; Row, S.; Deng, W.-M. Endoreplication: The Good, the Bad, and the Ugly. Trends Cell Biol. 2018, 28, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Belyaeva, E.S.; Goncharov, F.P.; Demakova, O.V.; Kolesnikova, T.D.; Boldyreva, L.V.; Semeshin, V.F.; Zhimulev, I.F. Late Replication Domains in Polytene and Non-Polytene Cells of Drosophila melanogaster. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Zhimulev, I.F.; Zykova, T.Y.; Goncharov, F.P.; Khoroshko, V.A.; Demakova, O.V.; Semeshin, V.F.; Pokholkova, G.V.; Boldyreva, L.V.; Demidova, D.S.; Babenko, V.N.; et al. Genetic organization of interphase chromosome bands and interbands in Drosophila melanogaster. PLoS ONE 2014, 9, e101631. [Google Scholar] [CrossRef] [PubMed]
- Kolesnikova, T.D.; Goncharov, F.P.; Zhimulev, I.F. Similarity in replication timing between polytene and diploid cells is associated with the organization of the Drosophila genome. PLoS ONE 2018, 13. [Google Scholar] [CrossRef] [PubMed]
- Kolesnikova, T.D.; Antonenko, O.V.; Makunin, I.V. Replication timing in Drosophila and its peculiarities in polytene chromosomes. Replication Timing Drosoph. Its Peculiarities Polytene Chromosom. 2019, 23, 140–151. [Google Scholar] [CrossRef]
- Lilly, M.A.; Duronio, R.J. New insights into cell cycle control from the Drosophila endocycle. Oncogene 2005, 24, 2765–2775. [Google Scholar] [CrossRef]
- Hassel, C.; Zhang, B.; Dixon, M.; Calvi, B.R. Induction of endocycles represses apoptosis independently of differentiation and predisposes cells to genome instability. Dev. Camb. Engl. 2014, 141, 112–123. [Google Scholar] [CrossRef]
- Maqbool, S.B.; Mehrotra, S.; Kolpakas, A.; Durden, C.; Zhang, B.; Zhong, H.; Calvi, B.R. Dampened activity of E2F1-DP and Myb-MuvB transcription factors in Drosophila endocycling cells. J. Cell Sci. 2010, 123, 4095–4106. [Google Scholar] [CrossRef]
- Andreyeva, E.N.; Kolesnikova, T.D.; Belyaeva, E.S.; Glaser, R.L.; Zhimulev, I.F. Local DNA underreplication correlates with accumulation of phosphorylated H2Av in the Drosophila melanogaster polytene chromosomes. Chromosome Res. Int. J. Mol. Supramol. Evol. Asp. Chromosome Biol. 2008, 16, 851–862. [Google Scholar] [CrossRef]
- Dialynas, G.; Delabaere, L.; Chiolo, I. Arp2/3 and Unc45 maintain heterochromatin stability in Drosophila polytene chromosomes. Exp. Biol. Med. Maywood Nj 2019, 244, 1362–1371. [Google Scholar] [CrossRef]
- Lakhotia, S.C.; Sinha, P. Replication in Drosophila chromosomes. X. Two kinds of active replicons in salivary gland polytene nuclei and their relation to chromosomal replication patterns. Chromosoma 1983, 88, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Steinemann, M. Chromosomal replication of Drosophila virilis. II. Organization of active origins in diploid brain cells. Chromosoma 1981, 82, 267–288. [Google Scholar] [CrossRef]
- Steinemann, M. Chromosomal replication in Drosophila virilis. III. Organization of active origins in the highly polytene salivary gland cells. Chromosoma 1981, 82, 289–307. [Google Scholar] [CrossRef] [PubMed]
