Update on Circulating Tumor Cells in Genitourinary Tumors with Focus on Prostate Cancer
Abstract
1. Introduction
2. Prostate Cancer
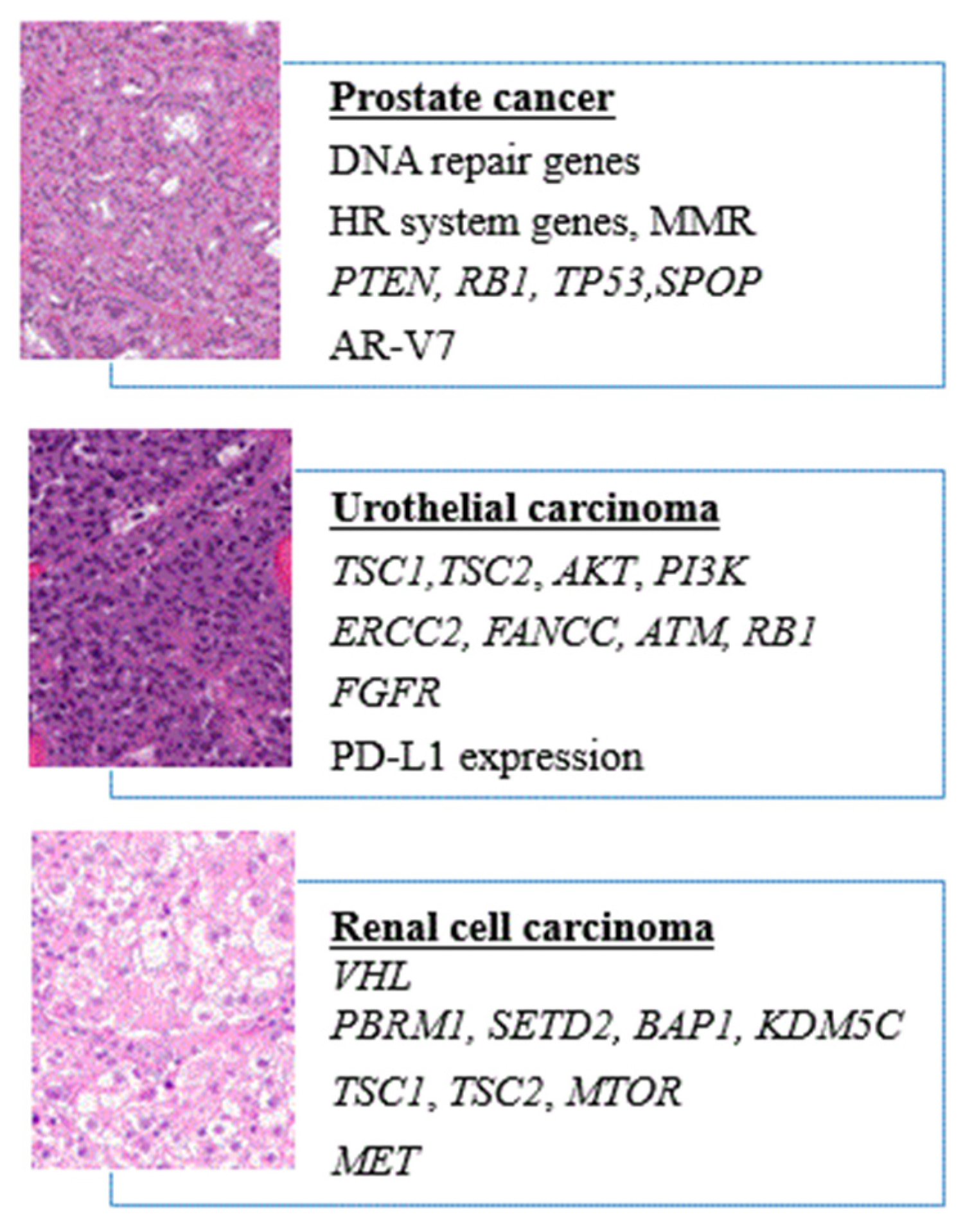
2.1. Genomic Landscape and Potential Targets
2.2. Selection of Published and Ongoing Clinical Trials
3. Bladder Cancer
3.1. Genomic Landscape and Potential Targets
3.2. Selection of Published Clinical Trials
4. Renal Cell Carcinoma
4.1. Genomic Landscape and Potential Targets
4.2. Selection of Published Clinical Trials
5. Strengths and Weaknesses of CTCs
6. Potential Application
Author Contributions
Funding
Conflicts of Interest
References
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef] [PubMed]
- Allard, W.J.; Matera, J.; Miller, M.C.; Repollet, M.; Connelly, M.C.; Rao, C.; Tibbe, A.G.; Uhr, J.W.; Terstappen, L.W. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 2004, 10, 6897–6904. [Google Scholar] [CrossRef] [PubMed]
- Molnar, B.; Ladanyi, A.; Tanko, L.; Sréter, L.; Tulassay, Z. Circulating tumor cell clusters in the peripheral blood of colorectal cancer patients. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2001, 7, 4080–4085. [Google Scholar]
- Brandt, B.; Junker, R.; Griwatz, C.; Heidl, S.; Brinkmann, O.; Semjonow, A.; Assmann, G.; Zänker, K.S. Isolation of prostate-derived single cells and cell clusters from human peripheral blood. Cancer Res. 1996, 56, 4556–4561. [Google Scholar]
- Hong, Y.; Li, Z.; Zhang, Q. A circulating tumor cell cluster-based model for tumor metastasis (Hypothesis). Oncol. Lett. 2016, 12, 4891–4895. [Google Scholar] [CrossRef][Green Version]
- Amintas, S.; Bedel, A.; Moreau-Gaudry, F.; Boutin, J.; Buscail, L.; Merlio, J.P.; Vendrely, V.; Dabernat, S.; Buscail, E. Circulating Tumor Cell Clusters: United We Stand Divided We Fall. Int. J. Mol. Sci. 2020, 21, 2653. [Google Scholar] [CrossRef]
- Lee, M.; Kim, E.J.; Cho, Y.; Kim, S.; Chung, H.H.; Park, N.H.; Song, Y.-S. Predictive value of circulating tumor cells (CTCs) captured by microfluidic device in patients with epithelial ovarian cancer. Gynecol. Oncol. 2017, 145, 361–365. [Google Scholar] [CrossRef]
- Patel, G.K.; Chugh, N.; Tripathi, M. Neuroendocrine Differentiation of Prostate Cancer-An Intriguing Example of Tumor Evolution at Play. Cancers Basel 2019, 11, 1405. [Google Scholar] [CrossRef]
- Beltran, H.; Oromendia, C.; Danila, D.C.; Montgomery, B.; Hoimes, C.; Szmulewitz, R.Z.; Vaishampayan, U.; Armstrong, A.J.; Stein, M.; Pinski, J.; et al. A phase II trial of the Aurora kinase A inhibitor Alistertib for patients with castration resistant and neuroendocrine prostate cancer: Efficacy and biomarkers. Clin. Cancer Res. 2019, 25, 43–51. [Google Scholar] [CrossRef]
- Robinson, D.; Van Allen, E.M.; Wu, Y.M.; Schultz, N.; Lonigro, R.J.; Mosquera, J.M.; Montgomery, B.; Taplin, M.E.; Pritchard, C.C.; Attard, G.; et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015, 161, 1215–1228. [Google Scholar] [CrossRef]
- Pritchard, C.C.; Mateo, J.; Walsh, M.F.; De Sarkar, N.; Abida, W.; Beltran, H.; Garofalo, A.; Gulati, R.; Carreira, S.; Eeles, R.; et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N. Engl. J. Med. 2016, 375, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Mateo, J.; Boysen, G.; Barbieri, C.E.; Bryant, H.E.; Castro, E.; Nelson, P.S.; Olmos, D.; Pritchard, C.C.; Rubin, M.A.; de Bono, J.S. DNA Repair in Prostate Cancer: Biology and Clinical Implications [figure presented]. Eur. Urol. 2017, 71, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Warner, E.W.; Yip, S.M.; Chi, K.N.; Wyatt, A.W. DNA repair defects in prostate cancer: Impact for screening, prognostication and treatment. BJU Int. 2019, 123, 769–776. [Google Scholar] [CrossRef]
- Mateo, J.; Carreira, S.; Sandhu, S.; Miranda, S.; Mossop, H.; Perez-Lopez, R.; Nava Rodrigues, D.; Robinson, D.; Omlin, A.; Tunariu, N.; et al. DNA-repair defects and olaparib in metastatic prostate cancer. N. Engl. J. Med. 2015, 373, 1697–1708. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.H.; Pritchard, C.C.; Boyd, T.; Nelson, P.S.; Montgomery, B. Biallelic Inactivation of BRCA2 in Platinum-sensitive Metastatic Castration-resistant Prostate Cancer. Eur. Urol. 2016, 69, 992–995. [Google Scholar] [CrossRef] [PubMed]
- Abida, W.; Cheng, M.L.; Armenia, J.; Middha, S.; Autio, K.A.; Vargas, H.A.; Rathkopf, D.; Morris, M.J.; Danila, D.C.; Slovin, S.F.; et al. Analysis of the Prevalence of Microsatellite Instability in Prostate Cancer and Response to Immune Checkpoint Blockade. JAMA Oncol. 2019, 5, 471–478. [Google Scholar] [CrossRef]
- Hamid, A.A.; Gray, K.P.; Shaw, G.; MacConaill, L.E.; Evan, C.; Bernard, B.; Loda, M.; Corcoran, N.M.; Van Allen, E.M.; Choudhury, A.D.; et al. Compound Genomic Alterations of TP53, PTEN, and RB1 Tumor Suppressors in Localized and Metastatic Prostate Cancer. Eur. Urol. 2019, 76, 89–97. [Google Scholar] [CrossRef]
- Mulholland, D.J.; Tran, L.M.; Li, Y.; Cai, H.; Morim, A.; Wang, S.; Plaisier, S.; Garraway, I.P.; Huang, J.; Graeber, T.G.; et al. Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell. 2011, 19, 792–804. [Google Scholar] [CrossRef]
- Carver, B.S.; Chapinski, C.; Wongvipat, J.; Hieronymus, H.; Chen, Y.; Chandarlapaty, S.; Arora, V.K.; Le, C.; Koutcher, J.; Scher, H.; et al. Reciprocal Feedback Regulation of PI3K and Androgen Receptor Signaling in PTEN-Deficient Prostate Cancer. Cancer Cell. 2011, 19, 575–586. [Google Scholar] [CrossRef]