- Belyakin, S.N.; Christophides, G.K.; Alekseyenko, A.A.; Kriventseva, E.V.; Belyaeva, E.S.; Nanayev, R.A.; Makunin, I.V.; Kafatos, F.C.; Zhimulev, I.F. Genomic analysis of Drosophila chromosome underreplication reveals a link between replication control and transcriptional territories. Proc. Natl. Acad. Sci. USA 2005, 102, 8269–8274. [Google Scholar] [CrossRef] [PubMed]
- Munden, A.; Rong, Z.; Sun, A.; Gangula, R.; Mallal, S.; Nordman, J.T. Rif1 inhibits replication fork progression and controls DNA copy number in Drosophila. eLife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Sher, N.; Bell, G.W.; Li, S.; Nordman, J.; Eng, T.; Eaton, M.L.; MacAlpine, D.M.; Orr-Weaver, T.L. Developmental control of gene copy number by repression of replication initiation and fork progression. Genome Res. 2012, 22, 64–75. [Google Scholar] [CrossRef]
- Kolesnikova, T.D.; Posukh, O.V.; Andreyeva, E.N.; Bebyakina, D.S.; Ivankin, A.V.; Zhimulev, I.F. Drosophila SUUR protein associates with PCNA and binds chromatin in a cell cycle-dependent manner. Chromosoma 2013, 122, 55–66. [Google Scholar] [CrossRef]
- Nordman, J.T.; Kozhevnikova, E.N.; Verrijzer, C.P.; Pindyurin, A.V.; Andreyeva, E.N.; Shloma, V.V.; Zhimulev, I.F.; Orr-Weaver, T.L. DNA Copy Number Control Through Inhibition of Replication Fork Progression. Cell Rep. 2014, 9, 841–849. [Google Scholar] [CrossRef]
- Seller, C.A.; O’Farrell, P.H. Rif1 prolongs the embryonic S phase at the Drosophila mid-blastula transition. Plos Biol. 2018, 16. [Google Scholar] [CrossRef]
- Andreyeva, E.N.; Bernardo, T.J.; Kolesnikova, T.D.; Lu, X.; Yarinich, L.A.; Bartholdy, B.A.; Guo, X.; Posukh, O.V.; Healton, S.; Willcockson, M.A.; et al. Regulatory functions and chromatin loading dynamics of linker histone H1 during endoreplication in Drosophila. Genes Dev. 2017, 31, 603–616. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, R.L.; Penke, T.J.R.; Chao, S.K.; Gentile, G.M.; Strahl, B.D.; Matera, A.G.; McKay, D.J.; Duronio, R.J. H3K9 Promotes Under-Replication of Pericentromeric Heterochromatin in Drosophila Salivary Gland Polytene Chromosomes. Genes 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Hoskins, R.A.; Carlson, J.W.; Kennedy, C.; Acevedo, D.; Evans-Holm, M.; Frise, E.; Wan, K.H.; Park, S.; Mendez-Lago, M.; Rossi, F.; et al. Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science 2007, 316, 1625–1628. [Google Scholar] [CrossRef]
- Hoskins, R.A.; Carlson, J.W.; Wan, K.H.; Park, S.; Mendez, I.; Galle, S.E.; Booth, B.W.; Pfeiffer, B.D.; George, R.A.; Svirskas, R.; et al. The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Res. 2015, 25, 445–458. [Google Scholar] [CrossRef]
- Lohe, A.R.; Brutlag, D.L. Multiplicity of satellite DNA sequences in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 1986, 83, 696–700. [Google Scholar] [CrossRef] [PubMed]
- Makunin, I.V.; Pokholkova, G.V.; Kholodilov, N.G.; Zakharkin, S.O.; Bonaccorsi, S.; Dimitri, P.; Zhimulev, I.F. A novel simple satellite DNA is colocalized with the Stalker retrotransposon in Drosophila melanogaster heterochromatin. Mol. Gen. Genet. Mgg 1999, 261, 381–387. [Google Scholar] [CrossRef]
- Lohe, A.R.; Hilliker, A.J.; Roberts, P.A. Mapping simple repeated DNA sequences in heterochromatin of Drosophila melanogaster. Genetics 1993, 134, 1149–1174. [Google Scholar] [CrossRef]