- Maughan, B.L.; Guedes, L.B.; Boucher, K.; Rajoria, G.; Liu, Z.; Klimek, S.; Zoino, R.; Antonarakis, E.S.; Lotan, T.L. P53 status in the primary tumor predicts efficacy of subsequent abiraterone and enzalutamide in castration-resistant prostate cancer. Prostate Cancer Prostatic Dis. 2018, 21, 260–268. [Google Scholar] [CrossRef]
- Tan, H.L.; Sood, A.; Rahimi, H.A.; Wang, W.; Gupta, N.; Hicks, J.; Mosier, S.; Gocke, C.D.; Epstein, J.I.; Netto, G.J.; et al. Rb loss is characteristic of prostatic small cell neuroendocrine carcinoma. Clin. Cancer Res. 2014, 20, 890–903. [Google Scholar] [CrossRef] [PubMed]
- Boysen, G.; Rodrigues, D.N.; Rescigno, P.; Seed, G.; Dolling, D.; Riisnaes, R.; Crespo, M.; Zafeiriou, Z.; Sumanasuriya, S.; Bianchini, D.; et al. SpoP-mutated/CHD1-deleted lethal prostate cancer and abiraterone sensitivity. Clin. Cancer Res. 2018, 24, 5585–5593. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Takhar, M.; Alshalalfa, M.; Erho, N.; Shoag, J.; Jenkins, R.B.; Karnes, R.J.; Ross, A.E.; Schaeffer, E.M.; Rubin, M.A.; et al. Impact of the SPOP Mutant Subtype on the Interpretation of Clinical Parameters in Prostate Cancer. JCO Precis. Oncol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Antonarakis, E.S.; Lu, C.; Wang, H.; Luber, B.; Nakazawa, M.; Roeser, J.C.; Chen, Y.; Mohammad, T.A.; Chen, Y.; Fedor, H.L.; et al. AR-V7 and Resistance to Enzalutamide and Abiraterone in Prostate Cancer. N. Engl. J. Med. 2014, 371, 1028–1038. [Google Scholar] [CrossRef]
- Scher, H.I.; Graf, R.P.; Schreiber, N.A.; Jayaram, A.; Winquist, E.; McLaughlin, B.; Lu, D.; Fleisher, M.; Orr, S.; Lowes, L.; et al. Assessment of the Validity of Nuclear-Localized Androgen Receptor Splice Variant 7 in Circulating Tumor Cells as a Predictive Biomarker for Castration-Resistant Prostate Cancer. JAMA Oncol. 2018, 4, 1179–1186. [Google Scholar] [CrossRef]
- Scher, H.I.; Lu, D.; Schreiber, N.A.; Louw, J.; Graf, R.P.; Vargas, H.A.; Johnson, A.; Jendrisak, A.; Bambury, R.; Danila, D.; et al. Association of AR-V7 on Circulating Tumor Cells as a Treatment-Specific Biomarker with Outcomes and Survival in Castration-Resistant Prostate Cancer. JAMA Oncol. 2016, 2, 1441–1449. [Google Scholar] [CrossRef]
- De Bono, J.S.; Scher, H.I.; Montgomery, R.B.; Parker, C.; Miller, M.C.; Tissing, H.; Doyle, G.V.; Terstappen, L.W.; Pienta, K.J.; Raghavan, D. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin. Cancer Res. 2008, 14, 6302–6309. [Google Scholar] [CrossRef]
- De Bono, J.S.; Logothetis, C.J.; Molina, A.; Fizazi, K.; North, S.; Chu, L.; Chi, K.N.; Jones, R.J.; Goodman, O.B., Jr.; Saad, F.; et al. Abiraterone and increased survival in metastatic prostate cancer. N. Engl. J. Med. 2011, 364, 1995–2005. [Google Scholar] [CrossRef]
- Scher, H.I.; Fizazi, K.; Saad, F.; Taplin, M.E.; Sternberg, C.N.; Miller, K.; de Wit, R.; Mulders, P.; Chi, K.N.; Shore, N.D.; et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N. Engl. J. Med. 2012, 367, 1187–1197. [Google Scholar] [CrossRef]
- Saad, F.; Fizazi, K.; Jinga, V.; Efstathiou, E.; Fong, P.C.; Hart, L.L.; Jones, R.; McDermott, R.; Wirth, M.; Suzuki, K.; et al. Orteronel plus prednisone in patients with chemotherapy-naive metastatic castration-resistant prostate cancer (ELM-PC 4): A double-blind, multicentre, phase 3, randomised, placebo-controlled trial. Lancet Oncol. 2015, 16, 338–348. [Google Scholar] [CrossRef]
- Fizazi, K.; Jones, R.; Oudard, S.; Efstathiou, E.; Saad, F.; de Wit, R.; De Bono, J.; Cruz, F.M.; Fountzilas, G.; Ulys, A.; et al. Phase III, randomized, double-blind, multicenter trial comparing orteronel (TAK-700) plus prednisone with placebo plus prednisone in patients with metastatic castration-resistant prostate cancer that has progressed during or after docetaxel-based therapy. J. Clin. Oncol. 2015, 33, 723–731. [Google Scholar] [CrossRef]
- Smith, M.; De Bono, J.; Sternberg, C.; Le Moulec, S.; Oudard, S.; De Giorgi, U.; Krainer, M.; Bergman, A.; Hoelzer, W.; De Wit, R.; et al. Phase III study of cabozantinib in previously treated metastatic castration-resistant prostate cancer: COMET-1. J. Clin. Oncol. 2016, 34, 3005–3013. [Google Scholar] [CrossRef] [PubMed]
- Heller, G.; McCormack, R.; Kheoh, T.; Molina, A.; Smith, M.R.; Dreicer, R.; Saad, F.; de Wit, R.; Aftab, D.T.; Hirmand, M.; et al. Circulating Tumor Cell Number as a Response Measure of Prolonged Survival for Metastatic Castration-Resistant Prostate Cancer: A Comparison With Prostate-Specific Antigen Across Five Randomized Phase III Clinical Trials. J. Clin. Oncol. 2017, 36, 572–580. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, A.J.; Halabi, S.; Luo, J.; Nanus, D.M.; Giannakakou, P.; Szmulewitz, R.Z.; Danila, D.C.; Healy, P.; Anand, M.; Rothwell, C.J.; et al. Prospective Multicenter Validation of Androgen Receptor Splice Variant 7 and Hormone Therapy Resistance in High-Risk Castration-Resistant Prostate Cancer: The PROPHECY Study. J. Clin. Oncol. 2019, 37, 1120–1129. [Google Scholar] [CrossRef] [PubMed]
- Okegawa, T.; Ninomiya, N.; Masuda, K.; Nakamura, Y.; Tambo, M.; Nutahara, K. AR-V7 in circulating tumor cells cluster as a predictive biomarker of abiraterone acetate and enzalutamide treatment in castration-resistant prostate cancer patients. Prostate 2018, 78, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Nagaya, N.; Nagata, M.; Lu, Y.; Kanayama, M.; Hou, Q.; Hotta, Z.U.; China, T.; Kitamura, K.; Matsushita, K.; Isotani, S.; et al. Prostate-specific membrane antigen in circulating tumor cells is a new poor prognostic marker for castration-resistant prostate cancer. PLoS ONE 2020, 15, e0226219. [Google Scholar] [CrossRef]
- Kratochwil, C.; Bruchertseifer, F.; Giesel, F.L.; Weis, M.; Verburg, F.A.; Mottaghy, F.; Kopka, K.; Apostolidis, C.; Haberkorn, U.; Morgenstern, A. 225Ac-PSMA-617 for PSMA-Targeted alpha-Radiation Therapy of Metastatic Castration-Resistant Prostate Cancer. J. Nucl. Med. 2016, 57, 1941–1944. [Google Scholar] [CrossRef]
- Lamb, A.D.; Bryant, R.J.; Mills, I.G.; Hamdy, F.C. First Report of Prostate-specific Membrane Antigen-targeted Immunotherapy in Prostate Cancer: The Future is Bright. Eur Urol. 2018, 73, 653–655. [Google Scholar] [CrossRef]
- Montironi, R.; Lopez-Beltran, A.; Cheng, L. Words of wisdom. Re: Antibody-drug conjugates targeting prostate-specific membrane antigen. Eur. Urol. 2014, 66, 1190–1193. [Google Scholar] [CrossRef]
- Cimadamore, A.; Cheng, M.; Santoni, M.; Lopez-Beltran, A.; Battelli, N.; Massari, F.; Galosi, A.B.; Scarpelli, M.; Montironi, R. New Prostate Cancer Targets for Diagnosis, Imaging, and Therapy: Focus on Prostate-Specific Membrane Antigen. Front. Oncol. 2018, 8, 653. [Google Scholar] [CrossRef]
- Takata, R.; Katagiri, T.; Kanehira, M.; Tsunoda, T.; Shuin, T.; Miki, T.; Namiki, M.; Kohri, K.; Matsushita, Y.; Fujioka, T.; et al. Predicting response to methotrexate, vinblastine, doxorubicin, and cisplatin neoadjuvant chemotherapy for bladder cancers through genome-wide gene expression profiling. Clin. Cancer Res. 2005, 11, 2625–2636. [Google Scholar] [CrossRef]
- Jung, Y.; Lippard, S.J. Direct cellular responses to platinum-induced DNA damage. Chem. Rev. 2007, 107, 1387–1407. [Google Scholar] [CrossRef] [PubMed]
- Guancial, E.A.; Werner, L.; Bellmunt, J.; Bamias, A.; Choueiri, T.K.; Ross, R.; Schutz, F.A.; Park, R.S.; O’Brien, R.J.; Hirsch, M.S.; et al. FGFR3 expression in primary and metastatic urothelial carcinoma of the bladder. Cancer Med. 2014, 3, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, J.N.; Akbani, R.; Broom, B.M.; Wang, W.; Verhaak, R.G.W.; McConkey, D.; Lerner, S.; Morgan, M.; Creighton, C.J.; Smith, C.; et al. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014, 507, 315–322. [Google Scholar] [CrossRef]
- Tomlinson, D.C.; Baldo, O.; Hamden, P.; Knowles, M.A. FGFR3 protein expression and its relationship to mutation status and prognostic variables in bladder cancer. J. Pathol. 2007, 213, 91–98. [Google Scholar] [CrossRef]
- Argani, P. MiT family translocation renal cell carcinoma. Semin. Diagn. Pathol. 2015, 32, 103–113. [Google Scholar] [CrossRef]
- Trpkov, K.; Hes, O. New and emerging renal entities: A perspective post-WHO 2016 classification. Histopathology 2019, 74, 31–59. [Google Scholar] [CrossRef]
- Lopez-Beltran, A.; Cheng, L.; Raspollini, M.R.; Montironi, R. SMARCB1/INI1 Genetic Alterations in Renal Medullary Carcinomas. Eur. Urol. 2016, 69, 1062–1064. [Google Scholar] [CrossRef]
- Faugeroux, V.; Lefebvre, C.; Pailler, E.; Pierron, V.; Marcaillou, C.; Tourlet, S.; Billiot, F.; Dogan, S.; Oulhen, M.; Vielh, P.; et al. An Accessible and Unique Insight into Metastasis Mutational Content Through Whole-exome Sequencing of Circulating Tumor Cells in Metastatic Prostate Cancer. Eur. Urol. Oncol. 2019. [Google Scholar] [CrossRef]
- Necchi, A.; Raggi, D.; Gallina, A.; Ross, J.S.; Farè, E.; Giannatempo, P.; Marandino, L.; Colecchia, M.; Lucianò, R.; Bianchi, M.; et al. Impact of Molecular Subtyping and Immune Infiltration on Pathological Response and Outcome Following Neoadjuvant Pembrolizumab in Muscle-invasive Bladder Cancer. Eur. Urol. 2020. [Google Scholar] [CrossRef]
- Seiler, R.; Ashab, H.A.D.; Erho, N.; van Rhijn, B.; Winters, B.; Douglas, J.; Van Kessel, K.E.; Fransen van de Putte, E.E.; Sommerlad, M.; Wang, N.Q.; et al. Impact of Molecular Subtypes in Muscle-invasive Bladder Cancer on Predicting Response and Survival after Neoadjuvant Chemotherapy. Eur. Urol. 2017, 72, 544–554. [Google Scholar] [CrossRef] [PubMed]
- Kamoun, A.; de Reyniès, A.; Allory, Y.; Sjödahl, G.; Robertson, A.G.; Seiler, R.; Hoadley, K.A.; Groeneveld, C.S.; Al-Ahmadie, H.; Choi, W.; et al. A Consensus Molecular Classification of Muscle-invasive Bladder Cancer. Eur. Urol. 2020, 77, 420–433. [Google Scholar] [CrossRef] [PubMed]
- Warrick, J.I.; Sjödahl, G.; Kaag, M.; Raman, J.D.; Merrill, S.; Shuman, L.; Chen, G.; Walter, V.; DeGraff, D.J. Intratumoral Heterogeneity of Bladder Cancer by Molecular Subtypes and Histologic Variants. Eur. Urol. 2019, 75, 18–22. [Google Scholar] [CrossRef] [PubMed]
- Faltas, B.M.; Prandi, D.; Tagawa, S.T.; Molina, A.M.; Nanus, D.M.; Sternberg, C.; Rosenberg, J.; Mosquera, J.M.; Robinson, B.; Elemento, O.; et al. Clonal evolution of chemotherapy-resistant urothelial carcinoma. Nat. Genet. 2016, 48, 1490–1499. [Google Scholar] [CrossRef] [PubMed]
- Iyer, G.; Hanrahan, A.J.; Milowsky, M.I.; Al-Ahmadie, H.; Scott, S.N.; Janakiraman, M.; Pirun, M.; Sander, C.; Socci, N.D.; Ostrovnaya, I.; et al. Genome sequencing identifies a basis for everolimus sensitivity. Science 2012, 338, 221. [Google Scholar] [CrossRef]