- Abad, J.P.; Carmena, M.; Baars, S.; Saunders, R.D.; Glover, D.M.; Ludeña, P.; Sentis, C.; Tyler-Smith, C.; Villasante, A. Dodeca satellite: A conserved G+C-rich satellite from the centromeric heterochromatin of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 1992, 89, 4663–4667. [Google Scholar] [CrossRef]
- Carlson, M.; Brutlag, D. Cloning and characterization of a complex satellite DNA from Drosophila melanogaster. Cell 1977, 11, 371–381. [Google Scholar] [CrossRef]
- Carvalho, A.B.; Clark, A.G. Efficient identification of Y chromosome sequences in the human and Drosophila genomes. Genome Res. 2013, 23, 1894–1907. [Google Scholar] [CrossRef]
- Krassovsky, K.; Henikoff, S. Distinct chromatin features characterize different classes of repeat sequences in Drosophila melanogaster. Bmc Genom. 2014, 15, 105. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Talbert, P.B.; Kasinathan, S.; Henikoff, S. Simple and Complex Centromeric Satellites in Drosophila Sibling Species. Genetics 2018, 208, 977–990. [Google Scholar] [CrossRef] [PubMed]
- Khost, D.E.; Eickbush, D.G.; Larracuente, A.M. Single-molecule sequencing resolves the detailed structure of complex satellite DNA loci in Drosophila melanogaster. Genome Res. 2017, 27, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, S.; Wei, K.H.-C.; Nalley, M.J.; Gibilisco, L.; Bachtrog, D. De novo assembly of a young Drosophila Y chromosome using single-molecule sequencing and chromatin conformation capture. Plos Biol. 2018, 16, e2006348. [Google Scholar] [CrossRef]
- Chang, C.-H.; Chavan, A.; Palladino, J.; Wei, X.; Martins, N.M.C.; Santinello, B.; Chen, C.-C.; Erceg, J.; Beliveau, B.J.; Wu, C.-T.; et al. Islands of retroelements are major components of Drosophila centromeres. Plos Biol. 2019, 17. [Google Scholar] [CrossRef]
- Chang, C.-H.; Larracuente, A.M. Heterochromatin-Enriched Assemblies Reveal the Sequence and Organization of the Drosophila melanogaster Y Chromosome. Genetics 2019, 211, 333–348. [Google Scholar] [CrossRef]
- Yamamoto, M.T.; Mitchelson, A.; Tudor, M.; O’Hare, K.; Davies, J.A.; Miklos, G.L. Molecular and cytogenetic analysis of the heterochromatin-euchromatin junction region of the Drosophila melanogaster X chromosome using cloned DNA sequences. Genetics 1990, 125, 821–832. [Google Scholar]
- Pimpinelli, S.; Berloco, M.; Fanti, L.; Dimitri, P.; Bonaccorsi, S.; Marchetti, E.; Caizzi, R.; Caggese, C.; Gatti, M. Transposable elements are stable structural components of Drosophila melanogaster heterochromatin. Proc. Natl. Acad. Sci. USA 1995, 92, 3804–3808. [Google Scholar] [CrossRef]
- Gatti, M.; Pimpinelli, S. Functional elements in Drosophila melanogaster heterochromatin. Annu. Rev. Genet. 1992, 26, 239–275. [Google Scholar] [CrossRef]
- Weiler, K.S.; Wakimoto, B.T. Heterochromatin and gene expression in Drosophila. Annu. Rev. Genet. 1995, 29, 577–605. [Google Scholar] [CrossRef]
- Miklos, G.L.; Yamamoto, M.T.; Davies, J.; Pirrotta, V. Microcloning reveals a high frequency of repetitive sequences characteristic of chromosome 4 and the beta-heterochromatin of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 1988, 85, 2051–2055. [Google Scholar] [CrossRef] [PubMed]