- Van Allen, E.M.; Mouw, K.W.; Kim, P.; Iyer, G.; Wagle, N.; Al-Ahmadie, H.; Zhu, C.; Ostrovnaya, I.; Kryukov, G.V.; O’Connor, K.W.; et al. Somatic ERCC2 mutations correlate with cisplatin sensitivity in muscle-invasive urothelial carcinoma. Cancer Discov. 2014, 4, 1140–1153. [Google Scholar] [CrossRef]
- Plimack, E.R.; Dunbrack, R.L.; Brennan, T.A.; Andrake, M.D.; Zhou, Y.; Serebriiskii, I.G.; Slifker, M.; Alpaugh, K.; Dulaimi, E.; Palma, N.; et al. Defects in DNA Repair Genes Predict Response to Neoadjuvant Cisplatin-based Chemotherapy in Muscle-invasive Bladder Cancer. Eur. Urol. 2015, 68, 959–967. [Google Scholar] [CrossRef]
- Casadei, C.; Dizman, N.; Schepisi, G.; Cursano, M.C.; Basso, U.; Santini, D.; Pal, S.K.; De Giorgi, U. Targeted therapies for advanced bladder cancer: New strategies with FGFR inhibitors. Ther. Adv. Med. Oncol. 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Loriot, Y.; Necchi, A.; Park, S.H.; Garcia-Donas, J.; Huddart, R.; Burgess, E.; Fleming, M.; Rezazadeh, A.; Mellado, B.; Varlamov, S.; et al. Erdafitinib in locally advanced or metastatic urothelial carcinoma. N. Engl. J. Med. 2019, 381, 338–348. [Google Scholar] [CrossRef]
- Kuroda, K. Significance of circulating tumor cells (CTCs) with fibroblast growth factor 2 expression in gastric cancer patients. Cancer Res. 2019, 79, 13. [Google Scholar] [CrossRef]
- Schehr, J.L.; Schultz, Z.D.; Warrick, J.W.; Guckenberger, D.J.; Pezzi, H.M.; Sperger, J.M.; Heninger, E.; Saeed, A.; Leal, T.; Mattox, K.; et al. High Specificity in Circulating Tumor Cell Identification Is Required for Accurate Evaluation of Programmed Death-Ligand 1. PLoS ONE 2016, 11, e0159397. [Google Scholar] [CrossRef]
- Eckstein, M.; Cimadamore, A.; Hartmann, A.; Lopez-Beltran, A.; Cheng, L.; Scarpelli, M.; Montironi, R.; Gevaert, T. PD-L1 assessment in urothelial carcinoma: A practical approach. Ann. Transl. Med. 2019, 7, 690. [Google Scholar] [CrossRef]
- Gevaert, T.; Cimadamore, A.; Eckstein, M.; Scarpelli, M.; Lopez-Beltran, A.; Cheng, L.; Montironi, R. Predictive biomarkers for immunotherapy in the treatment of advanced urothelial carcinoma: Where we stand and where we go. Future Oncol. 2019, 15, 2199–2202. [Google Scholar] [CrossRef]
- Gallagher, D.J.; Milowsky, M.I.; Ishill, N.; Trout, A.; Boyle, M.G.; Riches, J.; Fleisher, M.; Bajorin, D.F. Detection of circulating tumor cells in patients with urothelial cancer. Ann. Oncol. 2009, 20, 305–308. [Google Scholar] [CrossRef] [PubMed]
- Flaig, T.W.; Wilson, S.; Van Bokhoven, A.; Varella-Garcia, M.; Wolfe, P.; Maroni, P.; Genova, E.E.; Morales, D.; Lucia, M.S. Detection of circulating tumor cells in metastatic and clinically localized urothelial carcinoma. Urology 2011, 78, 863–867. [Google Scholar] [CrossRef] [PubMed]
- Rink, M.; Chun, F.K.; Dahlem, R.; Soave, A.; Minner, S.; Hansen, J.; Stoupiec, M.; Coith, C.; Kluth, L.A.; Ahyai, S.A.; et al. Prognostic role and HER2 expression of circulating tumor cells in peripheral blood of patients prior to radical cystectomy: A prospective study. Eur. Urol. 2012, 61, 810–817. [Google Scholar] [CrossRef] [PubMed]
- Gazzaniga, P.; De Berardinis, E.; Raimondi, C.; Gradilone, A.; Busetto, G.M.; De Falco, E.; Nicolazzo, C.; Giovannone, R.; Gentile, V.; Cortesi, E.; et al. Circulating tumor cells detection has independent prognostic impact in high-risk non-muscle invasive bladder cancer. Int. J. Cancer. 2014, 135, 1978–1982. [Google Scholar] [CrossRef] [PubMed]
- Nicolazzo, C.; Busetto, G.M.; Gradilone, A.; Sperduti, I.; Del Giudice, F.; Loreni, F.; Cortesi, E.; de Berardinis, E.; Gazzaniga, P.; Raimondi, C. Circulating Tumor Cells Identify Patients with Super-High-Risk Non-Muscle-Invasive Bladder Cancer: Updated Outcome Analysis of a Prospective Single-Center Trial. Oncologist 2019, 24, 612–616. [Google Scholar] [CrossRef] [PubMed]
- Naoe, M.; Ogawa, Y.; Morita, J.; Omori, K.; Takeshita, K.; Shichijyo, T.; Okumura, T.; Igarashi, A.; Yanaihara, A.; Iwamoto, S.; et al. Detection of circulating urothelial cancer cells in the blood using the CellSearch system. Cancer 2007, 109, 1439–1445. [Google Scholar] [CrossRef] [PubMed]
- Okegawa, T.; Hayashi, K.; Hara, H.; Nutahara, K.; Higashihara, E. Immunomagnetic quantification of circulating tumor cells in patients with urothelial cancer. Int. J. Urol. 2010, 17, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Fan, W.; Deng, Q.; Tang, S.; Wang, P.; Xu, P.; Wang, J.; Yu, M. The prognostic and diagnostic value of circulating tumor cells in bladder cancer and upper tract urothelial carcinoma: A metaanalysis of 30 published studies. Oncotarget 2017, 8, 59527–59538. [Google Scholar] [CrossRef] [PubMed]
- Fina, E.; Necchi, A.; Giannatempo, P.; Colecchia, M.; Raggi, D.; Daidone, M.G.; Cappelletti, V. Clinical significance of early changes in circulating tumor cells from patients receiving first-line cisplatin-based chemotherapy for metastatic urothelial carcinoma. Bladder Cancer 2016, 2, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Soave, A.; Riethdorf, S.; Dahlem, R.; Minner, S.; Weisbach, L.; Engel, O.; Fisch, M.; Pantel, K.; Rink, M. Detection and oncological effect of circulating tumour cells in patients with variant urothelial carcinoma histology treated with radical cystectomy. BJU Int. 2017, 119, 854–861. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, S.; Coym, A.; Ott, L.; Soave, A.; Rink, M.; Janning, M.; Stoupiec, M.; Coith, C.; Peine, S.; von Amsberg, G.; et al. Evaluation of PD-L1 expression on circulating tumor cells (CTCs) in patients with advanced urothelial carcinoma (UC). OncoImmunology 2020, 9, 1. [Google Scholar] [CrossRef] [PubMed]
- Anantharaman, A.; Friedlander, T.; Lu, D.; Krupa, R.; Premasekharan, G.; Hough, J.; Edwards, M.; Paz, R.; Lindquist, K.; Graf, R.; et al. Programmed death-ligand 1 (PD-L1) characterization of circulating tumor cells (CTCs) in muscle invasive and metastatic bladder cancer patients. BMC Cancer 2016, 16, 744. [Google Scholar] [CrossRef]
- Hsieh, J.J.; Purdue, M.P.; Signoretti, S.; Swanton, C.; Albiges, L.; Schmidinger, M.; Heng, D.Y.; Larkin, J.; Ficarra, V. Renal cell carcinoma. Nat. Rev. Dis. Prim. 2017, 3, 17009. [Google Scholar] [CrossRef]
- Pal, S.K.; Ali, S.M.; Yakirevich, E.; Geynisman, D.M.; Karam, J.A.; Elvin, J.A.; Frampton, G.M.; Huang, X.; Lin, D.I.; Rosenzweig, M.; et al. Characterization of Clinical Cases of Advanced Papillary Renal Cell Carcinoma via Comprehensive Genomic Profiling. Eur. Urol. 2018, 73, 71–78. [Google Scholar] [CrossRef]
- Davis, C.F.; Ricketts, C.J.; Wang, M.; Yang, L.; Cherniack, A.D.; Shen, H.; Buhay, C.; Kang, H.; Kim, S.C.; Fahey, C.C.; et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell. 2014, 26, 319–330. [Google Scholar] [CrossRef]
- Pal, S.K.; Choueiri, T.K.; Wang, K.; Khaira, D.; Karam, J.A.; Van Allen, E.; Palma, N.A.; Stein, M.N.; Johnson, A.; Squillace, R.; et al. Characterization of Clinical Cases of Collecting Duct Carcinoma of the Kidney Assessed by Comprehensive Genomic Profiling. Eur. Urol. 2016, 70, 516–521. [Google Scholar] [CrossRef]
- Creighton, C.; Morgan, M.; Gunaratne, P.; Wheeler, D.A.; Gibbs, R.A.; Robertson, A.G.; Chu, A.; Beroukhim, R.; Cibulskis, K.; Signoretti, S.; et al. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013, 499, 43–49. [Google Scholar] [CrossRef]
- Hakimi, A.A.; Ostrovnaya, I.; Reva, B.; Schultz, N.; Chen, Y.B.; Gonen, M.; Liu, H.; Takeda, S.; Voss, M.H.; Tickoo, S.K.; et al. Adverse outcomes in clear cell renal cell carcinoma with mutations of 3p21 epigenetic regulators BAP1 and SETD2: A report by MSKCC and the KIRC TCGA research network. Clin. Cancer Res. 2013, 19, 3259–3267. [Google Scholar] [CrossRef]
- Kwiatkowski, D.J.; Choueiri, T.K.; Fay, A.P.; Rini, B.I.; Thorner, A.R.; de Velasco, G.; Tyburczy, M.E.; Hamieh, L.; Albiges, L.; Agarwal, N.; et al. Mutations in TSC1, TSC2, and MTOR are associated with response to rapalogs in patients with metastatic renal cell carcinoma. Clin. Cancer Res. 2016, 22, 2445–2452. [Google Scholar] [CrossRef]
- Voss, M.H.; Hakimi, A.A.; Pham, C.G.; Brannon, A.R.; Chen, Y.B.; Cunha, L.F.; Akin, O.; Liu, H.; Takeda, S.; Scott, S.N.; et al. Tumor genetic analyses of patients with metastatic renal cell carcinoma and extended benefit from mTOR inhibitor therapy. Clin. Cancer Res. 2014, 20, 1955–1964. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.M.; Park, H.S.; Kim, S.; Kim, S.; Ali, S.M.; Greenbowe, J.R.; Yang, I.S.; Kwon, N.J.; Lee, J.L.; Ryu, M.H.; et al. Next-generation sequencing reveals somatic mutations that confer exceptional response to everolimus. Oncotarget 2016, 7, 10547–10556. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, J.J.; Chen, D.; Wang, P.I.; Marker, M.; Redzematovic, A.; Chen, Y.B.; Selcuklu, S.D.; Weinhold, N.; Bouvier, N.; Huberman, K.H.; et al. Genomic Biomarkers of a Randomized Trial Comparing First-line Everolimus and Sunitinib in Patients with Metastatic Renal Cell Carcinoma. Eur. Urol. 2017, 71, 405–414. [Google Scholar] [CrossRef]
- McDermott, D.F.; Huseni, M.A.; Atkins, M.B.; Motzer, R.J.; Rini, B.I.; Escudier, B.; Fong, L.; Joseph, R.W.; Pal, S.K.; Reeves, J.A.; et al. Clinical activity and molecular correlates of response to atezolizumab alone or in combination with bevacizumab versus sunitinib in renal cell carcinoma. Nat. Med. 2018, 24, 749–757. [Google Scholar] [CrossRef] [PubMed]
- Miao, D.; Margolis, C.A.; Gao, W.; Voss, M.H.; Li, W.; Martini, D.J.; Norton, C.; Bossé, D.; Wankowicz, S.M.; Cullen, D.; et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 2018, 359, 801–806. [Google Scholar] [CrossRef] [PubMed]
- Choueiri, T.K.; Plimack, E.; Arkenau, H.-T.; Jonasch, E.; Heng, D.; Powles, T.; Frigault, M.M.; Clark, E.A.; Handzel, A.A.; Gardner, H.; et al. Biomarker-Based Phase II Trial of Savolitinib in Patients With Advanced Papillary Renal Cell Cancer. J. Clin. Oncol. 2017, 35, 2993–3001. [Google Scholar] [CrossRef]
- Cimadamore, A.; Massari, F.; Santoni, M.; Mollica, V.; Di Nunno, V.; Cheng, L.; Lopez-Beltran, A.; Scarpelli, M.; Montironi, R.; Moch, H. Molecular characterization and diagnostic criteria of renal cell carcinoma with emphasis on liquid biopsies. Expert Rev. Mol. Diagn. 2020, 20, 141–150. [Google Scholar] [CrossRef]
- Gordan, J.D.; Lal, P.; Dondeti, V.R.; Letrero, R.; Parekh, K.N.; Oquendo, C.E.; Greenberg, R.A.; Flaherty, K.T.; Rathmell, W.K.; Keith, B.; et al. HIF-α Effects on c-Myc Distinguish Two Subtypes of Sporadic VHL-Deficient Clear Cell Renal Carcinoma. Cancer Cell. 2008, 14, 435–446. [Google Scholar] [CrossRef]