- Traverse, K.L.; Pardue, M.L. Studies of He-T DNA sequences in the pericentric regions of Drosophila chromosomes. Chromosoma 1989, 97, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Devlin, R.H.; Bingham, B.; Wakimoto, B.T. The organization and expression of the light gene, a heterochromatic gene of Drosophila melanogaster. Genetics 1990, 125, 129–140. [Google Scholar] [PubMed]
- Zhang, P.; Spradling, A.C. The Drosophila salivary gland chromocenter contains highly polytenized subdomains of mitotic heterochromatin. Genetics 1995, 139, 659–670. [Google Scholar] [PubMed]
- Berghella, L.; Dimitri, P. The heterochromatic rolled gene of Drosophila melanogaster is extensively polytenized and transcriptionally active in the salivary gland chromocenter. Genetics 1996, 144, 117–125. [Google Scholar]
- Koryakov, D.E.; Alekseyenko, A.A.; Zhimulev, I.F. Dynamic organization of the beta-heterochromatin in the Drosophila melanogaster polytene X chromosome. Mol. Gen. Genet. Mgg 1999, 260, 503–509. [Google Scholar] [CrossRef]
- Kolesnikova, T.D.; Koriakov, D.E.; Semeshin, V.F.; Beliaeva, E.S.; Zhimulev, I.F. Interline differences in morphology of the precentromeric region of polytene X-chromosome in Drosophila melanogaster salivary glands. Genetika 2001, 37, 1632–1641. [Google Scholar]
- Koryakov, D.E.; Domanitskaya, E.V.; Belyakin, S.N.; Zhimulev, I.F. Abnormal tissue-dependent polytenization of a block of chromosome 3 pericentric heterochromatin in Drosophila melanogaster. J. Cell Sci. 2003, 116, 1035–1044. [Google Scholar] [CrossRef]
- Andreyeva, E.N.; Kolesnikova, T.D.; Demakova, O.V.; Mendez-Lago, M.; Pokholkova, G.V.; Belyaeva, E.S.; Rossi, F.; Dimitri, P.; Villasante, A.; Zhimulev, I.F. High-resolution analysis of Drosophila heterochromatin organization using SuUR Su(var)3-9 double mutants. Proc. Natl. Acad. Sci. USA 2007, 104, 12819–12824. [Google Scholar] [CrossRef]
- He, B.; Caudy, A.; Parsons, L.; Rosebrock, A.; Pane, A.; Raj, S.; Wieschaus, E. Mapping the pericentric heterochromatin by comparative genomic hybridization analysis and chromosome deletions in Drosophila melanogaster. Genome Res. 2012, 22, 2507–2519. [Google Scholar] [CrossRef]
- Demakova, O.V.; Pokholkova, G.V.; Kolesnikova, T.D.; Demakov, S.A.; Andreyeva, E.N.; Belyaeva, E.S.; Zhimulev, I.F. The SU(VAR)3-9/HP1 Complex Differentially Regulates the Compaction State and Degree of Underreplication of X Chromosome Pericentric Heterochromatin in Drosophila melanogaster. Genetics 2007, 175, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Semeshin, F.; Belyaeva, S.; Zhimulev, F. Electron microscope mapping of the pericentric and intercalary heterochromatic regions of the polytene chromosomes of the mutant Suppressor of underreplication in Drosophila melanogaster. Chromosoma 2001, 110, 487–500. [Google Scholar] [CrossRef] [PubMed]
- Moshkin, Y.M.; Belyakin, S.N.; Rubtsov, N.B.; Kokoza, E.B.; Alekseyenko, A.A.; Volkova, E.I.; Belyaeva, E.S.; Makunin, I.V.; Spierer, P.; Zhimulev, I.F. Microdissection and sequence analysis of pericentric heterochromatin from the Drosophila melanogastermutant Suppressor of Underreplication. Chromosoma 2002, 111, 114–125. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, R.L.; Das, S.; Hill, C.A.; Duronio, R.J.; Nordman, J.T. Rif1 Functions in a Tissue-Specific Manner To Control Replication Timing Through Its PP1-binding Motif. Genetics 2020. [Google Scholar] [CrossRef]
- Cornacchia, D.; Dileep, V.; Quivy, J.P.; Foti, R.; Tili, F.; Santarella-Mellwig, R.; Antony, C.; Almouzni, G.; Gilbert, D.M.; Buonomo, S.B.C. Mouse Rif1 is a key regulator of the replication-timing programme in mammalian cells. Embo J. 2012, 31, 3678–3690. [Google Scholar] [CrossRef]
- Hayano, M.; Kanoh, Y.; Matsumoto, S.; Renard-Guillet, C.; Shirahige, K.; Masai, H. Rif1 is a global regulator of timing of replication origin firing in fission yeast. Genes Dev. 2012, 26, 137–150. [Google Scholar] [CrossRef]
- Sreesankar, E.; Bharathi, V.; Mishra, R.K.; Mishra, K. Drosophila Rif1 is an essential gene and controls late developmental events by direct interaction with PP1-87B. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef]
- Yamazaki, S.; Ishii, A.; Kanoh, Y.; Oda, M.; Nishito, Y.; Masai, H. Rif1 regulates the replication timing domains on the human genome. Embo J. 2012, 31, 3667–3677. [Google Scholar] [CrossRef]
- Tautz, D.; Hancock, J.M.; Webb, D.A.; Tautz, C.; Dover, G.A. Complete sequences of the rRNA genes of Drosophila melanogaster. Mol. Biol. Evol. 1988, 5, 366–376. [Google Scholar] [CrossRef]
- Tulin, A.V.; Kogan, G.L.; Filipp, D.; Balakireva, M.D.; Gvozdev, V.A. Heterochromatic Stellate gene cluster in Drosophila melanogaster: Structure and molecular evolution. Genetics 1997, 146, 253–262. [Google Scholar]
- Kim, M.; Ekhteraei-Tousi, S.; Lewerentz, J.; Larsson, J. The X-linked 1.688 Satellite in Drosophila melanogaster Promotes Specific Targeting by Painting of Fourth. Genetics 2018, 208, 623–632. [Google Scholar] [CrossRef]
- Ijdo, J.W.; Wells, R.A.; Baldini, A.; Reeders, S.T. Improved telomere detection using a telomere repeat probe (TTAGGG)n generated by PCR. Nucleic Acids Res. 1991, 19, 4780. [Google Scholar] [CrossRef] [PubMed]
- Lake, C.M.; Holsclaw, J.K.; Bellendir, S.P.; Sekelsky, J.; Hawley, R.S. The Development of a Monoclonal Antibody Recognizing the Drosophila melanogaster Phosphorylated Histone H2A Variant (γ-H2AV). G3 Genes Genomes Genet. 2013, 3, 1539–1543. [Google Scholar] [CrossRef] [PubMed]
- Zhimulev, I.F.; Vlassova, I.E.; Belyaeva, E.S. Cytogenetic analysis of the 2B3-4--2B11 region of the X chromosome of Drosophila melanogaster. III. Puffing disturbance in salivary gland chromosomes of homozygotes for mutation l(1)pp1t10. Chromosoma 1982, 85, 659–672. [Google Scholar] [CrossRef] [PubMed]
- Weißhart, K.D.; Fuchs, J.; Schubert, V. Structured illumination microscopy (SIM) and photoactivated localization microscopy (PALM) to analyze the abundance and distribution of RNA polymerase II molecules on flow-sorted Arabidopsis nuclei. Bio-Protocol 2016, 6, e1725. [Google Scholar]