- Gobé, G.; Rubin, M.; Williams, G.; Sawczuk, I.; Buttyan, R. Apoptosis and expression of Bcl-2, Bcl-XL, and Bax in renal cell carcinomas. Cancer Invest. 2002, 20, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Crawford, Y.; Kasman, I.; Yu, L.; Zhong, C.; Wu, X.; Modrusan, Z.; Kaminker, J.; Ferrara, N. PDGF-C Mediates the Angiogenic and Tumorigenic Properties of Fibroblasts Associated with Tumors Refractory to Anti-VEGF Treatment. Cancer Cell. 2009, 15, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.A.; Leong, H.S.; Pavia-Jimenez, A.; Fedyshyn, S.; Yang, J.; Kucejova, B.; Sivanand, S.; Spence, P.; Xie, X.J.; Peña-Llopis, S.; et al. Fibroblast Growth Factor Receptor-Dependent and -Independent Paracrine Signaling by Sunitinib-Resistant Renal Cell Carcinoma. Mol. Cell Biol. 2016, 36, 1836–1855. [Google Scholar] [CrossRef] [PubMed]
- Bluemke, K.; Bilkenroth, U.; Meye, A.; Fuessel, S.; Lautenschlaeger, C.; Goebel, S.; Melchior, A.; Heynemann, H.; Fornara, P.; Taubert, H. Detection of circulating tumor cells in peripheral blood of patients with renal cell carcinoma correlates with prognosis. Cancer Epidemiol. Biomark. Prev. 2009, 18, 2190–2194. [Google Scholar] [CrossRef]
- Haga, N.; Onagi, A.; Koguchi, T.; Hoshi, S.; Ogawa, S.; Akaihata, H.; Hata, J.; Hiraki, H.; Honda, R.; Tanji, R.; et al. Perioperative Detection of Circulating Tumor Cells in Radical or Partial Nephrectomy for Renal Cell Carcinoma. Ann. Surg. Oncol. 2020, 27, 1272–1281. [Google Scholar] [CrossRef]
- Verzoni, E.; Ratta, R.; Grassi, P.; Salvioni, R.; Stagni, S.; Montone, R.; Fucà, G.; Cappelletti, V.; Reduzzi, C.; De Giorgi, U.; et al. TARIBO trial: Targeted therapy with or without nephrectomy in metastatic renal cell carcinoma: Liquid biopsy for biomarkers discovery. Tumori 2017, 104, 401–405. [Google Scholar] [CrossRef]
- Cappelletti, V.; Verzoni, E.; Ratta, R.; Vismara, M.; Silvestri, M.; Montone, R.; Miodini, P.; Reduzzi, C.; Claps, M.; Sepe, P.; et al. Analysis of single circulating tumor cells in renal cell carcinoma reveals phenotypic heterogeneity and genomic alterations related to progression. Int. J. Mol. Sci. 2020, 21, 1475. [Google Scholar] [CrossRef]
- Heitzer, E.; Haque, I.S.; Roberts, C.E.S.; Speicher, M.R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 2019, 20, 71–88. [Google Scholar] [CrossRef]
- Cimadamore, A.; Gasparrini, S.; Massari, F.; Santoni, M.; Cheng, L.; Lopez-Beltran, A.; Scarpelli, M.; Montironi, R. Emerging Molecular Technologies in Renal Cell Carcinoma: Liquid Biopsy. Cancers Basel 2019, 11, 196. [Google Scholar] [CrossRef]
- Corcoran, R.B.; Chabner, B.A. Application of cell-free DNA analysis to cancer treatment. N. Engl. J. Med. 2018, 379, 1754–1765. [Google Scholar] [CrossRef]
- Liu, S.; Tian, Z.; Zhang, L.; Hou, S.; Hu, S.; Wu, J.; Jing, Y.; Sun, H.; Yu, F.; Zhao, L.; et al. Combined cell surface carbonic anhydrase 9 and CD147 antigens enable high-efficiency capture of circulating tumor cells in clear cell renal cell carcinoma patients. Oncotarget 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Nel, I.; Baba, H.A.; Ertle, J.; Weber, F.; Sitek, B.; Eisenacher, M.; Meyer, H.E.; Schlaak, J.F.; Hoffmann, A.C. Individual profiling of circulating tumor cell composition and therapeutic outcome in patients with hepatocellular carcinoma. Transl. Oncol. 2013, 6, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Zhuang, R.; Long, M.; Pavlovic, M.; Kang, Y.; Ilyas, A.; Asghar, W. Circulating tumor cell isolation, culture, and downstream molecular analysis. Biotechnol. Adv. 2018, 36, 1063–1078. [Google Scholar] [CrossRef] [PubMed]
- Franceschini, T.; Giunchi, F.; Montironi, R.; Scarpelli, M.; Lopez-Beltran, A.; Cheng, L.; Fiorentino, M. Liquid biopsies in urological cancers: What we need to know before starting using them. Expert Rev. Mol. Diagn. 2020, 20, 135–139. [Google Scholar] [CrossRef]
- Cheng, S.B.; Xie, M.; Chen, Y.; Xiong, J.; Liu, Y.; Chen, Z.; Guo, S.; Shu, Y.; Wang, M.; Yuan, B.F.; et al. Three-dimensional scaffold chip with thermosensitive coating for capture and reversible release of individual and cluster of circulating tumor cells. Anal. Chem. 2017, 89, 7924–7932. [Google Scholar] [CrossRef]
- Sarioglu, A.F.; Aceto, N.; Kojic, N.; Donaldson, M.C.; Hamza, B.; Engstrom, A.; Zhu, H.; Sundaresan, T.K.; David, T.; Luo, X.; et al. A microfuidic device for label-free, physical capture of circulating tumor cell-clusters. Nat. Methods 2015, 12, 685–691. [Google Scholar] [CrossRef]
- Kim, T.H.; Lim, M.; Park, J.; Oh, J.M.; Kim, H.; Jeong, H.; Lee, S.J.; Park, H.C.; Jung, S.; Kim, B.C.; et al. FAST: Size-selective, clog-free isolation of rare cancer cells from whole blood at a liquid-liquid interface. Anal. Chem. 2017, 89, 1155–1162. [Google Scholar] [CrossRef]
- Karabacak, N.M.; Spuhler, P.S.; Fachin, F.; Lim, E.J.; Pai, V.; Ozkumur, E.; Martel, J.M.; Kojic, N.; Smith, K.; Chen, P.I.; et al. Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nat. Protoc. 2014, 9, 694–710. [Google Scholar] [CrossRef]
- Campton, D.E.; Ramirez, A.B.; Nordberg, J.J.; Drovetto, N.; Clein, A.C.; Varshavskaya, P.; Friemel, B.H.; Quarre, S.; Breman, A.; Dorschner, M.; et al. High-recovery visual identification and single-cell retrieval of circulating tumor cells for genomic analysis using a dual-technology platform integrated with automated immunofluorescence staining. BMC Cancer 2015, 15, 360. [Google Scholar] [CrossRef]
- Gleghorn, J.P.; Pratt, E.D.; Denning, D.; Liu, H.; Bander, N.H.; Tagawa, S.T.; Nanus, D.M.; Giannakakou, P.A.; Kirby, B.J. Capture of circulating tumor cells from whole blood of prostate cancer patients using geometrically enhanced differential immunocapture (GEDI) and a prostate-specific antibody. Lab. Chip 2010, 10, 27–29. [Google Scholar] [CrossRef]
- Hvichia, G.E.; Parveen, Z.; Wagner, C.; Janning, M.; Quidde, J.; Stein, A.; Müller, V.; Loges, S.; Neves, R.P.; Stoecklein, N.H.; et al. A novel microfluidic platform for size and deformability based separation and the subsequent molecular characterization of viable circulating tumor cells. Int. J. Cancer. 2016, 138, 2894–2904. [Google Scholar] [CrossRef] [PubMed]
- Myung, J.H.; Gajjar, K.A.; Chen, J.; Molokie, R.E.; Hong, S. Differential detection of tumor cells using a combination of cell rolling, multivalent binding, and multiple antibodies. Anal. Chem. 2014, 86, 6088–6094. [Google Scholar] [CrossRef] [PubMed]
- Kirby, B.J.; Jodari, M.; Loftus, M.S.; Gleghorn, J.P.; Santana, S.M.; Liu, H.; Smith, J.P.; Santana, S.M.; Liu, H.; Smith, J.P.; et al. Functional characterization of circulating tumor cells with a prostate-cancer-specific microfluidic device. PLoS ONE 2012, 7, e35976. [Google Scholar] [CrossRef]
- Naoe, M.; Kusaka, C.; Ohta, M.; Hasebe, Y.; Unoki, T.; Shimoyama, H.; Nakasato, T.; Oshinomi, K.; Morita, J.; Fuji, K.; et al. Development of a Highly Sensitive Technique for Capturing Renal Cell Cancer Circulating Tumor Cells. Diagnostics 2019, 9. [Google Scholar] [CrossRef]
- Collins, F.S.; Varmus, H. A new initiative on precision medicine. N. Engl. J. Med. 2015, 372, 793–795. [Google Scholar] [CrossRef]
- Bailey, A.M.; Mao, Y.; Zeng, J.; Holla, V.; Johnson, A.; Brusco, L.; Chen, K.; Mendelsohn, J.; Routbort, M.J.; Mills, G.B.; et al. Implementation of biomarker-driven cancer therapy: Existing tools and remaining gaps. Discov. Med. 2014, 17, 101–114. [Google Scholar]
- Msaouel, P.; Koutsilieris, M. Diagnostic value of circulating tumor cell detection in bladder and urothelial cancer: Systematic review and meta-analysis. BMC Cancer 2011, 11, 336. [Google Scholar] [CrossRef]
- Massari, F.; Di Nunno, V.; Comito, F.; Cubelli, M.; Ciccarese, C.; Iacovelli, R.; Fiorentino, M.; Montironi, R.; Ardizzoni, A. Circulating tumor cells in genitourinary tumors. Ther. Adv. Urol. 2018, 10, 65–77. [Google Scholar] [CrossRef]
- Kim, Y.R.; Yoo, J.K.; Jeong, C.W.; Choi, J.W. Selective killing of circulating tumor cells prevents metastasis and extends survival. J. Hematol. Oncol. 2018, 11, 114. [Google Scholar] [CrossRef]
- Fumagalli, A.; Drost, J.; Suijkerbuijk, S.J.; van Boxtel, R.; de Ligt, J.; Offerhaus, G.J.; van Boxtel, R.; de Ligt, J.; Offerhaus, G.J.; Begthel, H.; et al. Genetic dissection of colorectal cancer progression by orthotopic transplantation of engineered cancer organoids. Proc. Natl. Acad. Sci. USA 2017, 114, E2357–E2364. [Google Scholar] [CrossRef]
- Sachs, N.; de Ligt, J.; Kopper, O.; Gogola, E.; Bounova, G.; Weeber, F.; Gogola, E.; Bounova, G.; Weeber, F.; Balgobind, A.V.; et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell 2018, 172, 373–386. [Google Scholar] [CrossRef] [PubMed]
| Reference | Study Design | Patients and Therapy | Results |
|---|---|---|---|
| de Bono et al. [20] | Multicenter prospective study | 231 mCRPC patients starting a new line of chemotherapy | Better OS in favorable group (<5 CTCs per 7.5 mL). Post-treatment decrease in CTC number correlated with a better OS compared to patients whose CTC number remained ≥ 5. |
| Heller et al. [26] | Analysis of 5 prospective randomized phase III trials | 6081 patients with mCRPC | CTC count before treatment start and CTC conversion from above to below 5 CTCs is a biomarker to differentiate OS for 13-week responders and non-responders. |
| Armstrong et al. [27] | Multicenter prospective validation study | 118 high-risk mCRPC patients treated with abiraterone or enzalutamide | CTC nuclear-specific AR-V7 protein assay is independently associated with worse PFS and OS. |
| Nagaya et al. [28] | Observational study | 56 CRPC patients who progressed on therapy and switched to new treatment | Shorter median PSA, PFS, and OS in the PSMA-positive CTC cohort. PSMA expression was associated with poorer response, and shorter PSA, PFS, and OS. |
| Rink et al. [41] | Prospective study | 100 consecutive UC patients treated with radical cystectomy | Higher risk of disease recurrence and cancer-specific and overall mortality in CTC-positive patients. |
| Gazzaniga et al. [42] | Prospective single center trial | 102 high-risk T1G3 bladder cancer | CTCs were detected in 20% of patients and predicted shorter time to first recurrence and time to progression. |
| Zhang et al. [43] | Meta-analysis of 30 studies | 2161 urothelial cancer patients | CTC-positive was significantly associated with tumor stage, histological grade, metastasis, regional lymph node metastasis, and poor OS, PFS/DFS, and CSS. |
| Gallagher et al. [44] | Observational study | 33 patients with metastatic UC | Higher number of CTCs was seen in patients with two or more sites of metastases. |