- Yang, F.; Trifonov, V.; Ng, B.L.; Kosyakova, N.; Carter, N.P. Generation of Paint Probes from Flow-Sorted and Microdissected Chromosomes. In Fluorescence In Situ Hybridization (FISH): Application Guide; Liehr, T., Ed.; Springer Protocols Handbooks; Springer: Berlin/Heidelberg, Germany, 2017; pp. 63–79. ISBN 978-3-662-52959-1. [Google Scholar]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Bao, W.; Kojima, K.K.; Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 2015, 6, 11. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Sidorenko, D.S.; Sidorenko, I.A.; Zykova, T.Y.; Goncharov, F.P.; Larsson, J.; Zhimulev, I.F. Molecular and genetic organization of bands and interbands in the dot chromosome of Drosophila melanogaster. Chromosoma 2019, 128, 97–117. [Google Scholar] [CrossRef]
- Hubley, R.; Finn, R.D.; Clements, J.; Eddy, S.R.; Jones, T.A.; Bao, W.; Smit, A.F.A.; Wheeler, T.J. The Dfam database of repetitive DNA families. Nucleic Acids Res. 2016, 44, D81–D89. [Google Scholar] [CrossRef]
- Losada, A.; Villasante, A. Autosomal location of a new subtype of 1.688 satellite DNA of Drosophila melanogaster. Chromosome Res. Int. J. Mol. Supramol. Evol. Asp. Chromosome Biol. 1996, 4, 372–383. [Google Scholar] [CrossRef]
- Wei, K.H.-C.; Grenier, J.K.; Barbash, D.A.; Clark, A.G. Correlated variation and population differentiation in satellite DNA abundance among lines of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2014, 111, 18793–18798. [Google Scholar] [CrossRef]
- Garavís, M.; Méndez-Lago, M.; Gabelica, V.; Whitehead, S.L.; González, C.; Villasante, A. The structure of an endogenous Drosophila centromere reveals the prevalence of tandemly repeated sequences able to form i-motifs. Sci. Rep. 2015, 5, 13307. [Google Scholar] [CrossRef] [PubMed]
- Zhimulev, I.F.; Belyaeva, E.S.; Makunin, I.V.; Pirrotta, V.; Volkova, E.I.; Alekseyenko, A.A.; Andreyeva, E.N.; Makarevich, G.F.; Boldyreva, L.V.; Nanayev, R.A.; et al. Influence of the SuUR gene on intercalary heterochromatin in Drosophila melanogaster polytene chromosomes. Chromosoma 2003, 111, 377–398. [Google Scholar] [CrossRef] [PubMed]
- Stormo, B.M.; Fox, D.T. Polyteny: Still a giant player in chromosome research. Chromosome Res. 2017, 25, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Hammond, M.P.; Laird, C.D. Control of DNA replication and spatial distribution of defined DNA sequences in salivary gland cells of Drosophila melanogaster. Chromosoma 1985, 91, 279–286. [Google Scholar] [CrossRef]
- Nordman, J.; Orr-Weaver, T.L. Regulation of DNA replication during development. Dev. Camb. Engl. 2012, 139, 455–464. [Google Scholar] [CrossRef]
- Hua, B.L.; Orr-Weaver, T.L. DNA Replication Control During Drosophila Development: Insights into the Onset of S Phase, Replication Initiation, and Fork Progression. Genetics 2017, 207, 29–47. [Google Scholar] [CrossRef]
- Kolesnikova, T.D.; Andreeva, E.N.; Pindiurin, A.V.; Anan’ko, N.G.; Beliakin, S.N.; Shloma, V.V.; Iurlova, A.A.; Makunin, I.V.; Pokholkova, G.V.; Volkova, E.I.; et al. Contribution of the SuUR gene to the organization of epigenetically repressed regions of Drosophila melanogaster chromosomes. Genetika 2006, 42, 1013–1028. [Google Scholar] [CrossRef]
- Hannibal, R.L.; Chuong, E.B.; Rivera-Mulia, J.C.; Gilbert, D.M.; Valouev, A.; Baker, J.C. Copy number variation is a fundamental aspect of the placental genome. PLoS Genet. 2014, 10, e1004290. [Google Scholar] [CrossRef]