| Fina et al. [45] | Single-center, prospective study | 31 patients mUC receiving first-line MVAC chemotherapy were collected at baseline (T0) and after 2 cycles (T2) | Changes in CTC better predicted 3-year PFS and OS compared to CTC status evaluated at single time points. No association was found between CTCs and objective response to MVAC. |
| Bluemke et al. [46] | Observational study | 154 RCC | Presence of CTCs correlates with lymph node metastasis, presence of synchronous metastases, and poor OS. |
| Haga et al. [47] | Single center study | 60 RCC patients underwent LRN, LPN, ORN, and OPN | ORN resulted in significantly perioperative changes in CTCs and in a greater number of postoperative CTCs compared to LRN, LPN, and OPN. |
| Cappelletti et al. [48] | Observational study | 21 blood samples serially collected from 10 patients with metastatic RCC entering the TARIBO trial | Two CTC subpopulations were identified: epithelial CTCs (eCTCs) and non-conventional CTCs (ncCTCs) lacking epithelial and leukocyte markers. With a threshold ≥1 CTC/10 mL of blood, eCTCs were found in 28% of samples, ncCTCs in 62%, and both CTC types in 71%. |
| Trial ID | Primary Outcome | Disease | Treatment | Method |
|---|---|---|---|---|
| NCT02978118 | Number of patients with detectable CTCs | UC and RCC | Immune checkpoint inhibitors | Not specified |
| NCT02552394 | Determine the effect of mAb Hu-J591 on reducing CTCs | Advanced prostate cancer (PCa) | J591 | CellSearch |
| NCT02456571 | Expression of immune checkpoint biomarkers (PD-L1, PD-L2, B7-H3, and CTLA-4) on CTCs | Metastatic PCa | Sipuleucel-T or abiraterone acetate or enzalutamide or androgen deprivation therapy (ADT) | CellSearch |
| NCT03712930 | Efficacy of pamiparib in patients with CTCs with homologous recombination deficiency (CTC-HRD) | mCRPC | Pamiparib | Not specified |
| NCT03700099 | Correlate AR-V7 status in CTCs and PSA response decline | mCRPC | Sequential treatment with docetaxel and enzalutamide | Not specified |
| NCT03050866 | Correlate AR-V7 CTCs with response to cabazitaxel | mCRPC | Cabazitaxel | Not specified |
| Technology | Advantages | Disadvantages | Potential Solutions |
|---|---|---|---|
| Size-based microfluidic isolation | Easy and rapid; feasible for epithelial cell adhesion molecule (EpCAM)-negative CTCs and for a wide range of tumors | Loss of smaller CTCs or clotting of filter pores by blood cells | Fluid-assisted separation technology, combined methods (CTC-iChip) [106,107,108] |
| Density gradient centrifugation | Operability; feasible for EpCAM-negative CTCs; Elimination of lymphocytes and mononuclear cells | Loss of some CTCs, lack of specificity | Combination with other methods (i.e., automated immunofluorescence staining) [109] |
| Immunoaffinity | High purity, visual confirmation of CTCs | Costly, absence of standardized markers | Use of multiple antibodies simultaneously [110] |
| Microfluidics sorting device | High recovery and efficiency; potential to recover CTCs for molecular or IHC characterization | Absence of standardized methods; high technical requirement | Combination with other methods (i.e., RT-PCR based selection) [111] |
| Antibodies for CTC Detection | Application | Findings |
|---|---|---|
| EpCAM and CD45 (CellSearch® System) | Epithelial tumors | EpCAM negative tumor cells may not be detected—lack of specificity for tumor cells. Nonmalignant epithelial cells are false positive |
| Citokeratins (CK8/18CK-19/CK-20) | Epithelial tumors | Cytokeratin (CK) negative tumor cells may not be detected—poor specificity for tumor cells |
| PSMA/HER2 (+size selection) | Prostate cancer | High cell capture efficiencies and highly pure captured cell [110] |
| EpCAM, HER-2 and PSA | Prostate cancer | High cell capture efficiency (tested on cell lines) [112] and |
| PSMA/CD45 | Prostate cancer | higher sensitivity compared to CellSearch [113] |
| CA9 and/or CD147 | Clear cell renal cell carcinoma (ccRCC) | CA9 and/or CD147 expression in 97.1% of patients with ccRCC tumors (EpCAM detected only 18.6%), poor specificity [101] |
| CA9 (mAbG250) | Clear cell renal cell carcinoma | Lack of specificity, CAIX can also be expressed in hypoxic or necrotic tissues regardless of their tumor origin [114] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cimadamore, A.; Aurilio, G.; Nolé, F.; Massari, F.; Scarpelli, M.; Santoni, M.; Lopez-Beltran, A.; Cheng, L.; Montironi, R. Update on Circulating Tumor Cells in Genitourinary Tumors with Focus on Prostate Cancer. Cells 2020, 9, 1495. https://doi.org/10.3390/cells9061495
Cimadamore A, Aurilio G, Nolé F, Massari F, Scarpelli M, Santoni M, Lopez-Beltran A, Cheng L, Montironi R. Update on Circulating Tumor Cells in Genitourinary Tumors with Focus on Prostate Cancer. Cells. 2020; 9(6):1495. https://doi.org/10.3390/cells9061495
Chicago/Turabian StyleCimadamore, Alessia, Gaetano Aurilio, Franco Nolé, Francesco Massari, Marina Scarpelli, Matteo Santoni, Antonio Lopez-Beltran, Liang Cheng, and Rodolfo Montironi. 2020. "Update on Circulating Tumor Cells in Genitourinary Tumors with Focus on Prostate Cancer" Cells 9, no. 6: 1495. https://doi.org/10.3390/cells9061495
APA StyleCimadamore, A., Aurilio, G., Nolé, F., Massari, F., Scarpelli, M., Santoni, M., Lopez-Beltran, A., Cheng, L., & Montironi, R. (2020). Update on Circulating Tumor Cells in Genitourinary Tumors with Focus on Prostate Cancer. Cells, 9(6), 1495. https://doi.org/10.3390/cells9061495