- Mehrotra, S.; Maqbool, S.B.; Kolpakas, A.; Murnen, K.; Calvi, B.R. Endocycling cells do not apoptose in response to DNA rereplication genotoxic stress. Genes Dev. 2008, 22, 3158–3171. [Google Scholar] [CrossRef]
- Dej, K.J.; Spradling, A.C. The endocycle controls nurse cell polytene chromosome structure during Drosophila oogenesis. Dev. Camb. Engl. 1999, 126, 293–303. [Google Scholar]
- Elgin, S.C.R.; Reuter, G. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb. Perspect. Biol. 2013, 5, a017780. [Google Scholar] [CrossRef] [PubMed]
- Schotta, G.; Ebert, A.; Reuter, G. SU(VAR)3-9 is a conserved key function in heterochromatic gene silencing. Genetica 2003, 117, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, R.L.; Penke, T.J.R.; Strahl, B.D.; Matera, A.G.; McKay, D.J.; MacAlpine, D.M.; Duronio, R.J. Chromatin conformation and transcriptional activity are permissive regulators of DNA replication initiation in Drosophila. Genome Res. 2018, 28, 1688–1700. [Google Scholar] [CrossRef]
- Bonaccorsi, S.; Lohe, A. Fine mapping of satellite DNA sequences along the Y chromosome of Drosophila melanogaster: Relationships between satellite sequences and fertility factors. Genetics 1991, 129, 177–189. [Google Scholar]
- Gorab, E. Triple-Helical DNA in Drosophila Heterochromatin. Cells 2018, 7. [Google Scholar] [CrossRef]
- Gatti, M.; Bonaccorsi, S.; Pimpinelli, S. Looking at Drosophila mitotic chromosomes. Methods Cell Biol. 1994, 44, 371–391. [Google Scholar] [CrossRef]
- Jagannathan, M.; Cummings, R.; Yamashita, Y.M. The modular mechanism of chromocenter formation in Drosophila. eLife 2019, 8. [Google Scholar] [CrossRef]
- Larson, A.G.; Elnatan, D.; Keenen, M.M.; Trnka, M.J.; Johnston, J.B.; Burlingame, A.L.; Agard, D.A.; Redding, S.; Narlikar, G.J. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 2017, 547, 236–240. [Google Scholar] [CrossRef]
- Strom, A.R.; Emelyanov, A.V.; Mir, M.; Fyodorov, D.V.; Darzacq, X.; Karpen, G.H. Phase separation drives heterochromatin domain formation. Nature 2017, 547, 241–245. [Google Scholar] [CrossRef]
- Boeynaems, S.; Alberti, S.; Fawzi, N.L.; Mittag, T.; Polymenidou, M.; Rousseau, F.; Schymkowitz, J.; Shorter, J.; Wolozin, B.; Van Den Bosch, L.; et al. Protein Phase Separation: A New Phase in Cell Biology. Trends Cell Biol. 2018, 28, 420–435. [Google Scholar] [CrossRef] [PubMed]
- Erdel, F.; Rippe, K. Formation of Chromatin Subcompartments by Phase Separation. Biophys. J. 2018, 114, 2262–2270. [Google Scholar] [CrossRef] [PubMed]
- Davé, A.; Cooley, C.; Garg, M.; Bianchi, A. Protein phosphatase 1 recruitment by Rif1 regulates DNA replication origin firing by counteracting DDK activity. Cell Rep. 2014, 7, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Hiraga, S.-I.; Alvino, G.M.; Chang, F.; Lian, H.-Y.; Sridhar, A.; Kubota, T.; Brewer, B.J.; Weinreich, M.; Raghuraman, M.K.; Donaldson, A.D. Rif1 controls DNA replication by directing Protein Phosphatase 1 to reverse Cdc7-mediated phosphorylation of the MCM complex. Genes Dev. 2014, 28, 372–383. [Google Scholar] [CrossRef] [PubMed]



| Genotype | Allele | Reference | |||
|---|---|---|---|---|---|
| Allele Name | Chromo- some | Mutation Description | Effect on Viability and Fertility | ||
| Rif1 mutants | |||||
| w118; Rif11 | Rif11 | 2 | Deletion obtained via CRISPR-based mutagenesis; frame shift mutations at amino acid position 14. No detectable Rif1 protein. FBal0343570 | Viable and fertile. A modest defect in the embryonic hatch rate relative to wild-type flies. | [27] |
| w118; Rif12 | Rif12 | 2 | Deletion obtained via CRISPR-based mutagenesis. Frame shift mutations at amino acid position 11. No detectable Rif1 protein. FBal0343571 | [27] | |
| w118; Rif11/Rif12 | Rif11/Rif12 | 2 | The heterozygous combination used in [27] as a Rif1-null mutant. | [27] | |
| w118; Rif1PP1 | Rif1PP1 | 2 | A Rif1 allele with a mutation in the PP1-interacting motif. FBal0343572 | Low viability and sterility of homozygotes, presumably owing to the off-target effect. | [27] |
| w118; Rif1KO | Rif1KO | 2 | A Rif1KO-null allele obtained by replacing the Rif1 open reading frame with a visible 3xP3-DsRed marker via CRISPR-based mutagenesis. FBal0344229 | A reduction in survival, a reduced male-to-female ratio. Embryos laid by rif1 mutant mothers have a reduced hatch rate. | [31] |
| Other | |||||
| In(1)wm4h; Rif11/Rif12 | In(1)wm4h | 1 | Inversion In(1)wm4h having break points 3C1-2 and 20F (h28). FBab0004257 | ||
| SuURES Su(var)3-906 | SuURES | 3 | FBal0091620 | Normal with respect to morphology, viability and fertility. C-heterochromatin staining does not reveal abnormalities in the karyotype [5]. | [60,62] |
| Su(var)3-906 | 3 | FBal0016562 | The SuURES Su(var)3-906 line is viable and fertile, but eventually loses its viability and the line is maintained in a laboratory fund in a heterozygous state. | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kolesnikova, T.D.; Kolodyazhnaya, A.V.; Pokholkova, G.V.; Schubert, V.; Dovgan, V.V.; Romanenko, S.A.; Prokopov, D.Y.; Zhimulev, I.F. Effects of Mutations in the Drosophila melanogaster Rif1 Gene on the Replication and Underreplication of Pericentromeric Heterochromatin in Salivary Gland Polytene Chromosomes. Cells 2020, 9, 1501. https://doi.org/10.3390/cells9061501
Kolesnikova TD, Kolodyazhnaya AV, Pokholkova GV, Schubert V, Dovgan VV, Romanenko SA, Prokopov DY, Zhimulev IF. Effects of Mutations in the Drosophila melanogaster Rif1 Gene on the Replication and Underreplication of Pericentromeric Heterochromatin in Salivary Gland Polytene Chromosomes. Cells. 2020; 9(6):1501. https://doi.org/10.3390/cells9061501
Chicago/Turabian StyleKolesnikova, Tatyana D., Alexandra V. Kolodyazhnaya, Galina V. Pokholkova, Veit Schubert, Viktoria V. Dovgan, Svetlana A. Romanenko, Dmitry Yu. Prokopov, and Igor F. Zhimulev. 2020. "Effects of Mutations in the Drosophila melanogaster Rif1 Gene on the Replication and Underreplication of Pericentromeric Heterochromatin in Salivary Gland Polytene Chromosomes" Cells 9, no. 6: 1501. https://doi.org/10.3390/cells9061501
APA StyleKolesnikova, T. D., Kolodyazhnaya, A. V., Pokholkova, G. V., Schubert, V., Dovgan, V. V., Romanenko, S. A., Prokopov, D. Y., & Zhimulev, I. F. (2020). Effects of Mutations in the Drosophila melanogaster Rif1 Gene on the Replication and Underreplication of Pericentromeric Heterochromatin in Salivary Gland Polytene Chromosomes. Cells, 9(6), 1501. https://doi.org/10.3390/cells9061501






